Abstract
Psoriasis is a human skin condition characterized by epidermal hyperproliferation and infiltration of multiple leukocyte populations. In characterizing a novel insulin growth factor (IGF)-like (IGFL) gene in mice (mIGFL), we found transcripts of this gene to be most highly expressed in skin with enhanced expression in models of skin wounding and psoriatic-like inflammation. A possible functional ortholog in humans, IGFL1, was uniquely and significantly induced in psoriatic skin samples. In vitro IGFL1 expression was up-regulated in cultured primary keratinocytes stimulated with tumor necrosis factor α but not by other psoriasis-associated cytokines. Finally, using a secreted and transmembrane protein library, we discovered high affinity interactions between human IGFL1 and mIGFL and the TMEM149 ectodomain. TMEM149 (renamed here as IGFLR1) is an uncharacterized gene with structural similarity to the tumor necrosis factor receptor family. Our studies demonstrate that IGFLR1 is expressed primarily on the surface of mouse T cells. The connection between mIGFL and IGFLR1 receptor suggests mIGFL may influence T cell biology within inflammatory skin conditions.
Keywords: Cell Surface Receptor, Cytokine, Immunology, Inflammation, Ligand-binding Protein, Protein Domains, Protein-Protein Interactions, Receptors, Tumor Necrosis Factor (TNF), TNF Receptors
Introduction
Psoriasis is an inflammatory skin condition in which infiltrating leukocytes, such as T cells and myeloid cells, promote the abnormal proliferation and differentiation of epidermal keratinocytes (1). Cytokine networks created by these inflammatory leukocytes have been proposed to play a critical role in orchestrating the pathogenic process in psoriasis. CD4+ T cells and their effector cytokines are essential for promoting psoriasis. Recently, a novel CD4+ T helper subset, Th17, has been linked with psoriasis (2, 3). Th17 cells produce IL-22 and IL-17, both of which target keratinocytes, promoting many downstream pathological features. IL-22 activates Stat3 in epithelial keratinocytes (4, 5), and transgenic overexpression of a dominant active form of Stat3 in mouse keratinocytes results in psoriasis-like phenotype-associated with leukocyte infiltration and activation in the skin (6). On the other hand, factors produced by keratinocytes also modulated various immune responses. For instance, chemokines and cytokines produced by keratinocytes can recruit and activate leukocytes during the pathogenesis of psoriasis (7).
Recently, a novel IGF-like (IGFL)4 gene family was identified consisting of four transcribed genes in humans (IGFL1–4) and one in mice (mIGFL) (8). IGFL proteins contain 11 regularly spaced cysteine residues, 6 of which are conserved within the IGF family, and a signal peptide but no transmembrane domain, indicating that they are likely secreted. The IGFL sequences are not well conserved between human and mice, and the identity of mIGFL and the human IGFLs varies from between 21% for IGFL1 to 38% for IGFL3. The IGFL genes were found to be expressed rarely and at low levels in human tissues and have no described biological function. In this study we analyzed the gene expression profile of mIGFL and discovered that it is primarily expressed in normal skin in mice. Interestingly, mIGFL expression is further up-regulated during inflammation responses in the skin and after skin wounding. Of the four human IGFL family members, only IGFL1 is up-regulated in human psoriatic skin samples. In vitro, IGFL1 is produced by keratinocytes, and TNFα enhances its expression. Finally, we found that both mIGFL and IGFL1 interact with specificity and high affinity to a novel receptor (formerly TMEM149, now renamed as IGFLR1 for “IGF-like family receptor 1”) that bears similarity to the tumor necrosis factor receptor (TNFR) family. IGFLR1 is expressed most abundantly on mouse T cells, suggesting mIGFL and IGFL1 produced in the skin may potentially exert regulatory functions on T cell responses.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Neonatal normal human epidermal keratinocytes were purchased from Lonza (Walkersville, MD), keratinocyte-SFM with supplements were from Invitrogen, and Collagen IV was purchased form Sigma. All transfections were performed with FuGENE 6 (Roche Applied Science) according to the manufacturer's protocol. Mouse total RNA adult tissue panel was purchased from Zyagen (San Diego, CA). Recombinant human cytokines were from Peprotech (Rocky Hill, NJ). The following anti-mouse antibodies were used for flow cytometry: anti-B220, anti-CD3, anti-CD11b, anti-CD11c, anti-DX5, anti-GR1, anti-NKG2D, and anti-F4/80 were all from BD Biosciences, anti-FLAG (M2) monoclonal antibody was purchased from Sigma and labeled with Alexa Fluor-647 monoclonal antibody labeling kit from Invitrogen, and anti-mouse IgG1-PE was from Jackson ImmunoResearch (West Grove, PA).
Cloning and Protein Purification
Full-length human and mouse IGFLs were cloned into the pRK5 vector containing an N-terminal FLAG epitope tag (underlined) following a signal peptide directing insertion into the secretory pathway (MGGTAARLGAVILFVVIVGLHGVRGKDYKDDDDKLE). The start and stop amino acid for each N-terminal FLAG fusion was: FLAG-IGFL1 (Gly-24 and Ser-110), FLAG-IGFL2 (Pro-26 and Pro-119), FLAG-IGFL3 (Gly-24 and Pro-125), FLAG-IGFL4 (Glu-20 and Ile-124), and FLAG-mIGFL (Ala-24 and Pro-140). The predicted extracellular domains of human and mouse IGFLR1 were cloned without the transmembrane domain into the pRK5 vector containing a C-terminal human IgG1-Fc or His8 tag or with the transmembrane domain into the pRK5 vector containing a C-terminal GFP tag. The start and stop amino acid for each ECD was: human IGFLR1 (Met-1 and Pro-159) and mouse IGFLR1 (Met-1 and Gly-158).
Soluble forms of these proteins were produced in CHO cells. Transient transfection of DNA into CHO cells was accomplished using DMRIE-C transfection reagent (Invitrogen catalog number 10459-014) or the cationic polymer, polyethyleneimine (Polyplus catalog 101-40N), as previously described (9, 10). FLAG-tagged, His8-tagged, and IgG1-Fc fusion proteins were purified from the transient conditioned media as follows. FLAG-tagged protein-containing media was batch-absorbed overnight at 4 °C to anti-FLAG (M2) agarose affinity gel (Sigma) preequilibrated in cold 0.1% Triton X-114, PBS. The resin was packed into a column and washed with PBS until a base line was reached. FLAG-tagged protein was eluted with 0.1 m acetic acid and immediately neutralized with a 4% volume of 1.5 m Tris, pH 8.6. His-tagged protein media was incubated for a minimum of 2 h at 4 °C with nickel-nitrilotriacetic acid-agarose (Qiagen) prewashed in PBS. Sequential washes were performed with: PBS, 0.3 m NaCl, 10 mm imidazole (10 column volumes); PBS, 0.3 m NaCl, 0.1% Triton X-114, 10 mm imidazole, pH 7.0 (10 column volumes); PBS, 0.3 m NaCl, 10 mm imidazole (>10 column volumes or until base line was reached); a final wash with PBS, 0.3 m NaCl, 25 mm imidazole, pH 8.0. Elution was performed in PBS, 0.3 m NaCl, 250 mm imidazole, pH 8.0. IgG1-Fc protein media was loaded onto a protein A-Sepharose (Amersham Biosciences) column and washed with Tris buffer (25 mm Tris, pH 7.5, 5 mm EDTA, 150 mm NaCl, 2 mm azide) with 0.1% Triton X-114 (5 column volumes), Tris buffer (10 column volumes), 0.4 m potassium phosphate, pH 7.0, 5 mm EDTA, 0.2% Tween 20 (3.5 column volumes), and Tris buffer (5 column volumes). IgG1-Fc protein was eluted with 100 mm acetic acid, pH 2.7, 1 mm azide, 0.5 mm PMSF and neutralized with a 10% volume of 1.5 m Tris, pH 8.6. In cases where further separation from aggregates and contaminants was necessary, gel filtration and/or ion exchange was used. Primary column pools were run on a Superdex 200 gel-filtration column in PBS containing 0.15 m NaCl until the desired peak was collected. Mono Q/S ion exchange (Amersham Biosciences) purifications were performed in 10 mm Tris, pH 8.0, or 10 mm sodium acetate, pH 5.0, respectively. A linear NaCl gradient was used to elute off either ion-exchange column. Protein purity was assessed by SDS-PAGE followed by SimplyBlue Safe Stain (Invitrogen), and purified proteins were aliquoted and frozen at −80 °C until needed.
