Abstract
Prostaglandin endoperoxide H synthases 1 and 2, also known as cyclooxygenases (COXs) 1 and 2, convert arachidonic acid (AA) to prostaglandin endoperoxide H2. Prostaglandin endoperoxide H synthases are targets of nonspecific nonsteroidal anti-inflammatory drugs and COX-2-specific inhibitors called coxibs. PGHS-2 is a sequence homodimer. Each monomer has a peroxidase and a COX active site. We find that human PGHS-2 functions as a conformational heterodimer having a catalytic monomer (Ecat) and an allosteric monomer (Eallo). Heme binds tightly only to the peroxidase site of Ecat, whereas substrates, as well as certain inhibitors (e.g. celecoxib), bind the COX site of Ecat. Ecat is regulated by Eallo in a manner dependent on what ligand is bound to Eallo. Substrate and nonsubstrate fatty acids (FAs) and some COX inhibitors (e.g. naproxen) preferentially bind to the COX site of Eallo. AA can bind to Ecat and Eallo, but the affinity of AA for Eallo is 25 times that for Ecat. Palmitic acid, an efficacious stimulator of human PGHS-2, binds only Eallo in palmitic acid/murine PGHS-2 co-crystals. Nonsubstrate FAs can potentiate or attenuate actions of COX inhibitors depending on the FA and whether the inhibitor binds Ecat or Eallo. Our studies suggest that the concentration and composition of the free FA pool in the environment in which PGHS-2 functions in cells, the FA tone, is a key factor regulating PGHS-2 activity and its responses to COX inhibitors. We suggest that differences in FA tone occurring with different diets will likely affect both base-line prostanoid synthesis and responses to COX inhibitors.
Keywords: Allosteric Regulation, Cyclooxygenase (COX) Pathway, Fatty Acid, Inflammation, Prostaglandins, NSAID, Arachidonic Acid, Coxib, Half-of-Sites, Palmitic Acid
Introduction
Prostaglandin endoperoxide H synthases (PGHSs),2 also known generically as cyclooxygenases (COXs), convert arachidonic acid (AA) to prostaglandin H2 (PGH2) in the committed step of prostanoid biosynthesis (1–6). PGHS-1 and PGHS-2 are products of different genes. In general, PGHS-1 is constitutively expressed, whereas PGHS-2 expression is inducible (4, 5, 7). The enzymes are embedded in the luminal monolayer of the endoplasmic reticulum and inner membrane of the nuclear envelope (8–11).
PGHSs catalyze two different reactions, a COX reaction and a peroxidase (POX) reaction. COX catalysis begins with abstraction of the 13-pro-S-hydrogen from AA by the enzyme in the rate-determining step to generate an arachidonyl radical (3, 6, 12, 13). Two molecules of O2 are then sequentially added to the arachidonyl chain with concomitant rearrangements to form PGG2. PGG2 can be reduced to PGH2 by the POX activity. The purified isoforms are about equally efficient in catalyzing the conversion of AA to PGH2 (1–6).
PGHSs are homodimers composed of tightly associated monomers with identical sequences (14). Each monomer comprising a PGHS homodimer has a physically distinct COX and POX active site. Dissociation of the dimers into monomers only occurs upon denaturation (14). Kulmacz and Lands (15, 16) provided the first evidence that the monomers of PGHS homodimers differ. They found that maximal COX activity of ovine PGHS-1 occurred with one heme per dimer (15) and that flurbiprofen and some other nonspecific nonsteroidal anti-inflammatory drugs (nsNSAIDs) inhibit PGHS-1 at a stoichiometry of one nsNSAID per dimer (16). More recent studies of both PGHS-1 and PGHS-2 using various fatty acids (FAs) and COX inhibitors have indicated that PGHSs function as conformational heterodimers during catalysis and inhibition (17–21). PGHS-2 and probably PGHS-1 exhibit half of sites' COX activity sites with AA, i.e. the COX active site of only one monomer of a PGHS-2 dimer can function at any given time (17). There is also evidence that the enzyme functions as an allosteric/catalytic couple in which one COX active site has a catalytic function that is modulated by the nature of the ligand occupying the COX site of the partner, allosteric monomer (17, 19, 21–23).
In this study, we examined the functioning of the human (hu) PGHS-2 conformational heterodimer by quantifying interactions involving huPGHS-2 and heme and different FAs and COX inhibitors. The data are consistent with a heme being bound to the POX active site of only one monomer, i.e. the catalytic monomer, of the huPGHS-2 dimer. Additional studies with FAs and COX inhibitors indicate that substrate FAs and one subset of COX inhibitors bind to the COX site of the catalytic monomer and that both substrate and nonsubstrate FAs and a second subset of COX inhibitors bind to the COX site of the allosteric monomer. The results of examining the interplay between FAs and COX inhibitors lead us to speculate that there can be important dietary effects on both base-line PG formation and responses to COX inhibitors. Diets leading to accumulation of different types of FAs in tissue have the potential to either potentiate or attenuate responses to COX inhibitors, depending on the diet and the inhibitor.
EXPERIMENTAL PROCEDURES
Materials
Arachidonic acid (AA), oleic acid (18:1ω9), palmitoleic acid (16:1ω7), elaidic acid (18:1ω9t), 15-hydroxyeicosatetraenoic acid, and 12-hydroxyheptadecaenoic acid were purchased from Cayman Chemical Co. Lauric acid (12:0), myristic acid (14:0), pentadecanoic acid (15:0), palmitic acid (16:0), margaric acid (17:0), stearic acid (18:0), nonadecylic acid (19:0), 11-cis-eicosaenoic acid (20:1ω9), FLAG peptide, and FLAG affinity resin were from Sigma. Hemin was from Frontier Scientific, Logan, UT. Palmitelaidic acid (16:1ω7t) was from MP Biomedicals LLC. Celebrex® (celecoxib) and Vioxx® (rofecoxib) were physician samples. Flurbiprofen, indomethacin, aspirin, and naproxen were from Sigma. Ibuprofen was from Tocris Bioscience. Co3+-protoporphyrin IX was purchased from Frontier Scientific (Logan, UT). Decyl maltoside, n-octyl β-d-glucopyranoside (β-OG), and C10E6 were purchased from Anatrace (Maumee, OH). BCA protein reagent was from Pierce. Polyacrylic acid (sodium salt) 5100 was from Hampton Research Corp. [1-14C]AA (1.85 GBq/mmol) was from American Radiolabeled Chemicals. Hexane, isopropyl alcohol, and acetic acid were HPLC grade from Thermo Fisher Scientific, Inc. Complete protease inhibitor was from Roche Applied Science. Restriction enzymes were from New England Biolabs, Inc. Nickel-nitrilotriacetic acid was from Qiagen. All other materials were analytical grade from Sigma.
Expression, Purification, and Assay of huPGHS-2
Procedures for the expression of huPGHS-2 in insect cells and purification of the enzyme were essentially the same as those reported previously (17, 24, 25). The purity of the recombinant huPGHS-2 was determined by SDS-PAGE and by Western blot analysis (24). The average specific activity of purified huPGHS-2 for the preparations used in the experiments reported here was 40 units/mg.
COX activity was measured polarographically at 37 °C in glass chambers containing 3 ml of 0.1 m Tris-HCl, pH 8.0, 100 μm AA, 1 mm phenol, and 5 μm hematin. A Yellow Springs Instruments model 53 oxygen monitor was used to monitor O2 consumption, and kinetic traces were recorded using DasyLab (DasyTec) software (24). Reactions were initiated by adding enzyme to the assay chamber unless otherwise indicated. In experiments involving COX inhibitors, the enzyme preparations were typically preincubated with the inhibitor plus or minus a FA, and an aliquot was added to the reaction chamber. The rates reported are maximal rates occurring after a lag phase. Under all the assay conditions reported here, the maximal rates are linearly related to the enzyme concentration. The lag time is defined as the time required for the COX activity to reach a maximum after initiating the reaction, which under optimal conditions was about 10 s. One unit of COX activity is defined as 1 μmol of O2 consumed per min at 37 °C in the assay standard mixture. A representative tracing obtained with a low concentration of AA is shown in supplemental Fig. S1.
COX assays performed at high enzyme/substrate ratios were conducted in a 100-μl reaction mixture containing 1 μm [1-14C]AA, 0.10–2.0 μm huPGHS-2, 5 μm hematin, and 1 mm phenol in 0.1 m Tris-HCl, pH 8.0, at 37 °C. The reactions were quenched with 100 μl of stop buffer consisting of ethyl acetate/acetic acid (95:5). The resultant samples were centrifuged at room temperature for 5 min at 4000 × g. An aliquot of the organic layer (100 μl) was subjected to HPLC on a Luna C18 (2) column (5 μm, 250 × 4.6 mm, Phenomenex) mounted on a Shimadzu HPLC system equipped with radio-HPLC detector (IN/US system, β-RAM model 4). The bound products were eluted in a gradient elution mode eluting with Solution A (acetonitrile/water; 30:70) for 5 min and then with a linear gradient of Solutions A and B (acetonitrile/water; 90:10) for 15 min, then Solution B for 8 min, and finally Solution A for 4 min; Solutions A and B both contained 0.1% acetic acid (v/v). The flow rate was 1 ml/min. Representative elution times for PGH2, 17-hydroxyheptadecatrienoic acid, and AA are 14, 18, and 30 min, respectively.
