Abstract
S-Palmitoylation of G protein-coupled receptors (GPCRs) is a prevalent modification, contributing to the regulation of receptor function. Despite its importance, the palmitoylation status of the β1-adrenergic receptor, a GPCR critical for heart function, has never been determined. We report here that the β1-adrenergic receptor is palmitoylated on three cysteine residues at two sites in the C-terminal tail. One site (proximal) is adjacent to the seventh transmembrane domain and is a consensus site for GPCRs, and the other (distal) is downstream. These sites are modified in different cellular compartments, and the distal palmitoylation site contributes to efficient internalization of the receptor following agonist stimulation. Using a bioorthogonal palmitate reporter to quantify palmitoylation accurately, we found that the rates of palmitate turnover at each site are dramatically different. Although palmitoylation at the proximal site is remarkably stable, palmitoylation at the distal site is rapidly turned over. This is the first report documenting differential dynamics of palmitoylation sites in a GPCR. Our results have important implications for function and regulation of the clinically important β1-adrenergic receptor.
Keywords: Adrenergic Receptor, G Protein-coupled Receptor (GPCR), Intracellular Trafficking, Post-translational Modification, Protein Acylation, Protein Palmitoylation, Bioorthogonal Labeling, Click Chemistry, Internalization
Introduction
The β1-adrenergic receptor (AR)3 is a G protein-coupled receptor (GPCR) critical to proper heart function and memory formation (1–3) and is the major cardiac target of β-blocker therapy for patients of chronic heart failure (4). Increasingly detailed molecular characterization of AR structure and signaling has led to novel treatment strategies, including rational drug design (5), targeting multiple components of the signaling complex (6, 7), and the potential to personalize treatment for patients based on genetic background (8).
The covalent addition of palmitic acid to cytoplasmic cysteine residues via a thioester bond is a prevalent modification of GPCRs. Unlike other acyl modifications, S-palmitoylation is reversible, and many proteins have regulated cycles of palmitoylation and depalmitoylation (9). Unlike soluble substrates, S-palmitoylation of integral membrane GPCRs is not required for membrane association, but instead changes their structure and contributes variably to receptor function (10, 11).
The palmitoylation status of β1AR has not yet been determined. Recently the crystal structure of turkey β1AR was solved, providing insight into the organization of the transmembrane domains, and the ligand binding pocket (12). This structure, however, lacked a large portion of the C-terminal tail and had a mutated putative palmitoylation site. Thus, unlike for the crystal structures of rhodopsin, which included palmitoylated cysteines (13, 14), no information on β1AR palmitoylation was gained.
We recently discovered that efficient delivery of β1AR to the cell surface required expression of a Golgi resident protein, golgin-160 (15). S-Palmitoylation is known to influence the trafficking and the specific subcellular localization of many substrates (16, 17). It has also been reported that golgin-160 interacts with GCP16 (18), which is a subunit of the Ras palmitoyltransferase (19). Thus, we reasoned that golgin-160 might influence the surface expression of β1AR by promoting proper palmitoylation at the Golgi. To test this hypothesis, we first investigated the palmitoylation of β1AR.
We report here that β1AR is S-palmitoylated on its C-terminal tail proximal to the seventh transmembrane domain at residues Cys392 and/or Cys393, which comprise a de facto consensus site for GPCR palmitoylation. Unexpectedly, we identified a second site of palmitoylation, further downstream on the tail at residue Cys414. These sites are modified in different subcellular compartments, and mutation of Cys414 but not Cys392 or Cys393 affects agonist-mediated internalization of β1AR. Interestingly, although the palmitate modification at the proximal site is quite stable, modification at the distal site is rapidly turned over. These results provide new information on β1AR modification and will inform future experiments that rely on an accurate structural understanding of this receptor.
EXPERIMENTAL PROCEDURES
Expression Constructs
A plasmid encoding human FLAG-tagged β1AR in pcDNA3 was provided by Randy Hall (Emory University, Atlanta, GA). Mutations were introduced using PCR-based QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). Nucleotide mutations introduced (individually, or in combination) were for Cys392 to Ser (nucleotides 1175 G to C); Cys393 to Ser (nucleotides 1178 G to C) and Cys414 to Ser (nucleotides 1241 G to C).
Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 10% fetal calf serum (FCS) and 0.1 mg/ml normocin-O (InvivoGen, San Diego, CA) (or lacking normocin-O for alkynyl-16 (alk-16) labeling experiments) at 37 °C in 5% CO2. For transient overexpression assays, cells were transfected with 3 μl of FuGENE6 (Roche Applied Science) or Lipofectamine 2000 (Invitrogen)/1 μg of cDNA.
[3H]Palmitic Acid Labeling
Transiently transfected cells were labeled for 30 min with 0.5 ml of 0.5 mCi/ml [3H]palmitic acid (PerkinElmer Life Sciences) in DMEM with 1% dimethyl sulfoxide, 2.5% FCS, 1 × nonessential amino acids, and 1 × sodium pyruvate (Invitrogen). Cells were lysed in detergent solution (50 mm Tris, pH 8.0, 62.5 mm EDTA, 1% Nonidet P-40, 0.4% deoxycholic acid) for 20 min at 0 °C, and debris was removed by centrifugation at 16,000 × g for 15 min. FLAG-β1AR was immunoprecipitated with FLAG-M2 beads (Sigma) as described (15). Protein was eluted with 2 × SDS-PAGE buffer (100 mm Tris, pH 6.8, 4% SDS, 30% glycerol, 0.1% bromphenol blue, 2% β-mercaptoethanol) for 20 min at room temperature. The sample was resolved by SDS-PAGE. The gel was equilibrated in dimethyl sulfoxide and incubated for 2 h in 2,5-diphenyloxazole, washed with water, dried, and exposed to film at −80 °C.