Hydrodynamic Tail Vein in Vivo Transfection and mIGFL ELISA
The technique for expressing exogenous genes in mice by rapidly injecting DNA dissolved in a saline solution equal to 8–12% of the body weight via the tail vein of the animal was adapted from Liu et al. (11). For hydrodynamic tail vein injection-induced expression of mIGFL, 8–10-week-old Balb/c mice were placed under a heat lamp for 5 min before the injection to dilate the tail veins. Mice were then restrained in an acrylic chamber to allow access to their tails, and 50 μg of empty pRK5 or pRK5 with N-terminal FLAG-tagged mIGFL in a volume of sterile Ringer's solution equal to 10% of the mouse body weight was injected into the tail vein over 5–8 s. Mice were bled 6 h after injections, euthanized via CO2 inhalation at 24 h after injections, and blood was collected via ventricular puncture.
mIGFL was detected in the serum via sandwich ELISA. To capture mIGFL, 384-well plates were coated with mIGFLR1-Fc overnight at 4 °C. After 3 washes, plates were blocked for nonspecific binding with 0.5% BSA in PBS for 1 h. Plates were again washed, and mouse serum was diluted with 50% assay buffer (PBS with 0.5% PBS and 0.05% Tween 20), or a dilution series of purified FLAG-mIGFL standards in assay buffer with 50% mouse serum were added to plates, incubated for 2 h at room temperature, and washed 6 times. To detect FLAG-mIGFL, plates were incubated with HRP-conjugated anti-FLAG, washed, and incubated with Moss substrate solution for development. The reaction was stopped with 1 m H3PO4, and plates were read at 450/650 nm.
Imiquimod-induced Psoriasis and Wounding in Mice
The Imiquimod-induced psoriasis like model was carried out in 8–12-week-old C57B/6 mice (Charles River). Three days before treatment, mice were anesthetized with isoflurane, and hair on their back hindquarters was removed with depilatory cream. Mice were anesthetized with isoflurane, and 62.5 mg of Imiquimod cream was administered to the shaved back and right ear daily. Ear thickness was monitored, and mice were scored for clinical signs of inflammation every 2 days according to the following scale: 0 = no disease; 1 = very mild erythema with very mild thickening and scaling involving a small area; 2 = mild erythema with mild thickening and scaling (irregular and patchy) involving a small area; 3 = moderate erythema with moderate thickening and scaling (irregular and patchy) involving a moderate area; 4 = severe erythema with marked thickening and scaling (irregular and patchy) involving a large area. One day after the last Imiquimod treatment, mice were euthanized via CO2 inhalation, and the skin covering the treated area was harvested for RNA purification.
Skin wounding assays were carried out in 8–10-week-old B6 mice. Briefly, mice were put under mild anesthesia, and using sterile conditions, the dorsal region of mice were shaved, excess hair was removed with hair removal lotion, and the region was prepped with Betadine followed by alcohol. Then, a 6-mm-diameter full thickness skin punch was removed at the midline between the scapulae, and a 0.5-mm silicone frame with a 10–12-mm diameter was placed around each wound, which was then dressed. Dressings were changed every other day. Mice were euthanized 7 days after wounding, and skin from the area of wounding was collected for RNA purification.
RT-PCR and RNA Microarray
Total RNA was purified using Qiagen (Valencia, CA) RNeasy (cells) or RNeasy Fibrous Tissue (skin) according to the manufacturer's protocol with DNase digest. The primer/probe set for mIGFL and IGFL4 were purchased from ABI (Foster City, CA), and primer/probes for IGFL1, -2, and -3 were synthesized in-house. One-step RT-PCR was performed on 25 or 50 ng of total RNA using TaqMan Gold with Buffer A kit on a Stratagene (La Jolla, CA) Mx3000P system. Copy numbers of mIGFL and mIGFLR1 in the mouse tissue panel were determined using a dilution series of mIGFL or mIGFLR1 cDNA and then normalized to the average expression level of all tissues examined. For RT-PCR performed on keratinocytes, skin, and leukocytes, results were normalized to the housekeeping gene RPL19 using comparative Ct method.
RNA microarrays were performed on two sets of normal control and psoriasis patient samples. Samples from normal skin, psoriatic skin, or adjacent nonlesion skin were run on HGU133A and HGU133B Genechips (Affymetrix, Santa Clara, CA) or WHG 44 × 44k microarray chips (Agilent Technologies, Foster City, CA). The following probes were used: 239430_at for IGFL1, 231148_at for IGFL2, and P_A51261_at for IGFL3 on HGU33 or HuGenen1 Affymetrix microarray chips (n = 5 normal and n = 11 psoriasis/PSNE, ±S.E.) and A_23_P119407 for IGFL4 on an Agilent WHG 4 × 44k microarray (n = 7 normal and n = 15 psoriasis/PSNE, ±S.E.). Expression values were measured by signal intensity from Affymetrix Microarray Analysis Suite Version 5, and samples were further normalized by using Robust Multi-chip Average quantile normalization.
In Situ Hybridization
In situ hybridization was performed on 5-μm-thick formalin-fixed, paraffin-embedded sections cut from lesion and non-lesion psoriatic skin. Radioactive in situ hybridization was performed as previously described (12). PCR primers were designed to amplify a 490-bp fragment of human IGFL1 spanning from nucleotides 104 to 594 of human IGFL1 reference sequence NM_198541 (upper, 5_-GGCCCCCATGACTCCTTACCT-3_; lower, 5_-CCCATCAGCACACGCATCTC-3_). Primers included extensions encoding 27-nucleotide T7 or T3 RNA polymerase initiation sites to allow in vitro transcription of sense or antisense probes, respectively, from the amplified products. Formalin-fixed, paraffin-embedded sections 5 μm thick were deparaffinized, deproteinated in 10 μg/ml Proteinase K for 30 min at 37 °C, and further processed for in situ hybridization as previously described (12). [33P]UTP-labeled sense and antisense probes were hybridized to the sections at 55 °C overnight. Unhybridized probe was removed by incubation in 20 μg/ml RNase A for 30 min at 37 °C followed by a high stringency wash at 55 °C in 0.1 × SSC for 2 h and dehydration through graded ethanol. The slides were dipped in NTB nuclear track emulsion (Eastman Kodak Co.), exposed in sealed plastic slide boxes containing desiccant for 4 weeks at 4 °C, developed, and counterstained with hematoxylin and eosin.
Cell Sorting and Flow Cytometry
Mouse leukocytes were isolated from female B6 spleens and lymph nodes. Briefly, spleens were disrupted by forcing them through a 40 μm nylon mesh, and red blood cells were lysed with RBC lysis solution (Qiagen). Cells were then labeled with lineage-specific antibodies and sorted on a FACSAria (BD Biosciences). Leukocyte populations were sorted according to the following antibody combinations: T cells, CD3+; B cells, CD19+; NK cells, NKG2D+DX5+; monocytes, CD11b+CD11c−GR1−; macrophages, CD11b+F4/80+; dendritic cells, CD11b+CD11c+. After sorting, cells were immediately lysed, and RNA was purified as described.
For flow cytometry, cells were washed and stained in PBS containing 2% BSA and 0.1 mm NaN3 and maintained at 4 °C throughout the procedure. HEK293T cells stably transfected with GFP-tagged human or mouse IGFLR1 or transiently transfected with TNFR were incubated for 30 min with FLAG-tagged purified proteins. Protein binding was detected with an Alexa Fluor-647-labeled anti-FLAG M2 antibody (Sigma). For primary cells, spleens and lymph nodes were prepared as described above, and nonspecific binding was blocked with anti-Fc receptor antibody. Cells were then incubated with anti-mouse IGFLR1 (IgG1κ) in the presence or absence or recombinant His-tagged mIGFLR1, washed, and then incubated with an anti-mouse IgG1-PE-labeled secondary antibody. Cells were then labeled with fluorescently labeled, lineage-specific antibodies (CD3 for T cells, B220 for B cells, DX5 for NK cells, CD11c for DC, F4/80 for macrophages, and CD11b/F480 for blood monocytes) and analyzed by FACS.