Titration of huPGHS-2 with Heme
Difference absorption spectra of apo-huPGHS-2 titrated with heme were obtained using a Shimadzu UV-2501 PC scanning spectrophotometer. Aliquots of a 200 μm hematin solution in 20 mm Tris-HCl, pH 8.3, containing 40 mm KCl, 0.1% C10E6, and 2% DMSO were added to a quartz cuvette containing 200 μl of 5 μm apo-huPGHS-2 in the same buffer solution without DMSO at room temperature (26, 27). The increase in absorbance at 408 nm was plotted in a linear form using the equation I0/α = (Kd/(1 − α) + e0), where I0 is the concentration of added heme; α, the fraction of enzyme sites occupied by heme, is given by α = Δ/Δmax, where Δ is the optical change measured (27); in this plot, the slope is equal to the Kd for heme binding, and the intercept (e0) represents the concentration of heme-binding sites. Titrations were also performed in the presence of palmitic acid and several COX inhibitors.
Crystallography
Recombinant His6 N580A murine (mu) PGHS-2 was overexpressed in insect cells and purified as described previously (23). For crystallization, apo-His6 N580A, muPGHS-2 were reconstituted with a 2-fold molar excess of Co3+-protoporphyrin IX, followed by dialysis against 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 0.6% (w/v) β-OG for 16 h at 4 °C. A 10-fold molar excess of PA was added to the Co3+-protoporphyrin IX reconstituted protein just prior to crystallization screening. Crystals were produced utilizing the sitting-drop vapor diffusion technique by combining 3 μl of protein solution with 3 μl of 23–34% polyacrylic acid 5100, 100 mm HEPES, pH 7.5, 20 mm MgCl2, and 0.6% (w/v) β-OG and equilibrating the resulting drop against reservoir solutions comprised of 23–34% polyacrylic acid 5100, 100 mm HEPES, pH 7.5, and 20 mm MgCl2. The crystals were transferred to a solution of 30% polyacrylic acid 5100, 100 mm HEPES, pH 7.5, 20 mm MgCl2, and 0.6% (w/v) β-OG supplemented with 10% ethylene glycol for cryoprotection 15–30 min prior to data collection. Data were collected on beamline A1 at the Cornell High Energy Synchrotron Source (Ithaca, NY) using an Area Detector Systems CCD Quantum-210 detector. The data were integrated and scaled using MOSFLM and SCALA, respectively, in the CCP4 suite of programs (28). Details of the data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics for muPGHS-2/palmitic acid co-crystal
| Crystallographic parameter | muPGHS-2:PA |
|---|---|
| Space group | I222 |
| No. in asymmetric units | 2 |
| Unit cell length | |
| a | 121.91 Å |
| b | 130.89 Å |
| c | 179.97 Å |
| α = β = γ | 90° |
| Wavelength | 0.9769 Å |
| Resolution | 20.0 to 2.10 Å |
| Highest resolution shella | 2.21 to 2.10 Å |
| Rmergeb | 9.6 (56.2) |
| Rpim | 4.4 (26.0) |
| Total observations | 457,344 (63,157) |
| Total uniquec | 82,053 (11,717) |
| I/σ(I) | 13.5 (2.9) |
| Completeness | 98.0% (96.8%) |
| Multiplicity | 5.6 (5.4) |
| Wilson B factor | 24.9 Å2 |
| No. of atoms in refinement | 10,038 |
| Rwork | 0.159 (0.211) |
| Rfreed | 0.199 (0.276) |
| Average B factor, protein | 27.1 Å2 |
| Average B factor, solvent | 28.1 Å2 |
| Average B factor, palmitate | 50.6 Å2 |
| Mean positional errore | 0.257 Å |
| r.m.s.d. in bond length | 0.011 Å |
| r.m.s.d. in bond angle | 1.329° |
a The values in parentheses represent the values in the outermost resolution shell.
b Rmerge and Rpim as defined in Ref. 54.
c Data represent reflections with F > 0 σF, which were used in the refinement.
d 5.0% of the total reflections was used to generate the test set.
e Coordinate error was calculated by Luzatti plot.
Protocols implemented during the structure solution of muPGHS-2 complexed with fatty acid substrates (23) were utilized to minimize model bias. Specifically, initial phases for the muPGHS-2/PA co-crystal structure were determined by molecular replacement methods using PHASER (29). The phases were then input into ARP/wARP (30) to build most of the deleted portions of the model. Iterative cycles of manual model building and refinement using COOT (31) and REFMAC5 (32) were then carried out to fit the remaining residues, adding waters, PA, and other ligand molecules. The final model includes two monomers in the crystallographic asymmetric unit comprised of protein residues 33–582 in monomer A and residues 33–583 in monomer B, Co3+-protoporphyrin IX, N-linked carbohydrates, and a β-OG detergent molecule. Minimized coordinates and stereochemical dictionary for PA were generated using the PRODRG server (33). TLS refinement was carried out during the final rounds of refinement (34). Final refinement statistics are summarized in Table 1.
To verify that PA bound with its carboxylate interacting with Arg-120 in only one monomer of the murine PGHS-2/PA co-crystal structure, we performed the following experiments. In the first test, PA was modeled into the COX channel of monomer A such that the carboxylate interacted with Tyr-385 and Ser-530. This conformation is similar to the nonproductive conformations of AA and eicosapentaenoic acid observed in monomer A of the co-crystal structures of muPGHS-2/AA and muPGHS-2/eicosapentaenoic acid (23). Cycles of REFMAC5 refinement were performed, followed by calculation of 2Fo − Fc and Fo − Fc electron density maps. Inspection of the resulting electron density maps clearly showed negative difference density around the improperly placed carboxylate of PA at the apex of the channel and excess positive difference density around the ω-end of PA near the side chain of Arg-120. In the second test, we placed PA into the COX channel of monomer B and modeled ethylene glycol into the COX channel of monomer A of the muPGHS-2/PA co-crystal structure. After REFMAC refinement and generation of electron density maps, there were negative density peaks around the carboxylate and ω-end of PA. Similarly, there were excess positive difference density peaks within the channel of monomer A that were not accounted for by the ethylene glycol molecules. Finally, in the third test, we modeled both PA and ethylene glycol molecules into the COX channel of monomer A and assigned occupancies of 50% to each. REFMAC5 refinement followed by electron density map calculations clearly showed positive electron density peaks for the ω-end of PA near the apex of the channel and for carbons 3–5 near the opening of the channel. Hence, these tests support the modeling of PA into monomer A only such that the carboxylate lies near the opening of the cyclooxygenase channel near the side chain of Arg-120.
Structural Analysis
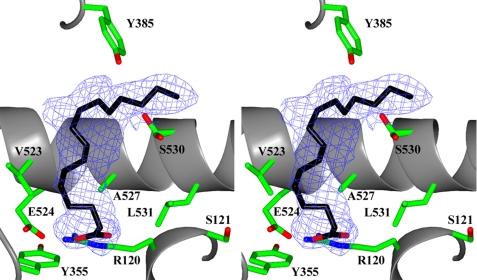
van der Waals and hydrogen bond interactions were calculated using the program COOT. The upper limit on distance for consideration as a van der Waals contact is 4.0 Å. Superposition of coordinates between structures was done using the program LSQAB within the CCP4 suite of programs and the coordinates for all Cα atoms unless otherwise stated. Simulated annealing omit maps were calculated using CNS (35). Model validation was performed using MOLPROBITY (36). Fig. 6 was created in CCP4MG (37). The coordinates and structure factors for Co3+-protoporphyrin IX reconstituted muPGHS-2 in complex with PA have been deposited in the Protein Data Bank, code 3QH0.
FIGURE 6.
Palmitic acid bound in the cyclooxygenase channel of monomer A of the murine PGHS-2/PA co-crystal structure. Stereo view of PA bound in the COX channel of monomer A. 2Fo − Fc simulated annealing omit map electron density, contoured at 4σ, is shown with the final refined model of PA (black carbon and red oxygen atoms). Residues lining the COX channel are labeled accordingly (green, red, and blue for carbon, oxygen, and nitrogen atoms, respectively).
RESULTS
Stoichiometry of Heme Binding to huPGHS-2
Titration of apo-huPGHS-2 with hematin yielded a major UV-visible spectral peak with a λmax at 408 nm indicative of formation of a heme·PGHS-2 complex (Fig. 1A) (15, 38). The data are consistent with one high affinity heme-binding site per PGHS-2 dimer (i.e. 0.92 mol of heme/mol of dimer) with a Kd = 88 ± 17 nm (n = 6) and a nonsaturable lower affinity site(s) (Fig. 1, B and C). The nonsaturable site(s) may represent lower affinity binding to the hexahistidine tag on huPGHS-2 and/or other nonspecific hydrophobic sites on the protein.
FIGURE 1.