Metabolic Labeling with Bioorthogonal Palmitate Reporter
Transiently transfected cells were labeled for 30 min with 0.5 ml of 50 μm alk-16 in DMEM with 10% FCS. To determine palmitate turnover, labeled cells were chased in normal growth medium for the indicated times. Cells were lysed in Brij lysis buffer (1% Brij-97, 150 mm NaCl, 50 mm triethanolamine, pH 7.4) with EDTA-free protease inhibitor mixture (Roche Applied Science) on ice. Cell lysates were collected following centrifuging at 16,000 × g for 20 min at 4 °C to remove cell debris. Immunoprecipitations were performed using anti-FLAG-M2 affinity resin as above. The beads were resuspended in 40 μl of SDS buffer (4% SDS, 50 mm triethanolamine, pH 7.4, 150 mm NaCl) and 3 μl of freshly prepared click-chemistry reaction mixture (azide-rhodamine (100 μm, 10 mm stock solution in dimethyl sulfoxide), TCEP (1 mm, 50 mm freshly prepared stock solution in deionized water), TBTA (100 μm, 10 mm stock solution in dimethyl sulfoxide), and CuSO4·5H2O (1 mm, 50 mm freshly prepared stock solution in deionized water)). Reactions were incubated with shaking for 1 h at 30 °C. The reactions were diluted with 5 × SDS-PAGE buffer (250 mm Tris, pH 6.8, 10% SDS, 50% glycerol, 0.5% bromphenol blue) and 0.5% β-mercaptoethanol and incubated for 20 min at 37 °C. Reactions were resolved by SDS-PAGE.
In-gel Fluorescence Imaging and Immunoblotting
After proteins were separated by SDS-PAGE, the gel was washed twice with deionized water for a total of 20 min. Palmitoylated β1AR was visualized by directly scanning the gel (excitation 532 nm, 580 nm filter, 30-nm band pass) on a Typhoon 9400 imager (GE Healthcare). No signal saturation was observed. Images were processed and analyzed using the ImageQuant TL software (GE Healthcare). Following in-gel fluorescence imaging, total β1AR was detected by either in-gel immunoblotting or traditional immunoblotting as described previously (15).
For in-gel immunoblotting, gels were washed in PBS with 0.1% Tween 20 (PBST) for 10 min at room temperature. Gels were incubated with anti-FLAG-M2 antibody (Sigma) in PBST followed by IRDye800-conjugated anti-mouse IgG secondary antibody (Rockland, Gilbertsville, PA) in PBST. In all cases, immunoblot images were collected using the Odyssey infrared imaging system (Licor, Lincoln, NE). For data analysis, the alk-16 signal was normalized to the relative amount of total β1AR detected by immunoblotting. For Fig. 5B, the normalized signal for the wild-type protein was set to 100% for each experiment, and the normalized signals from all mutants were compared with this signal. For Fig. 5C, the total signal for each β1AR construct in each experiment was set to 100%, and the contributions of mature and immature bands were calculated. For Fig. 6B, the 0 chase time point was set to 100%, and subsequent signals (normalized based on expression level) for each mutant from each experiment were compared. Variance was determined by one-way ANOVA, and p values were calculated with the Tukey test.
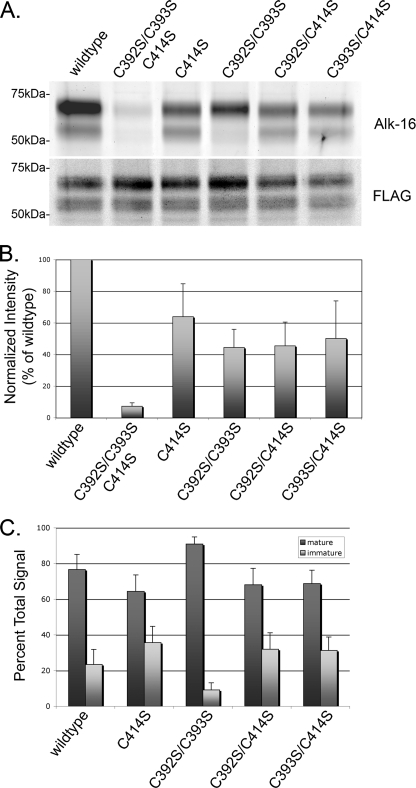
FIGURE 5.
Palmitoylation of the immature β1AR takes place at the proximal site. A, representative gel showing incorporation of alk-16 into the indicated β1AR construct (upper) and relative β1AR levels by immunoblotting with anti-FLAG antibody (lower). B, mean intensity of alk-16 signal from five separate labeling experiments, normalized to β1AR expression level and wild-type alk-16 signal. Error bars represent the S.D. C, mean contribution of mature and immature bands to the total signal for each construct. Error bars represent the S.D.
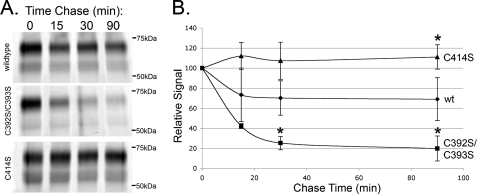
FIGURE 6.
β1AR palmitoylation sites are differentially regulated. A, representative gels for palmitoylation turnover experiments. Cells were pulse-labeled with alk-16 and chased for the indicated times. B, data points representing the mean intensity of signal relative to the 0 chase time point from three independent experiments. Error bars represent the S.D. *, p < 0.05 relative to wild type at that time point.
Measurement of β1AR Half-life
HEK293 cells grown in 35-mm dishes were transfected with 0.5 μg each of the indicated construct. 16 h later, the cells were starved for 15 min with DMEM lacking Met and Cys and labeled for 15 min in fresh Met/Cys-free DMEM with 0.2 mCi/ml Expre35S35S labeling mix (PerkinElmer Life Sciences). Medium was replaced with normal growth medium for the indicated times. Cells were lysed with detergent solution and immunoprecipitated as described above. SDS-polyacrylamide gels were dried, and radiolabeled proteins were detected by phosphorimaging (Molecular Imager FX, Bio-Rad). Bands were quantified using Quantity One software (Bio-Rad), and analysis was performed with Microsoft Excel.
Microscopy
HEK293 cells grown on poly-l-lysine-coated glass coverslips were fixed and permeabilized as described (20). Antibodies used were anti-FLAG-M2 (Sigma), sheep anti-TGN46 (Serotec, Raleigh, NC), Alexa Fluor 488 anti-mouse IgG (Molecular Probes, Eugene, OR), and Texas Red anti-sheep IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted in 1% fish skin gelatin. Images were collected on an Axioskop microscope (Zeiss, Thornwood, NY) equipped with epifluorescence and a Sensys CCD camera (Photometrics, Tucson, AZ) using IP Lab software (Signal Analytics, Vienna, VA).