Keratinocyte Stimulation
Primary human keratinocytes were cultured in supplemented Keratinocyte-SFM in tissue culture flasks coated with 0.67 μg/cm2 Collagen IV in a 37 °C incubator with 5% CO2. Medium was replaced every 2–3 days until cells reached 70–80% confluency at which time cells were subcultured or used for experiments. For stimulations, 3–5 × 105 cells were added to Collagen IV-coated 12-well tissue culture plates in 1 ml Keratinocyte-SFM and allowed to adhere overnight. The following day cytokines were added to a final concentration of 50 ng/ml TNFα, 50 ng/ml IL-1β, 50 ng/ml IFNγ, 40 ng/ml IL-17A, or 50 ng/ml IL-22. After 6 and 24 h, cells were harvested, snap-frozen on dry ice, and stored at −80 °C for RNA purification.
Protein Microarray
Proteins from the Genentech SPDI library (13) were printed and immobilized in duplicate on epoxy-derivatized slides (Schott, Elmsford, NY) using a NanoPrint microarrayer (Arrayit). Slides were blocked in 5% milk PBS-Tween and stained with Cy-5-labeled FLAG-IGFL1 or IGFLR1-Fc for 20 min (20 μg/ml) followed by 3 × 5 min washes with PBS-Tween using an A-Hyb hybridization station (Miltenyi). Fluorescent intensity at 635 nm minus local background was measured for each arrayed protein, and the scores were normalized by the average intensity over the entire slide.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) was run on a Biacore 3000 (GE Healthcare) at 25 °C using anti-human IgG F(ab′)2 (Jackson ImmunoResearch) amine-coupled to BiaCore CM5 sensor chips. For kinetic analysis of protein interactions, Fc-fusion proteins at 25 μg/ml were first captured onto sensor chips by injecting them for 3 min at 5 μl/min. Then, test proteins diluted to their indicated concentrations in HBS-P buffer (0.01 m Hepes, pH 7.4, 0.15 m NaCl, 0.005% Surfactant P20) were injected for 3 min at 30 μl/min. Dissociation in HBS-P buffer alone was then measured at a flow rate of 30 μl/min. Sensor chips surfaces were regenerated by injecting 10 mm glycine, pH 1.5, for 30 s after each cycle. Each concentration was measured sequentially in duplicate. Kinetic parameters were determined by fitting the data to a 1:1 Langmuir model using the Biacore 3000 evaluation software (GE Healthcare). For the TNF screen, EDA-A1, OX40L, FasL, 4–1BBL, and CD30L were purchased from R&D, and GITRL, ApoL, RANKL, TNFα, APRIL, LTα, LTβ, TL1A, TNFSF14, TRAIL, C40L, BAFF, and CD27L were produced in-house.
Radioligand Cell Binding Assay
To determine the affinity of IGFL with IGFLR1, ligands were iodinated with 125I using the lactoperoxidase method, and free 125I-Na was removed from the labeled protein by gel filtration using a NAP-5 column. 50-μl competition reaction mixtures containing a fixed concentration of iodinated ligand and decreasing concentrations of serially diluted, unlabeled ligand were placed into 96-well plates in triplicate. To each well, HEK293T cells stably expressing human or mouse IGFLR1 were added at a density of 100,000 cells in 0.2 ml of binding buffer (Dulbecco's modified Eagle's medium with 1% bovine serum albumin, 50 mm HEPES, pH 7.2, and 2 mm sodium azide). The final concentration of the iodinated ligand with cells in each competition reaction was 100 pm (100,000 cpm per 0.25 ml). The final concentration of the unlabeled ligand with cells in the competition reaction varied starting at 500 nm and then decreasing by 1–2-fold dilution for 10 concentrations and included a zero-added, buffer-only sample. Competition reactions were incubated for 2 h at room temperature, then transferred to a Millipore Multiscreen filter plate (Billerica, MA) and washed 4 times with binding buffer to separate the free from bound iodinated antibody. The filters were counted on a Wallac Wizard 1470 gamma counter (PerkinElmer Life Sciences). The binding data were evaluated using NewLigand software (Genentech), which uses the fitting algorithm of Munson and Rodbard to determine the binding affinity of the antibody (14).
Anti-IGFLR1 mAb
B6 mice were immunized by repeated footpad injections of recombinant human IGFLR1-Fc or mouse IGFLR1-His resuspended in monophosphoryl lipid A/trehalose corynomycolate adjuvant (Corixa Corp., Seattle, WA). Three days after the final boost, lymph nodes and spleens harvested were fused with SP2/0 myeloma cells (ATCC, Manassas, VA) using the Cyto Pulse CEEF-50 apparatus (Cyto Pulse Sciences, Glen Burnie, MD). After 7–10 days, single hybridoma clones were picked by ClonePix (Genetix), and 2–3 days later culture supernatants were collected and screened by ELISA for specific binding to recombinant human or murine IGFLR1. The selected hybridomas were then cloned by limiting dilution, and monoclonal antibodies were purified from hybridoma culture supernatants by affinity chromatography.
Protein Modeling
For structural predictions, multiple alignments of IGFLR1 orthologs were used to query the HHPRED and i-TASSER (15) fold recognition programs, which critically rely on HMM-HMM comparisons and PsiPRED-derived secondary structure predictions (15–17). The cytoplasmic death domain (DD) of the human IGFLR1 chain was first detected by the threading protein fold recognition program ProFIT (18) using fold superpositions of available DDs to construct structurally accurate sequence alignments (and derived profiles) that were used by ProFIT to scan a human sequence data bank. Full-atom structure of the human IGFLR1 DD-fold (amino acids 247–335) was derived using MODELLER (19) using superposed template structures from MyD88 (PDB ID 3MOP), Pelle (PDB ID 1D2Z), and FADD (PDB ID 1FAD).
RESULTS
mIGFL Is a Soluble Cytokine That Is Up-regulated in Skin with Inflammation and Wounding
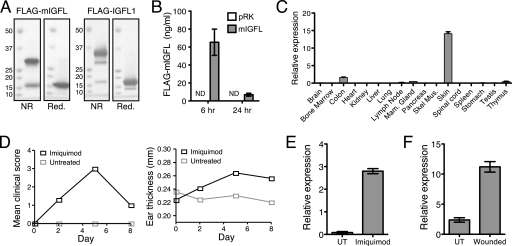
Recombinant FLAG-mIGFL purified from CHO supernatants migrated on a nonreducing SDS-PAGE gel as a major band at about ∼32 kDa, double the predicted molecular mass (Fig. 1A). Running the protein under reducing conditions caused the band to shift to ∼16 kDa, indicating that recombinant mIGFL is a disulfide-linked homodimer. To determine whether mIGFL can be produced as a soluble protein in vivo, a hydrodynamic tail vein injection-based transfection system was used to express an N-terminal FLAG epitope-tagged mIGFL transgene. FLAG-mIGFL was detectible by ELISA in serum from mice injected with the mIGFL transgene, but not control vector, as early as 6 h post-injection and persisted in the serum for at least 24 h (Fig. 1B). Therefore, mIGFL is likely produced as a secreted soluble dimeric protein.
FIGURE 1.
mIGFL is produced as a soluble dimeric protein and is up-regulated in inflammatory skin conditions in mice. A, Coomassie-stained PAGE gels of purified FLAG-mIGFL (left) and FLAG-IGFL1 (right) run under reducing (Red.) or non-reducing (NR) conditions. B, serum from mice injected with FLAG-mIGFL or control vector was assayed for mIGFL by ELISA at 6 and 24 h post-injection (n = 5, ±S.E.). No signal was detect (ND) for control vector pRK. C, RT-PCR analysis of mIGFL expression in normal mouse tissue is shown. mIGFL copy numbers were determined by comparing sample Ct values to Ct values generated with a dilution series of a known copy number of mIGFL cDNA, then normalized to the average mIGFL expression levels (±S.D.). D, clinical scores (left) and ear thickness (right) in the Imiquimod-induced psoriasis model in mice were monitored every other day during the treatment regimen. E, mIGFL expression, normalized to expression levels of RPL19, in skin RNA of mice treated daily for 5 days with Imiquimod (n = 5 treated and n = 3 untreated, ±S.E.) is shown. F, microarray analysis of mIGFL expression on day 7 after a full thickness skin punch (n = 5, ±S.E.) is shown. UT, untreated.