Stoichiometry of heme binding to huPGHS-2. A, difference absorption spectra of apo-huPGHS-2 (5 μm) titrated with heme at room temperature as described under “Experimental Procedures.” B, change in absorbance at 408 nm from A. C, titration data plotted in a linear form using the equation I0/α = (Kd/(1 − α) + e0), where I0 is the concentration of added heme; α, the fraction of enzyme sites occupied by heme, is given by α = Δ/Δmax, where Δ is the optical change measured (27). The slope is equal to the Kd value for heme binding, and the intercept (e0) represents the concentration of heme-binding sites. The data shown in A–C are from one experiment but representative of six separate experiments with different enzyme preparations. The Kd values varied over a 2–3-fold range among different enzyme preparations (e.g. supplemental Fig. S1). D, 22.5 nm huPGHS-2 and 100 μm [1-14C]AA was incubated with the indicated concentration of heme at 37 °C, and after a 30-s incubation, the reaction was stopped. The products and remaining AA were separated by HPLC. The amount of PGH2 formed was calculated based on the percentage of total radioactivity chromatographing with PGH2. Details of the HPLC separation and analyses are provided under “Experimental Procedures”. Values in D are from triplicate determinations. The error bars indicate ± S.E.
We presume that at least the higher affinity site represents binding of heme to a POX site of huPGHS-2. Heme titrations of huPGHS-2 were also performed in the presence of 20 μm palmitic acid (PA), 25 μm celecoxib, 50 μm naproxen, a combination of 20 μm palmitic acid and 25 μm celecoxib, and 1.0 and 2.5 mm aspirin. The Kd values for heme binding to huPGHS-2 were essentially the same in the presence of PA, celecoxib, naproxen, or aspirin (supplemental Tables S1 and S2).
In parallel with the spectroscopic titrations, COX assays were performed with huPGHS-2 in the presence of different concentrations of heme (Fig. 1D). Maximal COX activity occurred at a ratio of about one heme per dimer. These titrations, which for practical reasons were performed at 37 °C and at relatively low enzyme concentrations, predict a Kd value of 5–10 nm. This value is significantly less than the Kd value determined spectrophotometrically (i.e. 88 nm) at 25 °C in 0–0.15% DMSO with a 200 times higher concentration of enzyme. Nonetheless, the spectroscopic heme titration data indicate that there is only one high affinity heme-binding site per dimer, and the COX assay data suggest that huPGHS-2 functions optimally with one heme bound per mol of dimer.
Effects of FAs on COX Activity of huPGHS-2
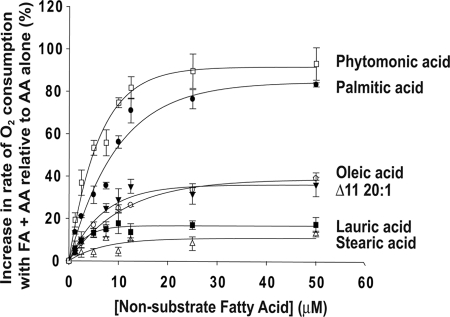
We previously reported that the COX activity of huPGHS-2 is stimulated by some nonsubstrate FAs (24). Fig. 2 compares the concentration-dependent effects of several different nonsubstrate FAs on the COX activities of huPGHS-2 with 5 μm AA as the substrate; the Km value of huPGHS-2 for AA in the absence of other FAs is about 8.5 μm. The rate of AA oxygenation by PGHS-2 was increased by many nonsubstrate FAs. The most abundant free FA in many mammalian cells (39, 40) is PA, and PA was the most efficacious of the FAs tested in stimulating AA oxygenation. Phytomonic acid (2-hexyl-cyclopropanedecanoic acid), which is produced by Lactobacillus arabinosus and is found in yogurt, also caused a particularly marked increase in the rate of AA oxygenation. FAs stimulated the rate of AA oxygenation in a concentration-dependent manner, and with each FA, the maximal effects occurred at concentrations of 10–12.5 μm. No inhibition of COX activity occurred at relatively high concentrations of the nonsubstrate FA concentrations; however, the critical micelle concentration (CMC) of saturated and monounsaturated FAs having 18 or more carbons was determined to be ≤10 μm, although the CMC of PA under the conditions of the assay is about 25 μm.3 When the effects of combinations of nonsubstrate FAs were tested for their effects on the rate of AA oxygenation, the results were approximately additive suggesting that the various FAs compete for the same site with similar affinities (supplemental Table S3). As reported previously, PA and 20:1ω9 decrease the Km value of huPGHS-2 for AA (24), and we presume this to be true for other nonsubstrate stimulatory FAs as well. The Km value for AA determined in the presence of 25 μm PA was 3.5 μm.
FIGURE 2.
Concentration-dependent effects of FAs that are not COX substrates on AA oxygenation by huPGHS-2. Results are shown as increases in the percentage of the rate of O2 consumption from O2 electrode assays of COX activities of huPGHS-2 with 5 μm AA plus the indicated concentrations of various nonsubstrate FAs relative to 5 μm AA alone. O2 consumption was measured using an O2 electrode as described under “Experimental Procedures.” The results are from a single experiment involving triplicate determinations. The error bars indicate ± S.E.
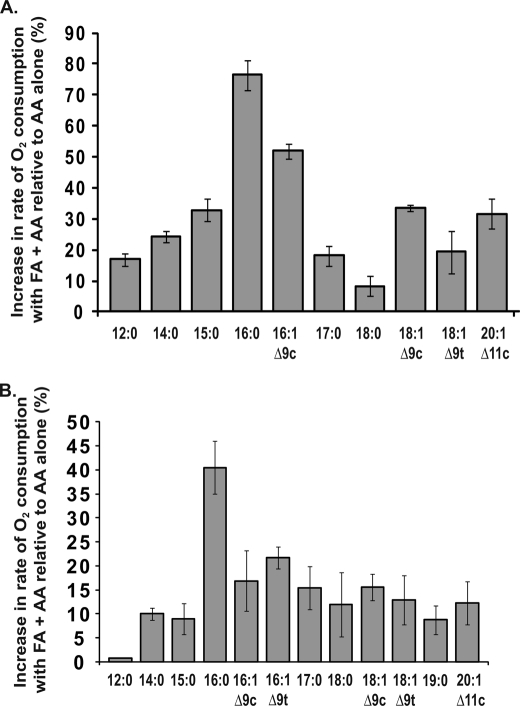
The relationships between the stimulatory effects of saturated and monounsaturated nonsubstrate FAs and their carbon chain length and degree of unsaturation are shown in Fig. 3, A and B. These experiments were performed with two different concentrations of AA and nonsubstrate FAs to minimize potential problems associated with the low CMCs of longer chain FAs. Maximal stimulation was observed with PA (Fig. 3). For saturated FAs with carbon chain lengths from 12 to 16, an increase in chain length leads to an increase in the stimulatory effect of the FA. For saturated FAs with chain lengths greater than 16 carbon atoms, the stimulatory effect decreases with increasing chain length. Palmitoleic acid stimulates the rate of AA oxygenation, but the stimulatory effect is less than that of PA. In related experiments, some of which are detailed below, HPLC or thin layer chromatography was used to separate the products formed upon oxygenation of [1-14C]AA by native huPGHS-2. The presence of nonsubstrate FAs in the reaction mixture did not change the profile of oxygenated products. PGH2 is the major oxygenation product of AA by huPGHS-2 in the reaction with or without nonsubstrate FAs, and PA increased the rate of production of 14C-labeled PGH2 and PGH2-derived products by 80% (data not shown).
FIGURE 3.
Effects of saturated and monounsaturated FAs on AA oxygenation by huPGHS-2. Results are shown as a percentage of the rate of increase for O2 electrode assays of COX activities of huPGHS-2 with FAs (25 μm) in combination with AA (5 μm) versus 5 μm AA alone (A) and FAs (5 μm) in combination with AA (2 μm) versus 2 μm AA alone (B). Rates were determined by measuring O2 consumption using an O2 electrode as described under “Experimental Procedures.” The results in A and B are each from a single experiment involving triplicate determinations. The error bars indicate ± S.D. FA designations and common names are as follows: 12:0, lauric acid; 14:0, myristic acid; 15:0, pentadecanoic acid; 16:0, palmitic acid; 16:1 Δ9c, palmitoleic acid; 16:1 Δ9c, palmitelaidic acid; 17:0, heptadecanoic acid; 18:0, stearic acid; 18:1 Δ9c, oleic acid; 18:1 Δ9t, elaidic acid; 20:1 Δ11c; Δ11-eicosaenoic acid.
We also tested the effects of PA on the rate of oxygenation by huPGHS-2 of substrates other than AA (Table 2). As reported earlier, PA caused a small increase in the rate of oxygenation of eicosapentaenoic acid but not docosahexaenoic acid (20). PA caused significant increases in the rate of oxygenation of several substrates having an acyl chain with an ω6 double bond, including adrenic acid, 2-arachidonoylglycerol ether, and dihomo-γ-linolenic acid, but there was no effect on the oxygenation of eicosadienoic acid. It should be noted that, except in the case of linoleate, PA either increased or had no effect on the rate of oxygenation of COX substrates. This implies that at the maximal concentrations of PA tested (25 μm), it does not compete directly with AA at the site at which AA is oxygenated.