Measurement of β1AR Surface Levels
HEK293 cells were grown on poly-l-lysine-coated wells in 12-well dishes and in 35-mm dishes for expression control. Each construct was transfected in triplicate in the 12-well dishes, plus one 35-mm dish, and one untransfected control to determine background binding. At 16 h after transfection, the cells in the 35-mm dishes were lysed as described (15) for analysis by Western blotting. Cells in the 12-well dishes were rinsed three times on ice with cold PBS, and incubated with 10 nm 3H-labeled CGP-12177 (PerkinElmer Life Sciences) in KRH buffer (136 mm NaCl, 4.7 mm KCl, 1.25 mm MgSO4, 1.25 mm CaCl2, 20 mm HEPES, pH 7.4, 2 mg/ml BSA) for 3 h at 4 °C. Cells were then rinsed three times on ice with cold PBS and lysed with detergent solution. Lysate was added to scintillation fluid and counted. For analysis, values were from samples where the maximum ligand binding was <30% of the input. Binding of ligand to nontransfected cells was <5% of that for cells expressing wild-type β1AR.
Internalization Assay
HEK293 cells grown on poly-l-lysine-coated glass coverslips transiently expressing the indicated FLAG-β1AR construct were fed anti-FLAG-M2 antibody at 1 μg/ml dilution and treated with or without 10 μm isoproterenol (Iso) at 37 °C (Sigma) for the times indicated. Following treatment, cells were washed with PBS and were untreated, or surface antibody was removed by an acid wash (0.5 m NaCl, 0.5% HOAc, pH 1) for 1 min at room temperature. Cells were washed with PBS, fixed, and permeabilized as above. Fixed cells were probed with rabbit anti-β1AR antibody (Santa Cruz Biotechnology, Santa Cruz, CA) with 1% bovine serum albumin (BSA), followed by incubation with Alexa Fluor 488 anti-mouse and Texas Red anti-sheep IgGs. All fields were selected for similar expression levels and expression profile in the anti-β1AR field before viewing in the anti-FLAG field. For each experiment, images were taken on the same day at the same shutter speed, and all manipulations of image intensity were applied consistently to all images. Average pixel intensity of internalized antibody was determined using ImageJ software (National Institutes of Health). Variance was determined by one-way ANOVA, and p values were calculated with the Tukey test.
RESULTS
β1AR Is Palmitoylated on Cysteines 392, 393, and 414
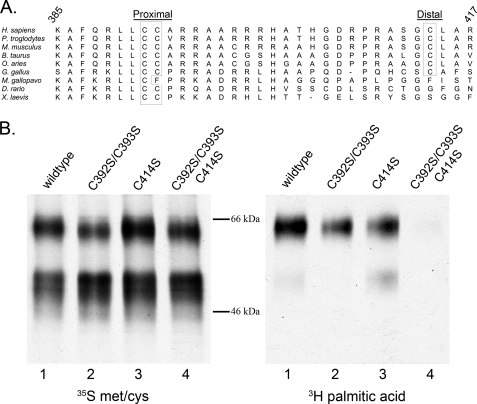
Many GPCRs are S-palmitoylated on their C-terminal tails, downstream of the seventh transmembrane domain (21). We used two programs to predict palmitoylation sites on β1AR, NBA-Palm, and CSS-Palm 2.0 (22, 23). Both programs predicted palmitoylation on cysteines 392 and 393, which reside at this position and are highly conserved across species (Fig. 1A). This position is also analogous to the palmitoylation site of the closely related β2AR, which has a single modified cysteine (24). To examine experimentally the palmitoylation state of β1AR, we incubated HEK293 cells transiently expressing FLAG-tagged β1AR with [3H]palmitic acid. Parallel dishes were incubated with [35S]methionine/cysteine to monitor protein expression levels. Lysates were immunoprecipitated with an anti-FLAG-M2 antibody and examined by fluorography. As described previously (15), two major bands were observed for β1AR: a faster migrating immature band (∼56 kDa) and a slower migrating mature band (∼64 kDa), representing the O-glycosylated mature form of β1AR (Fig. 1B, left) (25). Radiolabeled palmitic acid was incorporated into β1AR (Fig. 1B, right, lane 1), predominantly in the mature band. To confirm that the labeled palmitate was incorporated via a thioester bond, labeled β1AR was incubated with 1 m hydroxylamine. In-gel hydroxylamine treatment resulted in the loss of 3H signal, compared with a parallel gel treated with 1 m Tris (data not shown). These data indicate that, as expected, β1AR is modified with palmitic acid via a thioester bond. We next sought to identify the specific residues modified by palmitic acid. We expressed a construct with Cys392 and Cys393 mutated to serines (β1AR C392S/C393S) and found that although incorporation of [3H]palmitic acid was reduced, it was not eliminated (Fig. 1B, lane 2), suggesting additional sites of palmitoylation. We therefore introduced mutations at each of the other cytoplasmic facing cysteine residues (cysteines 261, 378, 414, 451, and 467) in combination with C392S/C393S. Only when Cys414 was mutated together with residues Cys392 and Cys393 was incorporation of [3H]palmitic acid abolished (Fig. 1B, lane 4, and data not shown). This site is highly, although not universally, conserved across species (Fig. 1A). Because of their relative positions on the C-terminal tail, we refer to residues 392 and 393 as the proximal palmitoylation site and amino acid 414 as the distal palmitoylation site. The triple cysteine mutant (C392S/C393S/C414S) is referred to as palmitoylation-null. Interestingly, the immature form of β1AR was labeled only when the proximal site cysteines were present (Fig. 1B), indicating that the proximal site is modified earlier in the secretory pathway than the distal site.
FIGURE 1.
β1AR is palmitoylated at cysteines 392, 393, and 414. A, BLAST alignment of residues 385–417 of the human β1AR with β1AR from the indicated species. B, [3H]palmitic acid signal revealed by fluorography (right), and similar protein expression demonstrated by [35S]methionine/cysteine label (left). The amino acids mutated from cysteine to serine are indicated.
Mutation of the Palmitoylation Sites Does Not Destabilize β1AR or Affect Its Steady-state Localization
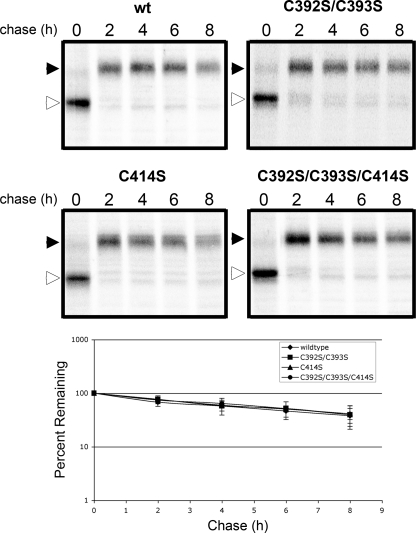
Mutation of the palmitoylation sites of several GPCRs leads to destabilization of the receptors, most likely due to misfolding. We found no significant difference in the half-lives or extent of maturation through the medial Golgi for any of the mutant proteins (Fig. 2).