We determined the tissue distribution of mIGFL expression by RT-PCR on a panel of normal mouse tissues and found that mIGFL mRNA had high relative expression in skin (Fig. 1C). Lower levels of mIGFL transcripts were also detectible in colon, thymus, mammary gland, lymph node, and lung. Because mIGFL was predominantly expressed in skin, we tested whether mIGFL expression could be regulated by inflammatory stimulations in skin. Imiquimod is a TLR7/8 agonist that can be topically applied to induce skin lesions that share many of the phenotypic and histological features of psoriasis, including epidermal alterations such as acanthosis, an inflammatory infiltration that includes T cells, neutrophils, and dendritic cells, and neoangiogenesis (20). In B6 mice, inflammatory alterations peaked after 5 days of daily application of Imiquimod (Fig. 1D). Skin RNA isolated on day 5 from Imiquimod-treated mice had levels of mIGFL that were increased 35-fold over untreated skin as analyzed by RT-PCR (Fig. 1E). mIGFL levels returned to normal by day 8, as inflammation was resolving (data not shown). In addition, mIGFL levels were also augmented in skin samples taken from healing full-thickness dorsal skin punches (Fig. 1F). Taken together, these data demonstrate that mIGFL is most highly expressed in skin, and its expression is further enhanced during inflammation and injury.
Human IGFL1 Is Up-regulated in Psoriatic Skin and May Be a Functional Ortholog of mIGFL
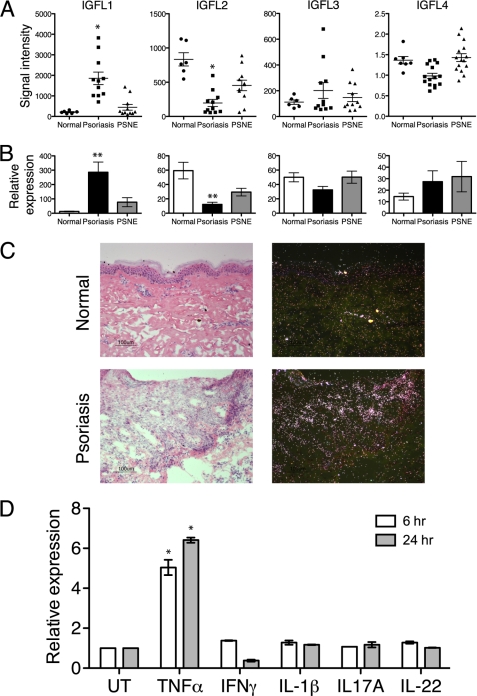
As sequence identity to the human IGFL genes are low (see Table 1), there is no clear human ortholog to mIGFL. It is not uncommon to have high sequence diversity between human and mouse homologs; for example, human and mouse OX40L share only 40% identity. We, therefore, sought to determine whether the expression of any human IGFL genes, like mIGFL, were altered during skin inflammation. First, microarray analysis of skin RNA isolated from affected or non-affected skin from psoriasis patients or normal controls revealed that IGFL1 was up-regulated in affected skin (Fig. 2A). A decrease in IGFL2 expression was also associated with psoriasis, whereas IGFL3 and -4 expression were not significantly altered. To confirm microarray data, we performed RT-PCR with primers specific for each of the human IGFL genes on skin RNA samples from psoriasis patients. Once again, IGFL1 was highly up-regulated, and IGFL2 was down-regulated (Fig. 2B), whereas there were no significant changes in the expression levels of IGFL3 or -4. In situ hybridization on psoriatic skin samples using a probe for IGFL1 revealed no detectible IGFL1 hybridization in normal skin, whereas in psoriatic skin there was a patchy distribution of IGFL1 hybridization in epithelial cells (Fig. 2C). Thus, IGFL1 and mIGFL expression levels were similarly enhanced during skin inflammation and, therefore, may represent functional orthologs.
TABLE 1.
Summary of IGFL identities and IGFLR1 interaction kinetics
ND, not determined.
| Gene name | Identity to mIGFL | IGFLR1 binding | KDa | SPR |
|||
|---|---|---|---|---|---|---|---|
| KD | χ2 | ka | kd | ||||
| % | nm | nm | m−1s−1 | s−1 | |||
| mIGFL | Yes (murine) | 0.73 | 0.15b | 4.9 | 1.02 × 106 | 1.57 × 10−4 | |
| IGFL1 | 21.2 | Yes (human) | 0.31 | 1.2b | 49.2 | 1.06 × 105 | 1.28 × 10−4 |
| IGFL2 | 33.1 | No | |||||
| IGFL3 | 37.7 | Yes (human) | ND | 0.085c | 3.5 | 1.10 × 106 | 9.31 × 10−5c |
| IGFL4 | 31.5 | No | |||||
a Determined via radioligand competition assay.
b Representative of assays done in duplicate.
c Value approaching the lower limit of SPR sensitivity.
FIGURE 2.
IGFL1 is up-regulated in psoriatic skin samples and in keratinocytes treated with TNFα. A, average intensity of IGFL family member expression was determined via RNA microarray on RNA from normal, psoriatic, or psoriatic adjacent non-effected (PSNE) skin samples. *, Student's t test p = 0.003 relative to normal control skin. B, shown is an RT-PCR analysis of IGFL family member expression in skin RNA from normal, psoriatic, or PSNE skin samples, normalized to RPL19 (n = 4, ±S.E.). **, Student's t test p = 0.03. C, hematoxylin and eosin-stained (left) and IGFL1 in situ hybridization (right) from normal and psoriatic skin samples (bars = 100 μm) is shown. D, RT-PCR analysis of IGFL1 expression in primary keratinocyte cultures treated with the indicated cytokines for 6 or 24 h is shown. Results are representative of three independent experiments (±S.D.). *, Student's t test p < 0.05.
TNFα Induces IGFL1 Expression from Cultured Primary Keratinocytes
Psoriatic skin is characterized as epidermal keratinocyte hyperplasia and leukocyte infiltration and activation. Both the infiltrating leukocyte populations and the keratinocytes are crucial sources for producing factors that enhance this tissue inflammation (1). To further identify the source of IGFL1, we analyzed mRNA expression levels and found that IGFL1 was expressed in primary keratinocytes but not in peripheral blood mononuclear cells (data not shown). Furthermore, activation of human peripheral blood mononuclear cells or mouse splenocytes by concanavalin A, phytohemagglutinin, or lipopolysaccharide did not induce IGFL1 or mIGFL expression (data not shown). To gain insights to the regulation of IGFL1 in skin, we used cultured primary keratinocytes to determine whether any of the cytokines known to play a major role in psoriasis altered IGFL1 expression. Treatment of cultured keratinocytes with TNFα for 6 h resulted in a 5-fold up-regulation of IGFL1 that remained elevated after 24 h of stimulation (Fig. 2D). Treatment of cells with IL-1β, IFNγ, IL-17A, or IL-22 did not induce any significant changes in IGFL1 mRNA levels. These results suggest that TNFα produced in the psoriatic lesion could contribute to the IGFL1 expression levels observed in patient skin.