TABLE 2.
Rates of oxygenation of various huPGHS-2 substrates in the presence and absence of PA
Results are shown as rates of O2 consumption under standard oxygen electrode assay conditions for measuring COX activity with huPGHS-2. Substrates were present at concentrations of 5 μm either with or without 25 μm PA. Details are provided under “Experimental Procedures.” Values are from at least triplicate assays and are shown as the means ± S.D.
| Substrate | COX activity |
Relative rate ((PA + substrate)/(substrate)) | |
|---|---|---|---|
| Substrate alone | Substrate+ 25 μm PA | ||
| units/mg | % | ||
| AA | 16 ± 0.73 | 28 ± 1.7 | 178b |
| Eicosapentaenoic acida | 5.9 ± 0.04 | 6.3 ± 0.21 | 107b |
| Docosahexaenoic acida | 2.0 ± 0.08 | 2.0 ± 0.11 | 100 |
| 2-Arachidonylglycerol ether | 10 ± 0.17 | 14 ± 0.42 | 137b |
| Adrenic acid | 8.1 ± 0.44 | 12 ± 0.06 | 153b |
| Dihomo-γ-linolenic acid | 18 ± 0.97 | 23 ± 1.2 | 127b |
| Linoleic acid | 2.6 ± 0.18 | 1.6 ± 0.16 | 62b |
| Eicosadienoic acid | 6.4 ± 0.09 | 6.5 ± 0.49 | 101 |
a Data are from Ref. 20.
b Significant difference was between substrate only and substrate plus PA in Student's t test (p < 0.005).
Oxygenation of AA by huPGHS-2 at High Enzyme to Substrate Ratios
The results of previous studies (24) and of the experiments described above suggest that FAs can bind the allosteric monomer of huPGHS-2 and modulate the catalytic efficiency of the partner, catalytic monomer toward AA and that nonsubstrate FAs do not bind the catalytic monomer. We reasoned that all FAs, including AA, must bind with higher affinity to the allosteric monomer than the catalytic monomer and, furthermore, that when a substrate FA like AA is bound to the allosteric monomer, it cannot be oxygenated. To test these concepts, we performed experiments pairing low concentrations of [1-14C]AA with relatively high concentrations of huPGHS-2 in the presence or absence of nonsubstrate FAs. As shown in Fig. 4, oxygenation of 1 μm [1-14C]AA by 1 μm huPGHS-2 plateaued at 4 min with about 6% of the original AA remaining. Simple Michaelis-Menten kinetic values for huPGHS-2 predict that AA should have decreased to undetectable levels in a few seconds.
FIGURE 4.
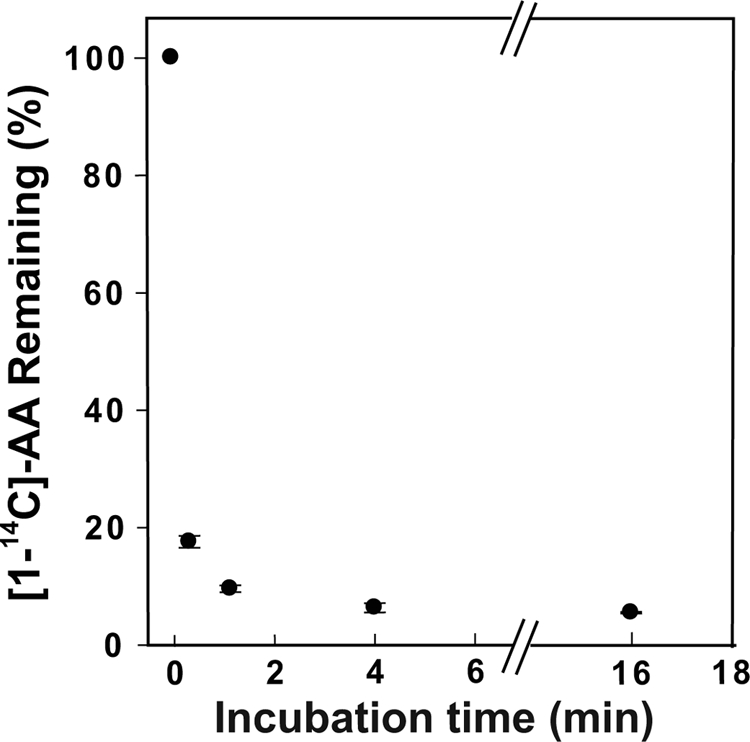
Effect of reaction time on oxygenation of [1-14C]AA measured by HPLC. [1-14C]AA (1 μm) was incubated with 1 μm huPGHS-2 at 37 °C, and after the indicated incubation times (0, 0.25, 1.0, 4.0, and 16 min), the reactions were stopped, and the products and remaining AA were separated and quantified by radio-HPLC as described under “Experimental Procedures.” Each time point represents the mean value ± S.E. for three separate incubations.
We also observed that the amount of AA remaining is directly dependent on the amount of huPGHS-2 in the reaction mixture (Table 3). When 1 μm [1-14C]AA was incubated for 15 s with 0.1, 0.5, 1.0, or 2.0 μm huCOX-2, ∼1, 7, 11, and 27% of the AA remains. However, when either PA or stearic acid was included in the reaction mixture, the AA was almost completely consumed (Table 4). PA is a more efficacious activator of huPGHS-2 than stearic acid, but both seem to have similar potencies (Fig. 2), and both appear to compete for the same sites (supplemental Table S3).
TABLE 3.
Oxygenation of AA at high huPGHS-2 to AA ratios
[1-14C]AA (1 μm) was incubated with the indicated concentrations of huPGHS-2 at 37 °C for 15 s; the reactions were stopped, and the radioactive products and unreacted AA were separated by radio-HPLC and quantified as described under “Experimental Procedures.” The results show the percentage of the original [1-14C]AA remaining.
| Reaction components | Unreacted [1-14C]AA remaining |
|||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | Average ± S.D. | |
| % | ||||
| 1 μm [1-14C]AA + 100 nm huPGHS-2 | 0.80 | 1.38 | 0.12 | 0.77 ± 0.63 |
| 1 μm [1-14C]AA + 0.5 μm huPGHS-2 | 6.27 | 9.74 | 7.11 | 7.7 ± 1.8a |
| 1 μm [1-14C]AA + 1 μm huPGHS-2 | 12.0 | 9.85 | 10.9 | 10.9 ± 1.1a |
| 1 μm [1-14C]AA + 2 μm huPGHS-2 | 23.3 | 30.5 | 25.6 | 26.5 ± 3.7a |
a Significant difference versus control was with 100 nm huPGHS-2 in Student's t test (p < 0.005).
TABLE 4.
Oxygenation of AA at high huPGHS-2 to AA ratios in the presence or absence of palmitic acid or stearic acid
[1-14C]AA (1 μm) was incubated with huPGHS-2 (1 μm) in the presence and absence of palmitic acid or stearic acid at 37 °C for 15 s or 4 min. The reactions were stopped, and the radioactive products and unreacted AA were separated and quantified by radio-HPLC as described under “Experimental Procedures.” The results are shown as the percentage of unreacted [1-14C]AA remaining. Values shown are means ± S.D. for triplicate determinations.
| Reaction components | Unreacted AA remaining after 15 s |
Unreacted AA remaining after 4 min |
||
|---|---|---|---|---|
| Palmitic acid | Stearic acid | Palmitic acid | Stearic acid | |
| % | % | |||
| 1 μm [1-14C]AA + 1 μm huPGHS-2 + no additional FA | 10.9 ± 1.1 | 10.9 ± 1.1 | 6.0 ± 0.8 | 6.0 ± 0.8 |
| 1 μm [1-14C]AA + 1 μm huPGHS-2 + 1.25 μm PA or SA | 3.5 ± 0.5a | 6.6 ± 1.5a | 2.9 ± 0.5a | 3.1 ± 0.4a |
| 1 μm [1-14C]AA + 1 μm huPGHS-2 + 2.5 μm PA or SA | 1.0 ± 0.4a | 5.2 ± 1.1a | 1.3 ± 0.8a | 1.3 ± 0.8a |
a Significant difference versus control was without palmitic acid or stearic acid in Student's t test (p < 0.015).