FIGURE 2.
The half-life of β1AR is unaffected by cysteine to serine mutations. Top two panels, autoradiographs are representative of four or five independent experiments. The immature (open arrowhead) and mature (closed arrowhead) forms of β1AR are indicated. Bottom panel, data points on the graph represent the mean intensity of signal relative to the 0 chase time point. Error bars represent S.D. Half-lives: wild type, 6.0 h; C392S/C393S, 6.2 h; C414S, 6.3 h; C392S/C393S/C414S, 6.2 h.
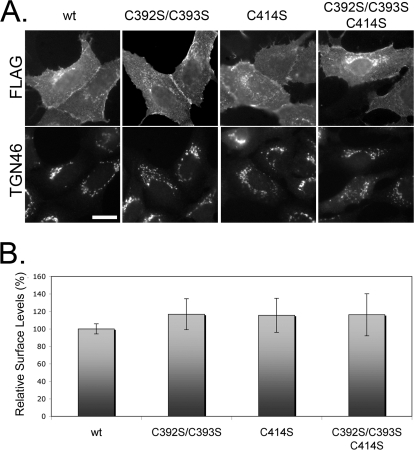
Because several GPCRs with mutated palmitoylation sites are trafficked inefficiently (26–29), we examined the steady-state distribution of β1AR palmitoylation mutants by indirect immunofluorescence microscopy using an antibody recognizing the N-terminal FLAG epitope. All mutant proteins were expressed at the plasma membrane, similar to the wild-type protein (Fig. 3A). The internal juxtanuclear staining co-localized with TGN46, a marker of the trans-Golgi network (Fig. 3A and supplemental Fig. S1), most likely representing β1AR en route to the plasma membrane. None of the mutant proteins accumulated in the endoplasmic reticulum, which together with the similar half-lives of the proteins, suggests that the mutant proteins were not misfolded.
FIGURE 3.
Mutation of β1AR palmitoylation sites does not affect steady-state surface levels. A, immunofluorescence micrograph showing the indicated β1AR constructs at cell surface and co-localized with Golgi marker, TGN46. Scale bar, 10 μm. B, surface levels of β1AR measured by radioactive ligand binding. Data represent the mean of at least three independent experiments. Error bars represent S.D.
To quantify the surface levels of β1AR, we assayed the binding of a radiolabeled ligand to the surfaces of cells expressing each of our constructs. We found no substantial difference in surface levels in cells expressing any of the mutants, compared with wild type (Fig. 3B). Nearly all binding was due to expression of the transfected β1AR constructs because untransfected controls bound less than 5% of ligand, relative to the wild-type β1AR-expressing samples (data not shown). Taken together, these data indicate that mutation of palmitoylated cysteines of β1AR does not disrupt the stability or steady-state distribution of the receptor. Thus, preventing palmitoylation of β1AR did not mimic the phenotype of reduced delivery to the cell surface observed in cells lacking golgin-160 (15). This observation along with the finding that overexpression of golgin-160 promotes palmitoylation-null β1AR surface expression similar to wild-type β1AR (data not shown) suggests that golgin-160 does not have a role in palmitoylation of β1AR.
Agonist-stimulated Internalization Is Impaired in Mutants Lacking a Distal Palmitoylation Site
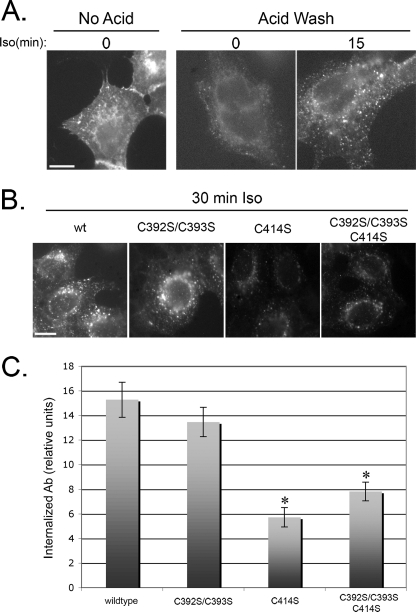
To evaluate the effect of β1AR palmitoylation on receptor internalization following agonist stimulation, we measured the surface levels of β1AR by binding radiolabeled ligand following stimulation with 10 μm Iso or vehicle control. However, we saw no significant change in surface levels (data not shown), consistent with previously published reports that β1AR has low levels of internalization following agonist stimulation in HEK293 cells (e.g. Ref. 30). To detect the low level of receptor internalized in these cells in a highly sensitive assay, we fed anti-FLAG-M2 antibody to live cells in the absence or presence of 10 μm Iso and visualized internalized antibody after removing surface antibody with an acid wash. Without an acid wash, the signal was primarily at the cell surface (Fig. 4A). However, when acid-washed, only weak, punctate staining representing internalized receptor was observed, indicating very low levels of basal internalization. This signal increased significantly for cells treated with Iso, although still represented only ∼6% of total fluorescent labeling (Fig. 4A and data not shown). Additionally, agonist stimulation did not lead to a significant loss of fluorescence signal in the absence of an acid wash, consistent with the results of the radioactive ligand binding experiment. Therefore, we examined the agonist stimulated internalization of β1AR by measuring the signal from internalized wild-type FLAG-β1AR or the indicated FLAG-β1AR mutants. We consistently observed that cells expressing FLAG-β1AR lacking the distal palmitoylation site internalized less antibody than when the distal site was intact (Fig. 4B). A quantification of the intensity of signal revealed that mutation of the distal site alone, or in combination with the proximal site, reduced internalization of β1AR by approximately half, whereas mutation of the proximal site alone caused no defect (Fig. 4C).
FIGURE 4.
Mutation of the distal palmitoylation site inhibits β1AR internalization following agonist stimulation. A, immunofluorescence micrographs of cells expressing FLAG-β1AR incubated with anti-FLAG antibody for 15 min in the absence or presence of 10 μm Iso. Prior to permeabilization and incubation with fluorescent secondary antibody, a brief acid wash (as indicated) was used to remove anti-FLAG bound to surface molecules for visualization of internalized antibody. Scale bar, 10 μm. B, representative immunofluorescence micrographs of acid-washed cells expressing the indicated constructs of FLAG-β1AR after 30 min of antibody uptake in the presence of 10 μm Iso. Images are representative of multiple fields photographed from three separate experiments. Scale bar, 10 μm. C, intensities of signal from experiments above quantified using ImageJ software. Error bars represent S.D. *, p < 0.01 relative to wild type.