IGFL1 and mIGFL Bind a Novel TNFR Family Member with High Affinity
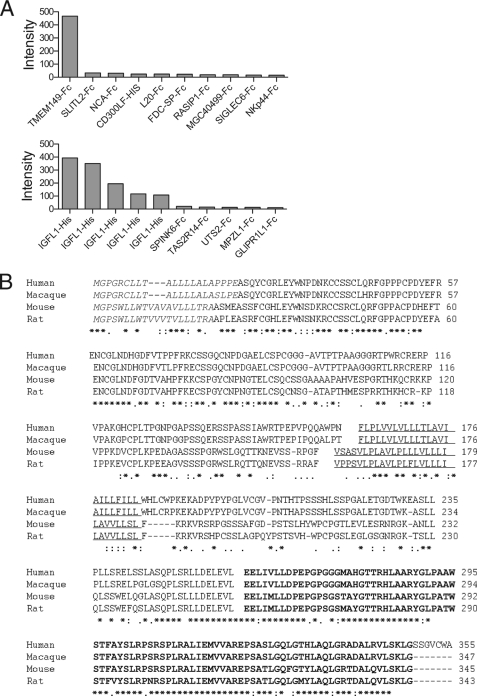
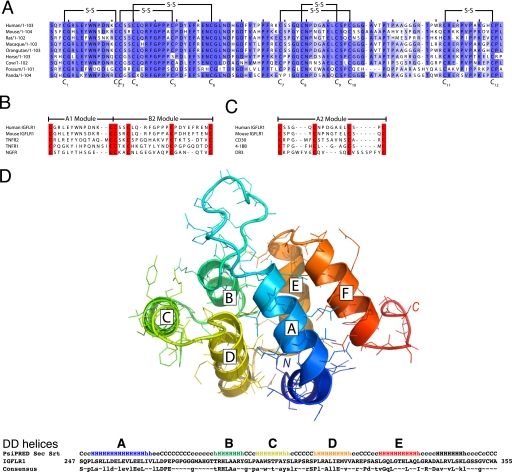
The IGFLs are orphan-secreted proteins with no known receptors. To identify a binding partner for IGFL1, we screened a protein microarray containing ∼700 unique mostly human proteins that are secreted or have transmembrane domains (13)5 and identified the top hit as IGFLR1 (formally named TMEM149) (Fig. 3A, top), a transmembrane protein with no described biology. In a reverse screen, IGFLR1 uniquely bound to five individual protein preparations of IGFL1 in the library (Fig. 3A, bottom). The IGFLR1 gene encodes a type 1a transmembrane protein that is conserved in mammalian species and fish. Sequence homology between mammalian species was moderate in the extracellular domain that contained 12 cysteines conserved among humans, macaque, mice, and rats (Fig. 3B). There was also a highly conserved region of ∼100 amino acids in the predicted cytoplasmic domain, with between 75–99% identity between all species analyzed.
FIGURE 3.
Protein microarray identification or a novel protein interaction between IGFL1 and TMEM149 (renamed here as IGFLR1). A, TMEM149-Fc (IGFLR1) was identified as an IGFL1-binding protein via protein microarray. Purified fluorescently labeled FLAG-IGFL1 (top) or IGFLR1-Fc (bottom) was used to probe a protein microarray slide. The proteins from spots with the top 10 intensity values are shown. B, shown is ClustalW2 alignment of the IGFLR1 gene from human, macaque, mouse, and rat. The predicted signal peptides are in italics, the transmembrane domains are underlined, and putative DD is in bold. Identical residues are indicated by asterisks (*), conserved residues by colons (:), and semi-conserved residues by periods (.).
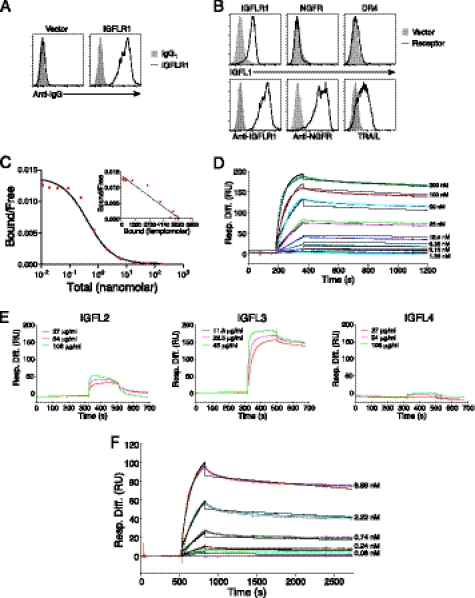
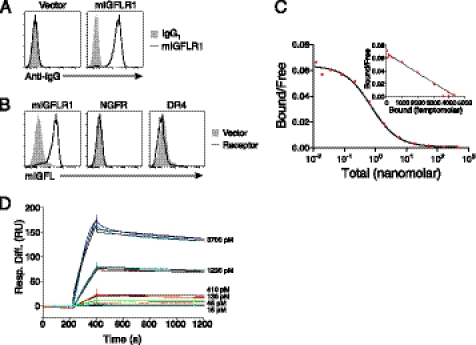
To further characterize the interaction between IGFLR1 and IGFL1, we analyzed the binding of recombinant IGFL1 to HEK293T cells expressing IGFLR1 or other TNFR family members. An antibody generated against the extracellular domain of IGFLR1 was used to demonstrate surface expression of IGFLR1 on transiently transfected cells (Fig. 4A). We then tested the ability of recombinant IGFL1 to interact with cell surface IGFLR1. Like mIGFL, the majority of recombinant human IGFL1 appeared to be a disulfide-linked dimer (Fig. 1B). We found specific binding of IGFL1 to cells expressing IGFLR1 but not the other TNFR family member NGFR or DR4 (Fig. 4B). A radioligand binding assay performed using IGFL1 with IGFLR1-expressing cells revealed a high affinity interaction with an equilibrium dissociation constant (KD) of 0.31 nm (Fig. 4C). This interaction was also analyzed by SPR, resulting in a KD of 1.2 nm (Fig. 4D, Table 1). Because the other members of the human IGFL family were not present on the protein microarray, we used SPR to determine whether they interacted with IGFLR1 and found that IGFL3 also tightly bound the receptor (Fig. 4E, middle). IGFL2 showed a low response and fast dissociation, whereas IGFL4 did not appear to interact with IGFLR1. The affinity of IGFL3 to IGFLR1 was determined to be subnanomolar by SPR (Fig. 4F and Table 1). To determine whether this interaction was conserved between species, we analyzed the ability of mIGFL to interact with mIGFLR1. Using an antibody directed against mIGFLR1, we could demonstrate surface expression of mIGFLR1 on transfected cells (Fig. 5A). mIGFL was also able to specifically bind cells expressing mIGFLR1 but not NGFR or DR4 (Fig. 5B). High affinity binding of mIGFL to mIGFLR1 was observed via radioligand binding assay (KD 0.73 nm) and SPR (KD ∼ 0.15 nm) (Fig. 5, C and D, Table 1).
FIGURE 4.
IGFL1 interacts with human IGFLR1 with high affinity. A, monoclonal anti-IGFLR1 binds to cells transfected with IGFLR1. HEK293T cells were transfected with IGFLR1 or vector and later stained with anti-IGFLR1 or control antibody. B, shown is FACS analysis of IFGL1 binding to cells expressing surface IGFLR1. HEK293T cells transfected control vector, or the indicated receptors were incubated with FLAG-IGFL1, and binding was demonstrated with fluorescently labeled anti-FLAG. NGFR expression was determined using a phycoerythrin-labeled anti-NGFR antibody, and DR4 expression was detected with soluble FLAG-TRAIL and anti-FLAG. C, a radioligand assay was used to determine the affinity of IGFL1 with IGFLR1. I125-labeled FLAG-IGFL1 was allowed to bind to HEK293T cells transfected with human IGFLR1 with increasing amounts of unlabeled ligand. Graphs are representative of two experiments. D, sensograms of FLAG-IGFL1 binding to immobilized IGFLR1-Fc analyzed by SPR (χ2 = 49.2) are shown. E, shown are sensograms of the indicated concentrations of FLAG-IGFL2, FLAG-IGFL3, and FLAG-IGFL4 binding to 7145 response units of immobilized IGFLR1-Fc. F, sensograms of FLAG-IGFL3 binding to immobilized IGFLR1-Fc analyzed by SPR (χ2 = 3.5) are shown. Black lines on D and F represent the fit to the data (colored lines). Response difference (Resp. Diff.) relative to a control reference flow cell is given in response units (RU).