Overall, the results are consistent with the idea that both AA and nonsubstrate FAs bind to the allosteric site of huPGHS-2 and that when added in excess nonsubstrate FAs can displace AA from the allosteric site. Displaced AA becomes available to the catalytic site where it is oxygenated. This concept is presented diagrammatically in Fig. 5, where Ecat designates the catalytic monomer and Eallo is the allosteric monomer. Using the model in Fig. 5 and the data in Table 4, we can estimate the Kd values for the binding of AA and nonsubstrate FAs to Ecat and Eallo. At concentrations tested, nonsubstrate FAs do not inhibit AA oxygenation (Figs. 2 and 3). Again, this implies that nonsubstrate FAs do not bind with appreciable affinities to the catalytic COX site. Accordingly, we estimate that for binding of nonsubstrate FAs to Ecat the Kd is greater than 50 μm. The data in Fig. 4 and Table 4 suggest that an approximate equilibrium is reached by 4 min and that at that time 4.7% (6.0 to 1.3%) of the AA is bound to Eallo (i.e. in the absence of a nonsubstrate FA). Thus, at 4 min there is 0.047 μm Eallo-AA/Ecat and 0.953 μm Eallo/Ecat and no Eallo-AA/Ecat-AA. We estimate that there is 0.013 μm unbound AA corresponding to the 1.3% AA that remains after adding either 2.5 μm stearic acid or palmitic acid (Table 4). The Kd value calculated from ([free AA] × [Eallo/Ecat]) ÷ (Eallo − AA/Ecat) is 0.26 μm for binding of AA to Eallo in the absence of other FAs. It can be seen that at a concentration of about 1 μm stearic acid or PA, about 50% of the original 6% or about 0.03 μm of unreacted AA remains. Thus, the molar ratio of stearic acid or PA to AA to achieve 50% displacement of AA from Eallo is about 30. From this, we estimate that the Kd value for binding of these nonsubstrates FAs to the Eallo site is about 30 times higher than that for AA or ∼7.5 μm.
FIGURE 5.
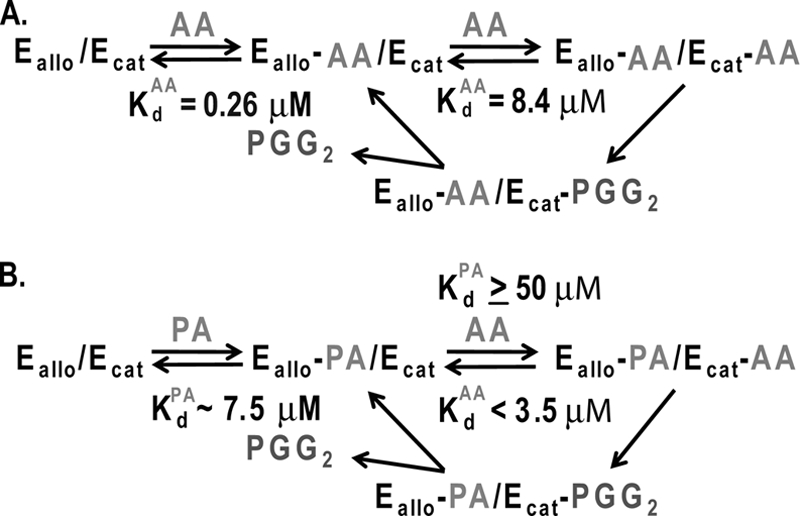
Model for allosteric interactions between monomers of the PGHS-2 homodimer caused by the binding of FAs. Ecat is the catalytic monomer to which heme is bound, and Eallo is the allosteric monomer that does not bind heme. A, binding of AA to Eallo and Ecat of huPGHS-2 and subsequent oxygenation of AA. The Kd value for Ecat is the Km value for AA (8.4 μm) in the absence of other FAs. The Kd value for AA binding to Eallo (0.26 μm) is calculated from the data in Fig. 4 and Tables 3 and 4 as outlined in the text. B, binding of AA to Eallo and Ecat in the presence of PA. We make the following assumptions: (a) the Kd value for binding of AA to the Ecat site is equal to the Km value in the presence of PA (i.e. < 3.5 μm); (b) the Kd value for PA binding to the Ecat site is high (≥50 μm) because, as shown in Fig. 2, PA does not inhibit AA oxygenation by huPGHS-2 even at low concentrations of AA. We estimate that the Kd values for the binding of palmitic acid and stearic acid to the Eallo site are ∼7.5 μm based on the concentration of the FAs to cause 50% inhibition of AA binding when the effective concentration of AA is about 0.025 μm.
As noted in Fig. 5, the Km values for AA oxygenation in the absence and presence of 25 μm PA were 8.4 and 3.5 μm, respectively (data not shown). We assume that the Kd value for AA to Ecat is 8.4 μm when AA is bound to Eallo and that the Kd value for AA binding to Ecat is something less than 3.5 μm when PA is bound to Eallo. We cannot ascertain an accurate value for the Kd for AA when Eallo is completely occupied by PA because the CMC for PA in the assay solution is about 25 μm, conditions under which more than 50% of Eallo would be occupied by AA.
Binding of PA to muPGHS-2 in muPGHS-2/PA Co-crystals
PA stimulated the COX activity of muPGHS-2. The potency of PA was similar with muPGHS-2 and huPGHS-2, but maximal stimulation was 43% with muPGHS-2 versus 76% with huPGHS-2 using 5 μm AA and 25 μm PA. We have elucidated the x-ray crystal structure of PA bound in the COX channel of murine PGHS-2 to 2.1 Å resolution utilizing synchrotron radiation. There are two monomers in the crystallographic asymmetric unit that include the canonical biological dimer. The overall tertiary structure of each monomer, which includes the epidermal growth factor domain, membrane-binding domain, and catalytic domain, is well resolved and conserved with respect to that observed for other murine PGHS-2/fatty acid co-crystal structures (23). The calculated root mean square deviation (r.m.s.d.) between monomer A and monomer B is 0.28 Å (for 549 Cα atom pairs) indicating that there are no significant structural differences between these monomers. Interpretable electron density for PA is only observed in monomer A of the muPGHS-2/PA crystal structure. In contrast, two molecules of ethylene glycol, which was utilized as a cryoprotectant during data collection, have been modeled into the COX channel of monomer B as detailed under “Experimental Procedures.”
PA is oriented in the COX channel with its carboxylate moiety located near Arg-120 and Tyr-355 at the opening of the channel and its ω-end near Tyr-385 and Ser-530 at the apex of the channel (Fig. 6). PA makes 36 contacts with residues that line the COX channel. Half of these contacts (19 of 36) are made between PA and the side chain atoms of Arg-120, Tyr-355, Tyr-385, and Ser-530 (supplemental Table S4). Five of these contacts are hydrophilic in nature. Specifically, the carboxylate oxygen atoms of PA form salt bridges to the Nϵ and Nη2 atoms of Arg-120 and hydrogen bonds to the phenolic oxygen atom of Tyr-355 and the Nϵ and Nη2 atoms of Arg-120. As PA is only 16 carbons in length, the ω-end of this fatty acid does not fully penetrate the hydrophobic groove above Ser-530 as is typically observed for productive binding of FA substrates to PGHS-1 and -2 (Fig. 6) (23, 41–43). Instead, two well ordered water molecules occupy the groove, which are analogous to the water molecules occupying the groove when AA and eicosapentaenoic acid bind in their nonproductive conformations to PGHS-2 (23). The side chain of Ser-530 exhibits alternate rotamer conformations, which has also been observed in crystal structures of muPGHS-2 bound with FA substrates (23). As a consequence of not fully extending into the hydrophobic groove, PA does not form interactions with the side chain atoms of Phe-209, Ile-377, Phe-381, Gly-533, and Leu-534. Contacts made between these residues are critical in the stabilization of the ω-end of fatty acid substrates within the hydrophobic groove when they are bound in a productive conformation (23). Hence, the increase in hydrophilic interactions observed between the carboxylate oxygen atoms of PA and the side chain atoms of Arg-120 and Tyr-355 serve as compensatory interactions to stabilize PA when it is bound in the COX channel.
Effects of FAs on the Inhibition of huPGHS-2 by nsNSAIDs and Coxibs
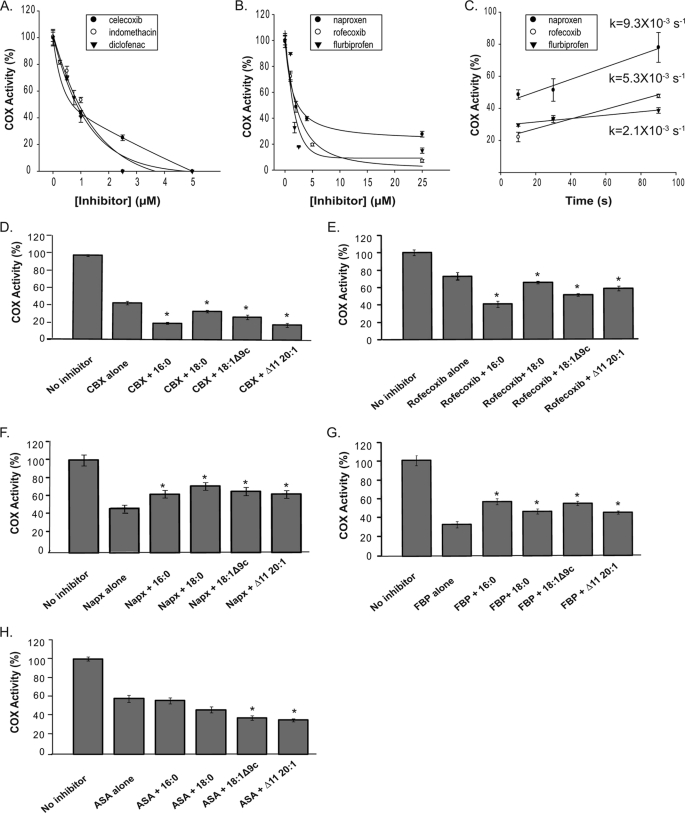
Many time-dependent COX inhibitors, both nsNSAIDs and coxibs, bind to one monomer of PGHSs to cause maximal COX inhibition (16, 17, 19–21). Naturally occurring nonsubstrate FAs also appear to bind preferentially to one huPGHS-2 monomer because these FAs do not cause enzyme inhibition (Figs. 2 and 3). Therefore, we investigated the interplay between these two groups of agents. For individual time-dependent inhibitors, we first examined the magnitudes of the inhibition following a fixed time of preincubation with different concentrations of an inhibitor (Fig. 7, A and B). Celecoxib, indomethacin, and diclofenac caused complete or near complete loss of activity (Fig. 7A). Naproxen, flurbiprofen, and rofecoxib caused incomplete COX inhibition even at relatively high inhibitor concentrations (Fig. 7B). In all of the experiments reported in Fig. 7, we report rates of O2 consumption determined using an O2 electrode. We also examined the product profiles with all combinations of FA and inhibitor, and there were no differences (data not shown). Because the same products were formed in all cases, a direct comparison of rates of O2 consumption across all of the experimental conditions is appropriate.