Use of Novel Palmitoylation Reporter to Quantify Extent of Palmitoylation Accurately
To investigate the level of labeling at each site and to characterize the dynamics of palmitoylation, we used a recently developed labeling method that could be quantified easily and accurately. Proteins labeled with [3H]palmitate must be detected by fluorography, and it is difficult to obtain an accurate quantitative signal on x-ray film due to the nonlinear exposure of silver grains by photons (31). We thus used bioorthogonal labeling and in-gel fluorescence for quantification (32). Transiently transfected HEK293 cells were incubated with medium containing the bioorthogonal palmitic acid reporter (alk-16) for 30 min. After lysis and immunoprecipitation, samples were reacted with azide-rhodamine (on-bead copper-catalyzed azide-alkyne cycloaddition). This labeling method has been shown to be more specific, sensitive, and efficient than radioactive methods (32, 33). Following SDS-PAGE, in-gel fluorescence analysis provided a linear, quantitative signal. The labeling pattern was similar to what we observed after [3H]palmitic acid labeling (compare Figs. 1B and 5A). To quantify the labeling, the fluorescent signals from five independent experiments were normalized to overall β1AR expression level as determined by Western blotting of the same gel (Fig. 5B). When Cys392 and Cys393 were mutated to serines, the label was 44% ± 11% of that obtained for the wild-type protein. When Cys414 was mutated to serine, the label was 64% ± 21% of that for the wild-type protein. When both sites were mutated, a low level (7% ± 2%) of labeling was observed. It is possible that when the normally palmitoylated cysteines are absent, additional cysteines can be S-palmitoylated to a minor extent. To examine the usage of each of the cysteines in the proximal site, we expressed β1AR with Cys392 and Cys414 mutated to serine, and β1AR with Cys393 and Cys414 mutated to serine. No significant difference (45% ± 15 and 50% ± 24% of wild type, respectively) was observed compared with the distal site mutant with both proximal cysteines available. This most likely indicates that most β1AR molecules expressed in HEK293 cells are palmitoylated on only one of the two cysteines in the proximal site, with only a small percentage of receptor, if any, modified simultaneously on both cysteines.
We also observed that the immature form of β1AR was labeled in all cases where a proximal site cysteine was available, but not when both were mutated to serine (Fig. 5A). We quantified the contribution of signal from mature and immature bands for each construct (Fig. 5C). Although labeling of the immature form accounted for 23% ± 8% of the wild-type signal, it contributed only 9% ± 4% of the signal in the C392S/C393S mutant, an amount that is similar to the labeling of palmitoylation-null described above. This indicates that the proximal site can be palmitoylated before the protein is processed in the medial Golgi, but the distal site is primarily or exclusively modified later in the secretory pathway.
Turnover at Distal Site Is Highly Dynamic
The previous experiments measured the steady-state levels of palmitoylation at each site of β1AR. The intensity of labeling is determined by both the extent of the incorporation of alk-16 as well as the rates of turnover. Palmitoylation sites with high rates of turnover will have increased signal, due to replacement of nonlabeled palmitic acid with the labeled analog. To study the dynamics of S-palmitoylation at each site, we examined the rates of turnover of palmitate at the proximal and distal sites using pulse-chase labeling. Cells expressing wild-type β1AR, C392S/C393S, or C414S were labeled with alk-16 for 30 min followed by chase in medium lacking alk-16 for various times. Although the signal from the proximal site showed no reduction after 90 min of chase, the label incorporated at the distal site was rapidly turned over, with very little palmitoylated protein left at 15 min of chase (Fig. 6). Because the half-lives of the proteins were all much longer than the loss of signal at the distal site (Fig. 2), the loss of signal is most likely due to palmitic acid turnover and not protein degradation. Taken together, these data indicate that the proximal site is modified early in the secretory pathway and turns over slowly, whereas the distal site is modified after trafficking through the medial Golgi and has a high rate of turnover. The surprising difference in turnover at the proximal and distal sites makes comparison of steady-state palmitoylation at each site difficult because only newly synthesized β1AR appears to be palmitoylated at the proximal site, whereas a larger pool of mature β1AR is likely available for modification at the distal site.
DISCUSSION
β1AR Is S-Palmitoylated at Two Sites in Its Cytoplasmic Tail
We report here that β1AR is palmitoylated at the two cysteines residing on the C-terminal tail proximal to the membrane (Cys392 and Cys393) and further downstream at Cys414. By primary sequence, β1AR is most closely related to β2AR (52% amino acid identity), which has a single palmitoylated cysteine, equivalent to the proximal site of β1AR (24). Based on this homology and the lack of a known sequence requirement for palmitoyltransferases, it has been assumed that β1AR is S-palmitoylated only at the cysteines residing at this proximal site (12, 34, 35). Our findings underscore the necessity to determine experimentally all of the residues that are modified on GPCRs to provide a complete understanding of receptor regulation and function. Making conserved mutations to a protein of interest is also the most direct way to examine the contribution of palmitoylation to the function of that protein because treatment with an inhibitor (such as 2-bromopalmitate) globally prevents palmitoylation and may indirectly impact the function of a protein of interest.
The finding that β1AR has an additional palmitoylation site relative to β2AR is surprising given the similarities regarding ligand binding and tissue distribution. However, these receptors have distinct activities. Although β2AR localizes to caveolae in unstimulated cardiomyocytes and relocalizes following ligand binding, β1AR is distributed throughout the plasma membrane and does not relocalize after ligand binding (36). Similarly, within cardiomyocytes co-cultured with sympathetic ganglion neurons, β1- and β2AR localize to contact sites, but only β2AR relocalizes away from the contact sites following sympathetic ganglion neuron stimulation, revealing differences in the spatial-temporal regulation of the receptors (37). The two receptors have unique binding partners (38) and form distinct signaling complexes through varied interactions with cAMP phosphodiesterases, which are differently regulated in response to agonist signaling (39). β1AR and β2AR also promote distinct downstream signaling events. β1AR couples only to Gαs, and excessive stimulation leads to apoptosis of cardiomyocytes. On the other hand, β2AR can couple to either Gαs or Gαi, and stimulation was found to protect cardiomyocytes against apoptosis (40). S-Palmitoylation has been shown for other proteins to regulate behavior such as protein localization (particularly regarding cholesterol-rich domains) and protein-protein interactions (11). It is possible that associated proteins regulate the function of these receptors by modulating the palmitoylation state at each site, allowing for a “fine-tuned” response.