FIGURE 5.
mIGFL interacts with murine IGFLR1 with high affinity. A, monoclonal anti-mIGFLR1 binds to cells expressing mIGFLR1. HEK293T cells were transfected with mIGFLR1 or vector and later stained with anti-mIGFLR1 or control IgG. B, FACS analysis of mIGFL binding to cells expressing surface mIGFLR1, NGFR, or DR4 is shown. HEK293T cell-transfected control vector or the indicated receptors were incubated with FLAG-mIGFL, and binding was demonstrated with fluorescently labeled anti-FLAG. C, a radioligand assay was used to determine the affinity of mIGFL1 with mIGFLR1. 125I-Labeled FLAG-mIGFL was allowed to bind to HEK293T cells transfected with murine IGFLR1 with increasing amounts of unlabeled ligand. Graphs are representative of two experiments. D, sensograms of FLAG-mIGFL binding to immobilized mIGFLR1-Fc analyzed by SPR (χ2 = 4.9) are shown. Black lines represent the fit to the data (colored lines). Response difference (Resp. Diff.) relative to a control reference flow cell is given in response units (RU).
Our sequence analysis of the ectodomain (ECD) of human IGFLR1 demonstrated protein homology to the TNFR family (21). The ECD of TNFRs are composed of cysteine-rich domains (CRDs) that can be further dissected into modular subdomains with conserved cysteine registers, disulfide bonding, and overall structures (22). Alignment of the ECD of IGFLR1 with these modules revealed that the first potential CRD was similar to A1-B2 CRD modules that are commonly observed in other TNFR family members (Fig. 6, A and B). A second potential CRD in IGFLR1 has sequence similarity to the A2 module (Fig. 6C). By the sensitive HHPRED fold recognition program (17) we found the structure of IGFLR1 is most closely matched to the structure of the human NGFR ECD. We also detected a death domain in the conserved cytoplasmic sequence of human IGFLR1 via the threading program ProFIT, (18). The HHPRED (17) and i-TASSER (15) fold recognition tools were used to more precisely designate the DD module in IGFLR1. The top 23 returns of an HHPRED fold search by the human IGFLR1 cytoplasmic domain are uniformly DD family members (with a top score of 97 and attached probability value of 99.3, E-value of 1.6 × 10−13, and p value of 7.1 × 10−18), indicating a highly significant match. Using MyD88, Pelle and FADD structures as templates, we constructed a theoretical three-dimensional model of the IGFLR1 cytoplasmic domain (Fig. 6D). Because IGFLR1 is an unrecognized TNFR family member, we screened TNF-like ligands for binding to IGFLR1 via SPR. There was no significant binding between 18 TNF family members screened and IGFLR1 (data not shown). Thus, we have positively identified a specific and high affinity protein interaction between mIGFL/IGFL1 and a novel TNFR with a putative cytoplasmic DD that has no described biological function.
FIGURE 6.
The ECD of IGFLR1 has structural characteristics of a TNFR and a putative cytoplasmic death domain. A, ClustalW2 alignment of the ECD of 9 mammalian IGFLR1 is shown. The predicted disulfide bonds indicated are based on the disulfide bonding pattern observed in other TNFR family members with similar CRD modules. Amino acids with more than 44% identity between species are highlighted in purple. B and C, shown is alignment of putative IGFLR1 CRD modules with known TNFR CRD modules. The consensus sequence of TNFR CRD modules and their structures are described in Naismith and Sprang (22). Cysteines are highlighted red. D, a three-dimensional model of IGFLR1 cytoplasmic domain is shown. The top panel shows a full-atom structure of the human IGFLR1 DD fold (amino acids 247–335) created with MODDLER using three superposed template structures from MyD88, Pelle, and FADD. The six α-helices of the DD fold are labeled A–E; the chain is color-ramped from the N (blue) to C terminus (red). The bottom panel shows the PsiPRED-derived helical prediction for human IGFLR1 as well as a consensus line marking the residue conservation of IGFLR1 orthologs (capital letters indicate nearly invariant residues, whereas lowercase letters are the most frequent amino acids).
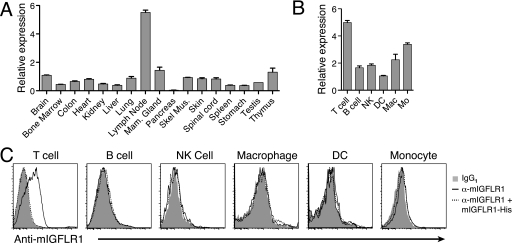
mIGFLR1 Is Highly Expressed in Mouse T Cells
To gain further insight for a potential function for IGFLR1, we used RT-PCR on RNA isolated from normal mice to determine the tissue distribution of mIGFLR1. RNA for mIGFLR1 was ubiquitously expressed throughout the tissue panel, with the highest expression levels observed in the lymph node (Fig. 7A). We also found that expression levels of mouse or human IGFLR1 were not altered in psoriasis models or psoriasis (data not shown). To better understand the expression pattern of mIGFLR1 within the immune system, we analyzed RNA from sorted leukocyte populations of mouse spleens and found mIGFLR1 was most highly expressed in T cells and monocytes, although it was expressed in all cell types analyzed (Fig. 7B). Antibody staining of leukocytes confirmed surface expression of mIGFLR1 on T cells but not other leukocytes examined (Fig. 7C). We could also compete anti-mIGFLR1 T cell surface staining with soluble recombinant mIGFLR1-His. No IGFLR1 was detectible on the surface of primary human keratinocytes (data not shown). These results indicate that mouse T cells could be the target cell population for mIGFL or IGFL1 produced in the skin.
FIGURE 7.
Murine IGFLR1 is expressed in lymph nodes and T cells. A, RT-PCR analysis of IGFLR1 expression in normal mouse tissue is shown. mIGFLR1 copy numbers were determined by comparing sample Ct values to Ct values generated with a dilution series of a known copy number of mIGFL cDNA, then normalized to the average mIGFLR1 expression levels (±S.D.). B, RT-PCR analysis of mIGFLR1 expression in purified leukocyte populations is shown. Leukocyte populations from spleens were sorted by flow cytometry based on surface expression of the lineage markers. Relative levels of IGFLR1 were then determined by RT-PCR performed on RNA isolated from sorted cells (±S.E.). Mac, macrophages; Mo, monocytes. C, shown is surface expression of IGFLR1. Single cell suspensions from lymph nodes, spleen, or peripheral blood mononuclear cells were blocked for nonspecific binding to antibodies and then labeled with anti-mIGFLR1 in the presence or absence of recombinant mIGFLR1-His followed by an isotype-specific phycoerythrin-labeled secondary antibody. Cells were then labeled with lineage-specific antibodies and analyzed by flow cytometry.
DISCUSSION
In this report we have demonstrated increased expression of mIGFL and IGFL1 in skin during inflammation associated with psoriasis. Furthermore, we have described a high affinity interaction between these proteins and a transmembrane protein, IGFLR1, that is primarily expressed on mouse T cells. In psoriasis, keratinocyte-derived proteins can enhance inflammatory infiltration and activation. Given the critical role T cells play in the pathogenesis of psoriasis, these IGFL family members may represent additional factors that augment the communication between keratinocytes and cells of the immune system.
Although we found a marked increase in mIGFL and IGFL1 expression in inflamed skin from Imiquimod-treated mice and psoriasis patients, we were unable to detect mIGFL or IGFL1 transcripts in primary human leukocytes or mouse splenocytes under resting or activating conditions. These data indicate that the source of IGFL in inflamed skin was most likely not the infiltrating leukocytes. There is ample evidence indicating that the abnormal behavior of keratinocytes is a key feature in the pathogenesis of psoriasis (7). IL-1β, IL-17A, IL-22, IFNγ, and TNFα have been identified as important factors that mediate these responses in keratinocytes, allowing them to modulate inflammation by up-regulating MHC expression, costimulatory ligands, chemokines, and adhesion molecules (3, 23–25). Considering these observations, we sought to determine whether these cytokines influenced IGFL1 expression by keratinocytes. Of the cytokines tested, TNFα was uniquely able to up-regulate IGFL1 in cultured primary keratinocytes, demonstrating that IGFL up-regulation was not a general response of keratinocytes to cytokine stimulation. The precise mechanism for TNFα-induced IGFL1 up-regulation remains to be identified, but the rapid up-regulation of IGFL1 in primary keratinocytes after cytokine treatment suggests a direct link with TNFα signaling and not dependence on TNFα target genes. TNFα is a proinflammatory cytokine with a well established link to psoriasis, as demonstrated both by the elevation of TNFα in psoriatic skin (26) and by the effectiveness of anti-TNF treatment for psoriasis patients (27–29). It is possible that a common pathway led to the up-regulation of mIGFL observed in the cutaneous skin-wounding model as TNFα is up-regulated after full thickness skin punches in mice (30, 31).