FIGURE 7.
Effects of nonsubstrate FAs on the responses of huPGHS-2 to different COX inhibitors. A, purified huPGHS-2 (1 μm) was pretreated with the indicated concentrations of celecoxib, indomethacin, or diclofenac at 37 °C for 30 min and then assayed for COX activity with 5 μm AA using an O2 electrode as described under “Experimental Procedures.” B, purified huPGHS-2 (1 μm) was pretreated with the indicated concentrations of naproxen, rofecoxib, or flurbiprofen at 37 °C for 30 min and then assayed for COX activity with 5 μm AA as described in A. C, purified huPGHS-2 (1 μm) was pretreated with 4 μm naproxen, 25 μm rofecoxib, or 25 μm flurbiprofen at 37 °C for 30 min, respectively. Pretreated enzyme (40 μl) was injected into an O2 electrode chamber containing 3 ml of assay mixture. AA (10 μl; 40 μm final concentration in the assay) was injected at the indicated incubation times after the addition of inhibitor-pretreated enzyme and O2 consumption was measured. D, effect of FAs on inhibition of huPGHS-2 by celecoxib. Purified native huPGHS-2 homodimer (1 μm) was incubated at 37 °C with or without celecoxib (1 μm) for 30 min in the presence or absence of various nonsubstrate FAs (5 μm) and then assayed for COX activity with 5 μm AA using an O2 electrode. E, effect of FAs on inhibition of huPGHS-2 by rofecoxib. The experiments were performed as in D except using rofecoxib (1 μm) instead of celecoxib. F, effect of FAs on inhibition of huPGHS-2 by naproxen (Napx). The experiments were performed as in D except using naproxen (2.0 μm) instead of celecoxib. G, effect of FAs on inhibition of huPGHS-2 by flurbiprofen (FBP). The experiments were performed as in D except using flurbiprofen (1.75 μm) instead of celecoxib. H, effect of FAs on inhibition of huPGHS-2 by aspirin. The experiments were performed as in D except using aspirin (100 μm) instead of celecoxib. Values in each panel are derived from the average of triplicate determinations ± S.D. Similar experiments were performed at least twice with different preparations of enzyme and yielded quantitatively similar results. D–H, the values denoted with an asterisk differ from the corresponding inhibitor treated control without FA as determined using a Student's t test (p < 0.005). CBX, celecoxib; ASA, aspirin.
The lack of complete inhibition by naproxen, flurbiprofen, and/or rofecoxib could have resulted from rapid recovery of time-dependent inhibited activity when the enzyme was diluted into the assay chamber to measure COX activity, particularly because there is about a 10-s lag before the maximal rate is achieved. To test for this, we measured the rate of recovery of COX activity with time by adding AA substrate to the assay chamber 0, 10, 30, or 90 s after the enzyme plus inhibitor were added to the assay chamber (Fig. 7C); when diluted into the assay chamber about 100-fold, the inhibitors are at concentrations that are insufficient to cause time-dependent inhibition. Enzyme treated with rofecoxib recovered sufficient activity relatively rapidly, enough to account for 5% being recovered during the first 10 s of the assay. Thus, in contrast to what is predicted from the results in Fig. 7B, rofecoxib appears to cause complete time-dependent enzyme inhibition when an appropriate correction is made for recovery in the assay chamber. However, the rates of recovery of activity of huPGHS-2 that had been treated with flurbiprofen or naproxen (Fig. 7C) were too slow to account for the 15 and 30% residual COX activity, respectively, seen with these inhibitors (Fig. 7B).
We next determined the effect of several nonsubstrate FAs on the time-dependent inhibition by the different inhibitors (Fig. 7, D–H). We found that the effects of celecoxib, rofecoxib, and aspirin could be potentiated by at least some FAs; importantly, none of the FAs tested attenuated the effects of any of these inhibitors. However, all of the FAs attenuated the actions of naproxen and flurbiprofen. FAs had no appreciable effects on responses to either diclofenac or indomethacin (data not shown).
Overall, time-dependent COX inhibitors appeared to fall into two broad classes. One class includes aspirin and those inhibitors that effectively cause complete COX inhibition (i.e. celecoxib, diclofenac, indomethacin, and rofecoxib); aspirin modifies the COX active site but the acetylated site retains the ability to bind and convert substrate to an unusual product (20). The actions of this class of time-dependent inhibitors are either potentiated or unaffected by FAs depending on the specific FA. The second class of time-dependent inhibitors includes flurbiprofen and naproxen. Neither inhibitor causes complete COX inhibition under standard assay conditions, and the actions of these inhibitors are attenuated by FAs.
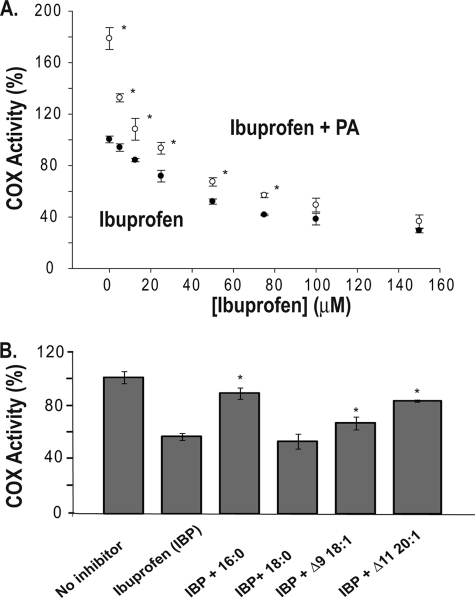
Ibuprofen is a time-independent COX inhibitor that binds both monomers of PGHS to cause COX inhibition (18). When ibuprofen was included at low concentrations in a reaction mixture containing PA and huPGHS-2, it caused an immediate drop in activity close to the level found in the absence of PA (Fig. 8). This is consistent with ibuprofen displacing PA from one COX site but without having an otherwise detectable effect on the other site (18). As the ibuprofen concentration was increased, it appeared that PA caused a minor interference with ibuprofen action by shifting the dose-response curve slightly to the right. When several FAs were compared for their abilities to affect ibuprofen actions, the effects approximately paralleled their abilities to stimulate huPGHS-2 activity.
FIGURE 8.
Effects of nonsubstrate FAs on the inhibition of huPGHS-2 by ibuprofen. A, concentration-dependent inhibition of native huPGHS-2 by ibuprofen (IBP). The indicated concentration of ibuprofen (closed circles) or the indicated concentration of ibuprofen plus 25 μm palmitic acid (open circles) were present in an O2 electrode assay chamber along with 5 μm AA. Purified huPGHS-2 was added to initiate AA oxygenation. B, effects of nonsubstrate FAs on the inhibition of huPGHS-2 by ibuprofen. Purified native huPGHS-2 homodimer was added to O2 electrode assays containing 5 μm AA and either no inhibitor or 50 μm ibuprofen and 5 μm of the indicated nonsubstrate FAs, and O2 consumption was measured. Values are derived from the average of triplicate determinations ± S.D. A, values denoted with an asterisk differ from the inhibitor-treated control without FA as determined using a Student's t test (p < 0.005); B, values denoted with an asterisk differ from the inhibitor-treated control without FA as determined using a Student's t test (p < 0.05).
DISCUSSION
We find that huPGHS-2 binds only one heme per dimer with high affinity (Kd = 88 nm) and that maximal COX activity occurs with a stoichiometry of one heme per subunit. These results conflict with those reported earlier for huPGHS-2 (38) but are consistent with what has been found for ovine (15) and human4 PGHS-1. We do not have an explanation for the discrepancy between our results and those reported previously for huPGHS-2. Heme binding is relatively unaffected by nonsubstrate FAs such as PA or by two COX inhibitors celecoxib or naproxen, which, as discussed below, appear to bind the Ecat and Eallo sites, respectively, of huPGHS-2. Adding confusion to the question of the heme stoichiometry are the results of x-ray studies of PGHS crystals. Crystallizations have generally been performed with ∼75 μm enzyme and 150 μm heme. These are conditions expected to foster occupancy of both monomers with heme.