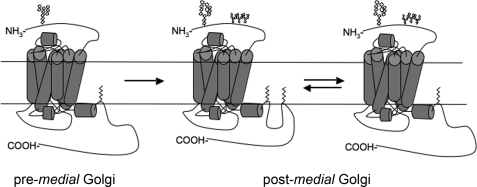
Some GPCRs are not S-palmitoylated, and the majority of those that are S-palmitoylated are modified only at the consensus site. The presence of an additional distal palmitoylation site on the tail of β1AR places it in a third group of GPCRs. The 5-hydroxytryptamine (HT) 4(a), 5-HT7(a), the TPβ isoform of thromboxane A2 (TPβ), and the follicle-stimulating hormone (FSH) receptors have all recently been reported to have distal palmitoylation sites, in addition to palmitoylation at the proximal site (41–44). Functionally, there is no obvious connection among these receptors, which have varied tissue distribution, signaling pathways, and are coupled to different G proteins. But they all likely adopt a conformation consisting of five intracellular loops when fully S-palmitoylated (Fig. 7), and the distal site may regulate receptor internalization similarly for all receptors (see below).
FIGURE 7.
Two potential cytoplasmic tail conformations dependent on palmitoylation at the distal site. Representation of β1AR with oligosaccharide and lipid modifications. Prior to arriving in the medial Golgi, β1AR is palmitoylated at the proximal site and has one immature (endoglycosidase H-sensitive) N-glycan. After trafficking through the medial Golgi, β1AR acquires O-linked glycans and is additionally palmitoylated at the distal site, generating a fifth cytoplasmic loop. Dynamic depalmitoylation and repalmitoylation of the distal site could allow a switch in tail conformation that would likely affect function of the receptor.
Distal Palmitoylation Site Contributes to Internalization following Agonist Stimulation
Following agonist binding and second messenger transduction, many GPCRs are desensitized, turning off signaling. The common route of desensitization involves phosphorylation by the GPCR kinase family and/or second messenger-regulated kinases PKA or PKC. Many receptors are then internalized and sequestered within the cell, destined for recycling to the surface, or down-regulation (for review, see Ref. 45). We observed a low level of β1AR internalization following agonist stimulation. This is consistent with several previous studies of β1AR internalization in HEK293 cells (e.g. 30), although not all (e.g. 46). In cell culture studies more closely resembling physiological conditions, Iso treatment of rat cardiac myocytes was found to cause internalization and down-regulation of β1AR. Interference with the endocytosis machinery caused surface retention of β1AR and deficient downstream signaling through Akt (47). In light of cell type differences, trafficking events other than internalization may be used to regulate β1AR function spatially (e.g. relocalization within the membrane; see above). We consider the internalization defect we observed for distal site mutants to be an intriguing preliminary observation, which may reflect a more specific paradigm of regulated relocalization that may only be observed in a relevant culture system and may be different for different cell types.
The involvement of a distal site in efficient internalization is analogous to observations made for the other GPCRs with acylation at distal sites. Mutation of the distal site reduced the rates of internalization of the FSH receptor (44), and although all three palmitoylation sites of TPβ contributed to ligand-induced internalization, only the distal site mutants were deficient in tonic internalization. By contrast, the proximal site alone was found to promote maximal coupling to Gαq (42). Studies also show a contribution of palmitoylation to internalization of 5-HT4(a). Unlike β1AR, FSH, or TPβ receptors, mutation of the 5-HT4(a) distal sites did not inhibit internalization. However, when the proximal site was mutated, there was a pronounced increase in ligand-stimulated internalization that was lost when the distal sites were mutated in combination (48), indicating the need for an intact distal site for the hyperinternalization phenotype. The consequence of mutation of the distal site to HT7(a) internalization was not reported (43). It is interesting to note that such a diverse group of GPCRs appear to have a similar use for a distal palmitoylation site, although the significance and universality of this feature are currently unclear. Characterization of additional GPCRs, as well as a better mechanistic understanding of the distal site usage, may reveal a common route.
Differences in Regulation at Two Palmitoylation Sites Suggest Unique Functions
To measure the relative levels of S-palmitoylation and the dynamics of turnover accurately, we needed a quantitative measure of palmitoylation. Fluorography is required to detect tritium on x-ray films, and densitometry of these films is imprecise due to the inherent nonlinearity of exposure of the silver grains by photons (31). To quantify S-palmitoylation accurately, we used a newly developed method of bioorthogonal labeling and in-gel fluorescence. This method allowed us to quantify levels of incorporation of the palmitic acid analog accurately and to normalize the signal to overall β1AR expression levels by immunoblotting of the same gel. This method provided a linear fluorescent signal that could be easily quantified and was more practical than the acyl-biotin switch assay or mass spectrometry (49). We observed that the sums of the signal obtained from mutations at the proximal and distal site (43 and 61%, respectively) nearly equaled the signal from the wild-type protein. This further suggests that the sites are independently modified and attests to the accuracy of the labeling method.
Additionally, we were able to use the bioorthogonal palmitate reporter in pulse-chase experiments to compare the rates of turnover for palmitic acid at each site. Unlike other acyl modifications, S-palmitoylation is reversible, and many proteins have regulated cycles of palmitoylation and depalmitoylation (9). We found palmitoylation of β1AR at the proximal site to be relatively stable. By contrast, palmitate at the distal site was rapidly turned over, with nearly all signal chased within 15 min. Because of the stable modification at the proximal site, it was not possible to calculate a half-life for the palmitoylation accurately with the chase times we used. However, because there was no loss of signal by 90 min, it is possible that the proximal site is modified once and only once during the course of the life of the protein. Thus, S-palmitoylation of the proximal site could cause a stable structural modification, as opposed to that at the distal site, which is more dynamic and therefore may function as a “switch” (Fig. 7). This is the first report of differential S-palmitoylation dynamics in a GPCR and predicts important functional consequences.