Microarray analysis of TNFα-treated primary keratinocytes revealed that this cytokine induces genes involved in immune and inflammatory responses and genes that are involved in healing (32). However, there are a number of experiments that indicate a negative role for TNFα in wound healing (30, 31, 33–35). We found the highest mIGFLR1 RNA expression in lymph nodes and T cells. T cells were also the only leukocyte population that had detectible mIGFLR1 on their surface, indicating that they are the apparent target cell type for IGFL produced in skin injury. However, more studies will be required to determine significance of IGFL up-regulation in inflammation and wounding. On one hand, the early responses of keratinocytes to TNFα stimulation in general stimulate inflammation and T cell activation via the induction of chemokines and cytokines, which are up-regulated in as little as 1 h. On the other hand, IGFL could regulate apoptosis or other inhibitory programs through the putative DD of IGFLR1. Further investigation will be necessary to determine which category the activities of mIGFL and IGFL1 fall, whether they can enhance the pathogenic process associated with psoriasis or suppress the immune system and promote healing. It is possible that other cytokines can regulate IGFL expression either alone or in concert with TNFα, and it would be important to determine how mIGFL and IGFL1 were regulated during other inflammatory conditions in the skin.
The altered or lack of gene expression changes associated with psoriasis suggest that IGFL2, -3, and -4 have biological roles that are unique and non-redundant to IGFLl. Many gene families involved in the immune response have diverged from mouse and humans presumably because of selective evolutionary pressure from environmental challenges. Examples of this occur in the TIM family, which has 3 members in humans and 8 in mice (35), and the Siglec family, which has 12 human members and 8 mouse members, many of which do not have a definite human homologs (36). In this case the IGFL family may have undergone expansion to provide a broader, pleiotropic function. Interestingly, despite the expansion of the human IGFL family compared with mouse, we find that two of the four human IGFL genes retain binding functionality for the putative receptor IGFLR1. Conversely, in humans there have been no clear IGFLR1 duplications. The fact that IGFL2 and IGFL4 failed to interact with IGFLR1 with high affinity would lead us to speculate that these two genes either interact with unidentified receptors or may serve to regulate IGFL1 and IGFL3. These possibilities are especially interesting considering the counterregulation of IGFL2 and IGFL1 in psoriatic skin samples. An important question for future study will be to determine whether any of the human IGFL proteins form heterodimeric complexes in vivo.
The binding of IGFL1 and IGFL3 to IGFLR1 despite only a 37% amino acid identity suggests a conservation of function. Furthermore, subnanomolar binding affinity is conserved in mice, where the extracellular domain of mIGFLR1 has only 51% identity with human IGFLR1. The diversity of the receptors employed by the IGF family demonstrates the adaptability of this family to signaling through different protein families. Interestingly, the insulin receptor and IGF-I receptor contain an extracellular domain that has a CRD with structural similarity to the CRD of the TNFR family (37). The CRD of IGF-I receptor is important for the high affinity binding of IGF-I (38, 39), providing some precedence for binding of an IGF family protein to a TNFR-like CRD. We are currently investigating the functional consequences of IGFL binding to IGFLR1. Given the observations that mIGFL and IGFL1 are both up-regulated during skin inflammation and have conserved binding to IGFLR1, it is highly likely that these interactions have a conserved functional role.
Analysis of the amino acid sequence of IGFLR1 indicated that structurally it likely comprises a novel member of the TNFR family. The TNFR family is a group of about 30 proteins that regulate cell survival, proliferation, differentiation, and apoptosis (40). TNFRs are most widely expressed in the immune system where they regulate both innate and adaptive immune responses but also have roles outside the immune system. Although their primary sequences are quite diverse, the tertiary folds of TNFR family member ECDs are similar and are composed of modular CRD subunits (22). The sequence of these modules have cysteine residues with a conserved register that form intrachain disulfide bridges and are strung together to form the elongated structure of TNFR family ECDs. Using fold recognition programs, the top hits for IGFLR1 were universally TNFR family members with high confidence, whereas sequence alignments indicated highest similarity between IGFLR1 and NGFR, ectodysplasin-A receptor, and X-linked ectodysplasin-A2 receptor. A three-dimensional model of IGFLR1 is consistent with an overall TNFR-like structure composed of distinct CRDs (data not shown). Fold prediction also strongly suggested that the highly conserved region in the cytoplasmic domain of IGFLR1 has a DD-like structure. DDs are globular folds consisting of six α-helical bundles that function as adaptors to recruit proteins to larger protein complexes (41). A number of the TNFR family members contain cytoplasmic DD, and activation of these receptors leads to recruitment of other DD-containing components of the death-inducing signaling complex that initiates signaling cascades resulting in programmed cell death. On the other hand, the DD-containing TNFR1 is a well established mediator of proinflammatory responses via NF-κB activation. Further experimentation will be needed to define the interaction partners and role of this domain in signaling.
Members of the TNFR family interact most commonly with the TNF family of ligands, although as protein interactions are mapped further, the number of non-TNF proteins that interact with TNFR family members has grown. For example, high affinity interactions have been demonstrated between herpes virus entry mediator (HVEM) and glycoprotein D of the herpesvirus (42), CD160 (43) and the B and T lymphocyte attenuator (44), between NGFR and members of the neurotrophin family (45), and between DR6 and β-amyloid precursor protein (46). Although these proteins, like IGFL, are not traditional TNF family members, they retain the ability to activate their respective receptors (45–47).
In conclusion, our data point to a role of IGFL in skin biology and a possible connection to a novel TNFR family member expressed on T cells in mice. Further studies will aim to evaluate the relationship between these two genes in regulating skin and immune responses.
Acknowledgments
We thank Ganesh Kolumam and Mercedesz Balasz for wound healing and Imiquimod experiments; Zora Modrusan and Matt Bauer for microarray experiments and data analysis; Lauri Diehl and Gretchen Frantz for in situ hybridization; Anan Chuntharapai, Xinhua Wang, Tao Sai, and Thresa Shek for monoclonal anti-IGFLR1 antibodies. We thank Sarah Hymowitz for helpful discussions on TNFR structural homology and CRD sequence alignments and Athena Wong and Phil Hass for protein production.
All authors are (or were) employees of Genentech at the time this work was conducted.
S. R. Ramani, I. Tom, N. Lewin-Koh, B. Wranik, L. DePalatis, J. Zhang, D. Eaton, and L. Gonzalez, manuscript in preparation.
- IGFL
- IGF-like
- IGFLR1
- IGF-like family receptor 1
- mIGFL
- mouse IGFL
- CRD
- cysteine-rich domain
- ECD
- ectodomain
- NGFR
- nerve growth factor receptor
- SPR
- surface plasmon resonance
- TNFR
- TNF receptor
- PSNE
- psoriatic adjacent non-effected
- DD
- death domain
- FADD
- FAS-associated death domain protein.