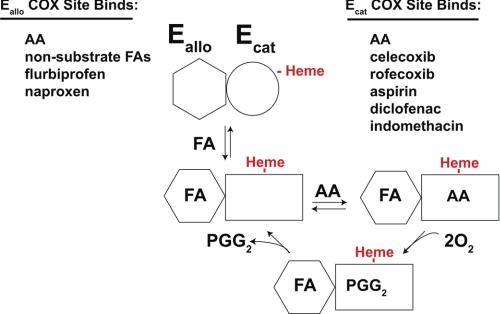
An examination of huPGHS-2 heterodimers having a defective POX active site indicated that the COX site of a monomer cannot function unless the POX site of that monomer is functional (25). This finding implies that the monomer of native huPGHS-2 to which heme is bound is the COX catalytic monomer. Accordingly, we have designated this heme-binding monomer as Ecat and the partner monomer, i.e. the allosteric monomer, as Eallo (Figs. 5 and 9). It is unclear whether a conformational huPGHS-2 heterodimer with one monomer having a higher affinity for heme exists prior to heme binding or whether heme binding to one monomer of a conformational homodimer induces structural changes that lead to different affinities of the POX sites for the heme (i.e. an Ecat with a higher affinity for heme and an Eallo with a lower affinity for heme). The fact that occupancy of the Ecat and Eallo sites does not appreciably affect the affinity of huPGHS-2 for heme suggests that apo-huPGHS-2 pre-exists in solution as a conformational heterodimer. Whatever the mechanism, our model (Figs. 5 and 9) is that heme binding defines one monomer as Ecat and the other as Eallo.
FIGURE 9.
FA, nsNSAID, and coxib modulators of huPGHS-2. As noted, different ligands bind preferentially to either Eallo or to Ecat. In the illustration, Eallo is shown binding a nonsubstrate, activating FA (e.g. PA). Eallo with bound FA activates Ecat by decreasing the Km for AA.
Another important aspect of the model is that the form of the enzyme (Eallo-FA/Ecat) having FA bound to Eallo and heme bound to Ecat is relatively stable, i.e. neither FA nor heme dissociate quickly compared with the rates of AA binding to Ecat and subsequent oxygenation. This is consistent with the relative affinities of various FAs for the Eallo and Ecat sites and of the heme for Ecat. Having a stable Eallo-FA/Ecat is necessary for half of sites for COX activity.
A confusing question that arose from earlier studies of half of sites of COX activity was how a heterodimer like G533A/native huPGHS-2 could have the same activity as the native huPGHS-2 homodimer if there was a rapid equilibrium between the monomers (17). A reasonable explanation in the context of the model in Fig. 5 is that the predominant form of G533A/native huPGHS-2 in solution is one in which the G533A monomer is Eallo and the native monomer is Ecat.
We previously reported that various substrate and nonsubstrate FAs affect the activities of PGHS-1 and PGHS-2 with AA as substrate (24). The earlier results indicated that nonsubstrate FAs like PA and 20:1ω9 bind to one monomer, an allosteric monomer Eallo, and affect the affinity of AA for the partner, catalytic monomer Ecat. Several new lines of evidence provide further support for this concept. For example, we observed in kinetic experiments performed with high huPGHS-2 to AA ratios that AA is incompletely oxygenated, but when nonsubstrate FAs are added to the reaction mixture, essentially complete oxygenation of AA occurs. These studies have also allowed us to estimate the relative affinities of AA and PA for Ecat and Eallo. The results are consistent with the idea that AA binds to Eallo with a significantly higher affinity than to Ecat, and when bound to Eallo, AA is not oxygenated. Adding a nonsubstrate FA like PA displaces AA from Eallo making AA available for oxygenation by Ecat. Nonsubstrate FAs do not bind with appreciable affinities to Ecat at least when a nonsubstrate FA is bound to Eallo, and so nonsubstrate FAs do not interfere with AA oxygenation by Ecat.
Our crystallographic results also provide support for the model in Fig. 5 as well as insights into how nonsubstrate FAs can bind to Eallo. Interestingly, only one monomer of the muPGHS-2 dimer is occupied by PA in the muPGHS-2·PA complex. This finding is consistent with the predicted affinities of PA for Eallo and Ecat that come from the kinetic studies discussed above. In the muPGHS-2/PA structure, the carboxylate group of PA is observed to interact with Arg-120 and Tyr-355 of what we predict is Eallo. In a previous structure of AA complexed with muPGHS-2, AA is observed in two different conformations (23). In one, which looks to be a catalytically productive conformation based on studies with ovine PGHS-1 (41), AA is bound with its carboxylate group interacting with the hydrogen bonding network involving Arg-120 and Tyr-355 (23). AA is bound in a catalytically nonproductive conformation in the partner monomer with the carboxylate group of AA interacting with the phenolate oxygen of Tyr-385; a related, nonproductive conformation of AA was first suggested by results obtained in examining a complex of AA with apo-muPGHS-2 (44). Mutagenesis studies will be important in evaluating the predictions of the crystallographic studies of the complexes of AA and of PA with muPGHS-2. For example, it would be important to know if the AA or PA binding is affected in a native/Y385F huPGHS-2 heterodimer.
We have found that there is a specificity to the actions of nonsubstrate FAs on huPGHS-2. PA is the most efficacious FA activator of the enzyme, although nonsubstrate FAs having either fewer or greater numbers of double bonds are less effective activators of the enzyme. We hypothesize that prostanoid production in vivo is directly and importantly influenced by the effective concentration and FA composition of the milieu in which PGHSs operate, what we term the FA tone. We presume that every individual establishes a fairly constant FA tone as a consequence of dietary habits and genetic profile (45, 46). We imagine that having a FA tone outside some normal range would change prostanoid formation to an extent sufficient to affect the health of an individual. For example, we speculate that having a FA tone that enhances PG formation via PGHS-2 would exacerbate inflammatory conditions believed to underlie many chronic diseases.
At the concentrations tested, the effects of nonsubstrate FAs are less than 2-fold. The effects are on the Km value for AA (24) and that the Km value is in the range of the expected apparent concentrations of free AA following cell stimulation. Although the magnitudes of the FA effects are relatively small, they are in the range of physiological significance. Studies with hypomorphic PGHS-1 mice and with COX inhibitors in mice and humans have shown that near complete suppression of PG formation is required for effects on PGHS-2-dependent reproductive responses and on PGHS-1-dependent platelet aggregation (47–49). However, incomplete (50–75%) suppression of PG formation correlates with diminution of some PG-mediated inflammatory and analgesic responses (50–52). Thus, seemingly modest changes in PG synthesis can lead to significant clinical changes. One can imagine that a small decrease in PG formation occurring via PGHS-2 in affected joints could diminish the need for anti-inflammatory/analgesic therapy (50–52).
Knowing that there are interactions between PGHSs and nonsubstrate FAs and between PGHSs and COX inhibitors, it was not unexpected that nonsubstrate FAs would affect responses to COX inhibitors. The time-independent competitive inhibitor ibuprofen must bind to both COX sites of a PGHS-2 dimer to cause inhibition of AA oxygenation (18). Binding to one site occurs with high affinity but does not affect the rate of oxygenation. Oxygenation of AA is only affected when ibuprofen binds the second COX site. We presume that the latter site corresponds to Ecat and the former site to Eallo in the model depicted in Fig. 9. Consistent with this idea is that relatively low concentrations of ibuprofen essentially eliminate the stimulatory effect of PA on AA oxygenation. In fact, this form of competition (i.e. for Eallo) may underlie part of the pharmacological effect of inhibitors that bind this monomer. Secondarily, when the ibuprofen concentration is raised further, there is a dose-dependent decrease in AA oxygenation paralleling that seen in the absence of PA.
Unlike time-independent inhibitors, time-dependent inhibitors appear to bind to a single monomer to elicit effects (16, 17, 19, 20). Interactions between nonsubstrate FAs and time-dependent inhibitors seem to fall into two broad categories depending on whether the inhibitor functions by interacting with Eallo or Ecat as depicted in Fig. 9. Time-dependent inhibitors that bind to Eallo are allosteric regulators and at least in the cases of naproxen and flurbiprofen cause incomplete COX inhibition. Nonsubstrate FAs, which bind Eallo with reasonably high affinities, are observed to attenuate the actions of these inhibitors. We propose that this occurs because nonsubstrate FAs compete with inhibitors like naproxen and flurbiprofen for binding to Eallo (Fig. 9). Naproxen is the one COX inhibitor the use of which appears to be devoid of adverse cardiovascular side effects (53). It would be of interest to determine whether this relates to the ability of naproxen to be a modestly efficacious COX-2 inhibitor that functions via Eallo.
Inhibitors in the second group of time-dependent inhibitors bind to Ecat and compete directly with AA (Fig. 9). Not surprisingly, COX inhibitors in this group can cause complete enzyme inhibition. The effects of these inhibitors, including celecoxib and rofecoxib, are frequently potentiated by the presence of a nonsubstrate FA. Presumably, this involves having the FA bind to Eallo and causing an allosteric effect on Ecat that facilitates inhibitor binding and/or the time-dependent transformation of huPGHS-2 to an inhibited form (or stabilization in an inhibited form).