Consequences of β1AR Palmitoylation
We observed that S-palmitoylation at the two sites in newly synthesized β1AR likely occurs in different cellular compartments. The immature form of β1AR was palmitoylated only when the proximal site was intact. This indicates that the proximal site can be palmitoylated prior to trafficking through the Golgi. Because of the relatively long labeling time (compared with the rate of trafficking), we cannot currently determine whether the proximal site can also be palmitoylated in a later compartment or whether the mature signal represents β1ARs that were palmitoylated pre-Golgi or in the early Golgi and chased into the mature fraction. Because of the long half-life of the proximal modification, it does not appear likely that this signal comes from dynamic turnover of palmitic acid. This suggests that the two sites in β1AR may be modified by different protein palmitoyltransferases, allowing for a greater flexibility of receptor activity and additional pathways of regulation.
Due to early acquisition of palmitate at the proximal site of β1AR and its low rate of turnover, this modification may play a structural role contributing to the proper folding of β1AR. The proximal palmitoylation site is directly downstream of the eighth helix (H8), a structural component known to contribute to G protein coupling of many GPCRs, including rhodopsin (50, 51) and β1AR (52). We observed that the proximal site is palmitoylated early in the secretory pathway. It is possible that palmitoylation contributes to the structural stability of H8, strongly anchoring it to the membrane, thus contributing to G protein coupling. Although proteins mutated at this site do not have a shorter half-life in our cell culture system, it is possible that there is a long term consequence to an animal expressing β1AR mutated at this site, particularly under stress conditions. For example, a recent report describes the photoreceptor cell degeneration of mice expressing a palmitoylation-null rhodopsin, but only when the mice were exposed to bright light. Under normal laboratory lighting conditions, no defect was noted (53). It is possible that β1AR mutated at the proximal site would similarly have a dramatic defect in experiments testing stress conditions over a long term experiment in animal models.
We found that palmitoylation at the distal site of β1AR was highly dynamic, and mutation of this site impaired agonist stimulated internalization. It is therefore possible that regulation of this site contributes to desensitization following signaling. Despite their sequence similarities, β1AR does not behave like β2AR following ligand binding. β1AR internalizes at a higher rate in cardiomyocytes and does not relocalize away from contact sites with sympathetic ganglion neurons or out of caveolar fractions following ligand binding (36, 37). Therefore, regulation of S-palmitoylation at the distal site by a specific subset of palmitoyltransferases could contribute to discrimination between these receptors. This would provide additional control of the signaling response. β1AR could be desensitized by increased phosphorylation, arrestin binding, and/or decreased coupling to G proteins. It is possible that modulation of distal site palmitoylation can provide a rapid mechanism for control of these phenomena, contributing to desensitization, down-regulation, or recycling.
The prediction of palmitoylation sites is inexact and can only be conclusively demonstrated experimentally. We have identified three cysteines at two sites on β1AR that are palmitoylated. S-Palmitoylation at the two sites is apparently regulated independently because modification occurs in different compartments and is turned over at different rates. This study provides information necessary to investigate further the contribution of each palmitoylation site to the function of β1AR in specific contexts, cell culture, and animal systems.
Supplementary Material
Acknowledgments
We thank members of the Machamer laboratory and Egbert Hoiczyk for helpful discussions and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM42522 (to C. E. M.) and 1R01GM087544 (to H. C. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- AR
- adrenergic receptor
- alk-16
- alkynyl-16
- GPCR
- G protein-coupled receptor
- HT
- hydroxytryptamine
- Iso
- isoproterenol
- TPβ
- TPβ isoform of thromboxane A2
- TBTA
- Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- TCEP
- Tris (2-carboxyethyl) phosphine hydrochloride.
REFERENCES
- 1. Cahill L., Prins B., Weber M., McGaugh J. L. (1994) Nature 371, 702–704 [DOI] [PubMed] [Google Scholar]
- 2. Rohrer D. K., Desai K. H., Jasper J. R., Stevens M. E., Regula D. P., Jr., Barsh G. S., Bernstein D., Kobilka B. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7375–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winder D. G., Martin K. C., Muzzio I. A., Rohrer D., Chruscinski A., Kobilka B., Kandel E. R. (1999) Neuron 24, 715–726 [DOI] [PubMed] [Google Scholar]
- 4. Kaumann A. J., Molenaar P. (2008) Pharmacol. Ther. 118, 303–336 [DOI] [PubMed] [Google Scholar]
- 5. Lundstrom K. (2009) Methods Mol. Biol. 552, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel P. A., Tilley D. G., Rockman H. A. (2009) J. Mol. Cell. Cardiol. 46, 300–308 [DOI] [PubMed] [Google Scholar]
- 7. Van Tassell B. W., Radwanski P., Movsesian M., Munger M. A. (2008) Pharmacotherapy 28, 1523–1530 [DOI] [PubMed] [Google Scholar]
- 8. Muthumala A., Drenos F., Elliott P. M., Humphries S. E. (2008) Eur. J. Heart. Fail. 10, 3–13 [DOI] [PubMed] [Google Scholar]
- 9. Resh M. D. (2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 10. Qanbar R., Bouvier M. (2003) Pharmacol. Ther. 97, 1–33 [DOI] [PubMed] [Google Scholar]
- 11. Chini B., Parenti M. (2009) J. Mol. Endocrinol. 42, 371–379 [DOI] [PubMed] [Google Scholar]