REFERENCES
- 1. Tonel G., Conrad C. (2009) Int. J. Biochem. Cell Biol. 41, 963–968 [DOI] [PubMed] [Google Scholar]
- 2. Lowes M. A., Kikuchi T., Fuentes-Duculan J., Cardinale I., Zaba L. C., Haider A. S., Bowman E. P., Krueger J. G. (2008) J. Investig. Dermatol. 128, 1207–1211 [DOI] [PubMed] [Google Scholar]
- 3. Teunissen M. B., Koomen C. W., de Waal Malefyt R., Wierenga E. A., Bos J. D. (1998) J. Investig. Dermatol. 111, 645–649 [DOI] [PubMed] [Google Scholar]
- 4. Wolk K., Haugen H. S., Xu W., Witte E., Waggie K., Anderson M., Vom Baur E., Witte K., Warszawska K., Philipp S., Johnson-Leger C., Volk H. D., Sterry W., Sabat R. (2009) J. Mol. Med. 87, 523–536 [DOI] [PubMed] [Google Scholar]
- 5. Wolk K., Witte E., Wallace E., Döcke W. D., Kunz S., Asadullah K., Volk H. D., Sterry W., Sabat R. (2006) Eur. J. Immunol. 36, 1309–1323 [DOI] [PubMed] [Google Scholar]
- 6. Sano S., Chan K. S., Carbajal S., Clifford J., Peavey M., Kiguchi K., Itami S., Nickoloff B. J., DiGiovanni J. (2005) Nat. Med. 11, 43–49 [DOI] [PubMed] [Google Scholar]
- 7. Albanesi C., De Pità O., Girolomoni G. (2007) Clin. Dermatol. 25, 581–588 [DOI] [PubMed] [Google Scholar]
- 8. Emtage P., Vatta P., Arterburn M., Muller M. W., Park E., Boyle B., Hazell S., Polizotto R., Funk W. D., Tang Y. T. (2006) Genomics 88, 513–520 [DOI] [PubMed] [Google Scholar]
- 9. Wong A. W., Scales S. J., Reilly D. E. (2007) J. Biol. Chem. 282, 22953–22963 [DOI] [PubMed] [Google Scholar]
- 10. Wong A. W., Baginski T. K., Reilly D. E. (2010) Biotechnol. Bioeng. 106, 751–763 [DOI] [PubMed] [Google Scholar]
- 11. Liu F., Song Y., Liu D. (1999) Gene Ther. 6, 1258–1266 [DOI] [PubMed] [Google Scholar]
- 12. Jubb A. M., Pham T. Q., Frantz G. D., Peale F. V., Jr., Hillan K. J. (2006) Methods Mol. Biol. 326, 255–264 [DOI] [PubMed] [Google Scholar]
- 13. Clark H. F., Gurney A. L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Currell B., Deuel B., Dowd P., Eaton D., Foster J., Grimaldi C., Gu Q., Hass P. E., Heldens S., Huang A., Kim H. S., Klimowski L., Jin Y., Johnson S., Lee J., Lewis L., Liao D., Mark M., Robbie E., Sanchez C., Schoenfeld J., Seshagiri S., Simmons L., Singh J., Smith V., Stinson J., Vagts A., Vandlen R., Watanabe C., Wieand D., Woods K., Xie M. H., Yansura D., Yi S., Yu G., Yuan J., Zhang M., Zhang Z., Goddard A., Wood W. I., Godowski P., Gray A. (2003) Genome Res. 13, 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munson P. J., Rodbard D. (1980) Anal. Biochem. 107, 220–239 [DOI] [PubMed] [Google Scholar]
- 15. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 17. Söding J. (2005) Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 18. Flöckner H., Braxenthaler M., Lackner P., Jaritz M., Ortner M., Sippl M. J. (1995) Proteins 23, 376–386 [DOI] [PubMed] [Google Scholar]
- 19. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 20. van der Fits L., Mourits S., Voerman J. S., Kant M., Boon L., Laman J. D., Cornelissen F., Mus A. M., Florencia E., Prens E. P., Lubberts E. (2009) J. Immunol. 182, 5836–5845 [DOI] [PubMed] [Google Scholar]
- 21. Zhang G. (2004) Curr. Opin. Struct. Biol. 14, 154–160 [DOI] [PubMed] [Google Scholar]
- 22. Naismith J. H., Sprang S. R. (1998) Trends Biochem. Sci. 23, 74–79 [DOI] [PubMed] [Google Scholar]
- 23. Banno T., Adachi M., Mukkamala L., Blumenberg M. (2003) Antivir. Ther. 8, 541–554 [PubMed] [Google Scholar]
- 24. Sa S. M., Valdez P. A., Wu J., Jung K., Zhong F., Hall L., Kasman I., Winer J., Modrusan Z., Danilenko D. M., Ouyang W. (2007) J. Immunol. 178, 2229–2240 [DOI] [PubMed] [Google Scholar]
- 25. Wolk K., Witte E., Warszawska K., Schulze-Tanzil G., Witte K., Philipp S., Kunz S., Döcke W. D., Asadullah K., Volk H. D., Sterry W., Sabat R. (2009) Eur. J. Immunol. 39, 3570–3581 [DOI] [PubMed] [Google Scholar]
- 26. Ettehadi P., Greaves M. W., Wallach D., Aderka D., Camp R. D. (1994) Clin. Exp. Immunol. 96, 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaudhari U., Romano P., Mulcahy L. D., Dooley L. T., Baker D. G., Gottlieb A. B. (2001) Lancet 357, 1842–1847 [DOI] [PubMed] [Google Scholar]
- 28. Leonardi C. L., Powers J. L., Matheson R. T., Goffe B. S., Zitnik R., Wang A., Gottlieb A. B. (2003) N. Engl. J. Med. 349, 2014–2022 [DOI] [PubMed] [Google Scholar]
- 29. Reich K., Nestle F. O., Papp K., Ortonne J. P., Evans R., Guzzo C., Li S., Dooley L. T., Griffiths C. E. (2005) Lancet 366, 1367–1374 [DOI] [PubMed] [Google Scholar]
- 30. Lai J. J., Lai K. P., Chuang K. H., Chang P., Yu I. C., Lin W. J., Chang C. (2009) J. Clin. Investig. 119, 3739–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mori R., Kondo T., Ohshima T., Ishida Y., Mukaida N. (2002) FASEB J. 16, 963–974 [DOI] [PubMed] [Google Scholar]
- 32. Banno T., Gazel A., Blumenberg M. (2004) J. Biol. Chem. 279, 32633–32642 [DOI] [PubMed] [Google Scholar]
- 33. Bettinger D. A., Pellicane J. V., Tarry W. C., Yager D. R., Diegelmann R. F., Lee R., Cohen I. K., DeMaria E. J. (1994) J. Trauma 36, 810–814 [DOI] [PubMed] [Google Scholar]
- 34. Buck M., Houglum K., Chojkier M. (1996) Am. J. Pathol. 149, 195–204 [PMC free article] [PubMed] [Google Scholar]
- 35. Freeman G. J., Casasnovas J. M., Umetsu D. T., DeKruyff R. H. (2010) Immunol. Rev. 235, 172–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Varki A., Angata T. (2006) Glycobiology 16, 1R–27R [DOI] [PubMed] [Google Scholar]
- 37. Ward C. W., Hoyne P. A., Flegg R. H. (1995) Proteins 22, 141–153 [DOI] [PubMed] [Google Scholar]
- 38. Kjeldsen T., Andersen A. S., Wiberg F. C., Rasmussen J. S., Schäffer L., Balschmidt P., Møller K. B., Møller N. P. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4404–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vashisth H., Abrams C. F. (2010) J. Mol. Biol. 400, 645–658 [DOI] [PubMed] [Google Scholar]
- 40. Locksley R. M., Killeen N., Lenardo M. J. (2001) Cell 104, 487–501 [DOI] [PubMed] [Google Scholar]
- 41. Park H. H., Lo Y. C., Lin S. C., Wang L., Yang J. K., Wu H. (2007) Annu. Rev. Immunol. 25, 561–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whitbeck J. C., Peng C., Lou H., Xu R., Willis S. H., Ponce de Leon M., Peng T., Nicola A. V., Montgomery R. I., Warner M. S., Soulika A. M., Spruce L. A., Moore W. T., Lambris J. D., Spear P. G., Cohen G. H., Eisenberg R. J. (1997) J. Virol. 71, 6083–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai G., Anumanthan A., Brown J. A., Greenfield E. A., Zhu B., Freeman G. J. (2008) Nat. Immunol. 9, 176–185 [DOI] [PubMed] [Google Scholar]
- 44. Gonzalez L. C., Loyet K. M., Calemine-Fenaux J., Chauhan V., Wranik B., Ouyang W., Eaton D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1116–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barker P. A. (2004) Neuron 42, 529–533 [DOI] [PubMed] [Google Scholar]
- 46. Nikolaev A., McLaughlin T., O'Leary D. D., Tessier-Lavigne M. (2009) Nature 457, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Cheung T. C., Steinberg M. W., Oborne L. M., Macauley M. G., Fukuyama S., Sanjo H., D'Souza C., Norris P. S., Pfeffer K., Murphy K. M., Kronenberg M., Spear P. G., Ware C. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6244–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]