- 12. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 14. Shimamura T., Hiraki K., Takahashi N., Hori T., Ago H., Masuda K., Takio K., Ishiguro M., Miyano M. (2008) J. Biol. Chem. 283, 17753–17756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hicks S. W., Horn T. A., McCaffery J. M., Zuckerman D. M., Machamer C. E. (2006) Traffic 7, 1666–1677 [DOI] [PubMed] [Google Scholar]
- 16. Greaves J., Chamberlain L. H. (2007) J. Cell Biol. 176, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 18. Ohta E., Misumi Y., Sohda M., Fujiwara T., Yano A., Ikehara Y. (2003) J. Biol. Chem. 278, 51957–51967 [DOI] [PubMed] [Google Scholar]
- 19. Swarthout J. T., Lobo S., Farh L., Croke M. R., Greentree W. K., Deschenes R. J., Linder M. E. (2005) J. Biol. Chem. 280, 31141–31148 [DOI] [PubMed] [Google Scholar]
- 20. Hicks S. W., Machamer C. E. (2002) J. Biol. Chem. 277, 35833–35839 [DOI] [PubMed] [Google Scholar]
- 21. Escribá P. V., Wedegaertner P. B., Goñi F. M., Vögler O. (2007) Biochim. Biophys. Acta 1768, 836–852 [DOI] [PubMed] [Google Scholar]
- 22. Xue Y., Chen H., Jin C., Sun Z., Yao X. (2006) BMC Bioinformatics 7, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Dowd B. F., Hnatowich M., Caron M. G., Lefkowitz R. J., Bouvier M. (1989) J. Biol. Chem. 264, 7564–7569 [PubMed] [Google Scholar]
- 25. Hakalahti A. E., Vierimaa M. M., Lilja M. K., Kumpula E. P., Tuusa J. T., Petäjä-Repo U. E. (2010) J. Biol. Chem. 285, 28850–28861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blanpain C., Wittamer V., Vanderwinden J. M., Boom A., Renneboog B., Lee B., Le Poul E., El Asmar L., Govaerts C., Vassart G., Doms R. W., Parmentier M. (2001) J. Biol. Chem. 276, 23795–23804 [DOI] [PubMed] [Google Scholar]
- 27. Fukushima Y., Saitoh T., Anai M., Ogihara T., Inukai K., Funaki M., Sakoda H., Onishi Y., Ono H., Fujishiro M., Ishikawa T., Takata K., Nagai R., Omata M., Asano T. (2001) Biochim. Biophys. Acta 1539, 181–191 [DOI] [PubMed] [Google Scholar]
- 28. Percherancier Y., Planchenault T., Valenzuela-Fernandez A., Virelizier J. L., Arenzana-Seisdedos F., Bachelerie F. (2001) J. Biol. Chem. 276, 31936–31944 [DOI] [PubMed] [Google Scholar]
- 29. Petäjä-Repo U. E., Hogue M., Leskelä T. T., Markkanen P. M., Tuusa J. T., Bouvier M. (2006) J. Biol. Chem. 281, 15780–15789 [DOI] [PubMed] [Google Scholar]
- 30. Shiina T., Kawasaki A., Nagao T., Kurose H. (2000) J. Biol. Chem. 275, 29082–29090 [DOI] [PubMed] [Google Scholar]
- 31. Laskey R. A., Mills A. D. (1975) Eur. J. Biochem. 56, 335–341 [DOI] [PubMed] [Google Scholar]
- 32. Charron G., Zhang M. M., Yount J. S., Wilson J., Raghavan A. S., Shamir E., Hang H. C. (2009) J. Am. Chem. Soc. 131, 4967–4975 [DOI] [PubMed] [Google Scholar]
- 33. Charron G., Wilson J., Hang H. C. (2009) Curr. Opin. Chem. Biol. 13, 382–391 [DOI] [PubMed] [Google Scholar]
- 34. Mason D. A., Moore J. D., Green S. A., Liggett S. B. (1999) J. Biol. Chem. 274, 12670–12674 [DOI] [PubMed] [Google Scholar]
- 35. Small K. M., McGraw D. W., Liggett S. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 381–411 [DOI] [PubMed] [Google Scholar]
- 36. Rybin V. O., Xu X., Lisanti M. P., Steinberg S. F. (2000) J. Biol. Chem. 275, 41447–41457 [DOI] [PubMed] [Google Scholar]
- 37. Shcherbakova O. G., Hurt C. M., Xiang Y., Dell'Acqua M. L., Zhang Q., Tsien R. W., Kobilka B. K. (2007) J. Cell Biol. 176, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hall R. A. (2004) Semin. Cell Dev. Biol. 15, 281–288 [DOI] [PubMed] [Google Scholar]
- 39. Richter W., Day P., Agrawal R., Bruss M. D., Granier S., Wang Y. L., Rasmussen S. G., Horner K., Wang P., Lei T., Patterson A. J., Kobilka B., Conti M. (2008) EMBO J. 27, 384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao R. P., Zhu W., Zheng M., Chakir K., Bond R., Lakatta E. G., Cheng H. (2004) Trends Pharmacol. Sci. 25, 358–365 [DOI] [PubMed] [Google Scholar]
- 41. Ponimaskin E. G., Heine M., Joubert L., Sebben M., Bickmeyer U., Richter D. W., Dumuis A. (2002) J. Biol. Chem. 277, 2534–2546 [DOI] [PubMed] [Google Scholar]
- 42. Reid H. M., Kinsella B. T. (2007) Cell. Signal. 19, 1056–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kvachnina E., Dumuis A., Wlodarczyk J., Renner U., Cochet M., Richter D. W., Ponimaskin E. (2009) Biochim. Biophys. Acta 1793, 1646–1655 [DOI] [PubMed] [Google Scholar]
- 44. Uribe A., Zariñán T., Pérez-Solis M. A., Gutiérrez-Sagal R., Jardón-Valadez E., Piñeiro A., Dias J. A., Ulloa-Aguirre A. (2008) Biol. Reprod. 78, 869–882 [DOI] [PubMed] [Google Scholar]
- 45. Claing A., Laporte S. A., Caron M. G., Lefkowitz R. J. (2002) Prog. Neurobiol. 66, 61–79 [DOI] [PubMed] [Google Scholar]
- 46. Rapacciuolo A., Suvarna S., Barki-Harrington L., Luttrell L. M., Cong M., Lefkowitz R. J., Rockman H. A. (2003) J. Biol. Chem. 278, 35403–35411 [DOI] [PubMed] [Google Scholar]
- 47. Morisco C., Marrone C., Galeotti J., Shao D., Vatner D. E., Vatner S. F., Sadoshima J. (2008) Cardiovasc. Res. 78, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ponimaskin E., Dumuis A., Gaven F., Barthet G., Heine M., Glebov K., Richter D. W., Oppermann M. (2005) Mol. Pharmacol. 67, 1434–1443 [DOI] [PubMed] [Google Scholar]
- 49. Drisdel R. C., Alexander J. K., Sayeed A., Green W. N. (2006) Methods. 40, 127–134 [DOI] [PubMed] [Google Scholar]
- 50. Ernst O. P., Meyer C. K., Marin E. P., Henklein P., Fu W. Y., Sakmar T. P., Hofmann K. P. (2000) J. Biol. Chem. 275, 1937–1943 [DOI] [PubMed] [Google Scholar]
- 51. König B., Arendt A., McDowell J. H., Kahlert M., Hargrave P. A., Hofmann K. P. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 6878–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Delos Santos N. M., Gardner L. A., White S. W., Bahouth S. W. (2006) J. Biol. Chem. 281, 12896–12907 [DOI] [PubMed] [Google Scholar]
- 53. Maeda A., Okano K., Park P. S., Lem J., Crouch R. K., Maeda T., Palczewski K. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8428–8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.