We think that aspirin is an exception, unique to PGHS-2, to the case of a time-dependent inhibitor causing complete enzyme inhibition even though aspirin acts on Ecat. We propose that aspirin acetylates Ecat but that this does not completely interfere with AA binding and that bound AA is converted to (15R)-hydroxyeicosatetraenoic acid instead of the normal product PGG2 (20).
We speculate that both diclofenac and indomethacin bind to Ecat to elicit their effects. Binding of one diclofenac molecule per huPGHS-2 (20) or one indomethacin to PGHS-1 (16) causes complete inhibition. The FAs we tested did not potentiate inhibition by these agents. More importantly, nonsubstrate FAs did not interfere with inhibition as would have been expected were diclofenac or indomethacin binding to Eallo.
As discussed above, FA tone could importantly influence basal prostanoid formation. Similarly, the FA tone would be expected to affect responses to COX inhibitors. These drugs are the most widely used pharmaceutical agents in the United States, but they have adverse side effects responsible for an estimated 20,000 deaths in the United States annually (53). We envisage that some side effects, which are generally of unknown origin, are related to diet/drug interactions and could be ameliorated by altering diet or drug. For example, we speculate that responses of patients to nsNSAIDs or COX-2-specific inhibitors would vary among groups of individuals on typical Western versus Mediterranean versus ω3 fish oil-enriched diets. The differences may well be drug-specific. Dietary studies need to be performed to address these issues.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM 068848 (to W. L. S.) and GM 077176 (to M. G. M.). This work was based on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by National Science Foundation Award DMR-0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by National Institutes of Health Award RR-01646 from NCRR.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1, Tables S1–S4, and additional references.
The atomic coordinates and structure factors (code 3QH0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
CMC values were determined in 100 mm sodium phosphate, pH 7.6, containing 5 μm 1,6-diphenyl-1,3,5-hexatriene (DPH) as described previously (24). CMC values for various FAs tested under these conditions are as follows: lauric acid (12:0), 400 μm; pentadecanoic acid (15:0), 75 μm; palmitic acid (16:0), 30 μm; heptadecanoic acid (17:0), 10 μm; oleic acid (Δ9 cis18:1), ∼5 μm.
H. Zou and W. L. Smith, unpublished observations.
- PGHS
- prostaglandin endoperoxide H synthase
- AA
- arachidonic acid
- β-OG
- n-octyl β-d-glucopyranoside
- CMC
- critical micelle concentration
- COX
- cyclooxygenase
- FA
- fatty acid
- hu
- human
- mu
- murine
- nsNSAID
- nonspecific nonsteroidal anti-inflammatory drug
- PA
- palmitic acid
- PG
- prostaglandin
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Smith W. L., DeWitt D. L., Garavito R. M. (2000) Annu. Rev. Biochem. 69, 145–182 [DOI] [PubMed] [Google Scholar]
- 2. van der Donk W. A., Tsai A. L., Kulmacz R. J. (2002) Biochemistry 41, 15451–15458 [DOI] [PubMed] [Google Scholar]
- 3. Schneider C., Pratt D. A., Porter N. A., Brash A. R. (2007) Chem. Biol. 14, 473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith W. L. (2008) Trends Biochem. Sci. 33, 27–37 [DOI] [PubMed] [Google Scholar]
- 5. Rouzer C. A., Marnett L. J. (2009) J. Lipid Res. 50, S29–S34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai A. L., Kulmacz R. J. (2010) Arch. Biochem. Biophys. 493, 103–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang Y. J., Mbonye U. R., DeLong C. J., Wada M., Smith W. L. (2007) Prog. Lipid Res. 46, 108–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rollins T. E., Smith W. L. (1980) J. Biol. Chem. 255, 4872–4875 [PubMed] [Google Scholar]
- 9. Otto J. C., Smith W. L. (1994) J. Biol. Chem. 269, 19868–19875 [PubMed] [Google Scholar]
- 10. Spencer A. G., Thuresson E., Otto J. C., Song I., Smith T., DeWitt D. L., Garavito R. M., Smith W. L. (1999) J. Biol. Chem. 274, 32936–32942 [DOI] [PubMed] [Google Scholar]
- 11. Spencer A. G., Woods J. W., Arakawa T., Singer I. I., Smith W. L. (1998) J. Biol. Chem. 273, 9886–9893 [DOI] [PubMed] [Google Scholar]
- 12. Hamberg M., Samuelsson B. (1967) J. Biol. Chem. 242, 5336–5343 [PubMed] [Google Scholar]
- 13. Tsai A. L., Palmer G., Wu G., Peng S., Okeley N. M., van der Donk W. A., Kulmacz R. J. (2002) J. Biol. Chem. 277, 38311–38321 [DOI] [PubMed] [Google Scholar]
- 14. Xiao G., Chen W., Kulmacz R. J. (1998) J. Biol. Chem. 273, 6801–6811 [DOI] [PubMed] [Google Scholar]
- 15. Kulmacz R. J., Lands W. E. (1984) J. Biol. Chem. 259, 6358–6363 [PubMed] [Google Scholar]
- 16. Kulmacz R. J., Lands W. E. (1985) J. Biol. Chem. 260, 12572–12578 [PubMed] [Google Scholar]
- 17. Yuan C., Rieke C. J., Rimon G., Wingerd B. A., Smith W. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6142–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prusakiewicz J. J., Duggan K. C., Rouzer C. A., Marnett L. J. (2009) Biochemistry 48, 7353–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rimon G., Sidhu R. S., Lauver D. A., Lee J. Y., Sharma N. P., Yuan C., Frieler R. A., Trievel R. C., Lucchesi B. R., Smith W. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma N. P., Dong L., Yuan C., Noon K. R., Smith W. L. (2010) Mol. Pharmacol. 77, 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sidhu R. S., Lee J. Y., Yuan C., Smith W. L. (2010) Biochemistry 49, 7069–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mbonye U. R., Yuan C., Harris C. E., Sidhu R. S., Song I., Arakawa T., Smith W. L. (2008) J. Biol. Chem. 283, 8611–8623 [DOI] [PubMed] [Google Scholar]
- 23. Vecchio A. J., Simmons D. M., Malkowski M. G. (2010) J. Biol. Chem. 285, 22152–22163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan C., Sidhu R. S., Kuklev D. V., Kado Y., Wada M., Song I., Smith W. L. (2009) J. Biol. Chem. 284, 10046–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J., Seibold S. A., Rieke C. J., Song I., Cukier R. I., Smith W. L. (2007) J. Biol. Chem. 282, 18233–18244 [DOI] [PubMed] [Google Scholar]
- 26. Duncan T., Osawa Y., Kutty R. K., Kutty G., Wiggert B. (1999) J. Lipid Res. 40, 1222–1228 [PubMed] [Google Scholar]
- 27. Huber W. J., 3rd, Backes W. L. (2008) Anal. Biochem. 373, 167–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dodson E. J., Winn M., Ralph A. (1997) Methods Enzymol. 277, 620–633 [DOI] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 34. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 35. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 36. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 38. Percival M. D., Ouellet M., Vincent C. J., Yergey J. A., Kennedy B. P., O'Neill G. P. (1994) Arch. Biochem. Biophys. 315, 111–118 [DOI] [PubMed] [Google Scholar]
- 39. Górski J., Nawrocki A., Murthy M. (1998) Mol. Cell. Biochem. 178, 113–118 [DOI] [PubMed] [Google Scholar]
- 40. Lagarde M., Drouot B., Guichardant M., Dechavanne M. (1984) Biomed. Biochim. Acta 43, S319–322 [PubMed] [Google Scholar]
- 41. Malkowski M. G., Ginell S. L., Smith W. L., Garavito R. M. (2000) Science 289, 1933–1937 [DOI] [PubMed] [Google Scholar]
- 42. Malkowski M. G., Thuresson E. D., Lakkides K. M., Rieke C. J., Micielli R., Smith W. L., Garavito R. M. (2001) J. Biol. Chem. 276, 37547–37555 [DOI] [PubMed] [Google Scholar]
- 43. Thuresson E. D., Malkowski M. G., Lakkides K. M., Rieke C. J., Mulichak A. M., Ginell S. L., Garavito R. M., Smith W. L. (2001) J. Biol. Chem. 276, 10358–10365 [DOI] [PubMed] [Google Scholar]
- 44. Kiefer J. R., Pawlitz J. L., Moreland K. T., Stegeman R. A., Hood W. F., Gierse J. K., Stevens A. M., Goodwin D. C., Rowlinson S. W., Marnett L. J., Stallings W. C., Kurumbail R. G. (2000) Nature 405, 97–101 [DOI] [PubMed] [Google Scholar]
- 45. Mathias R. A., Vergara C., Gao L., Rafaels N., Hand T., Campbell M., Bickel C., Ivester P., Sergeant S., Barnes K. C., Chilton F. H. (2010) J. Lipid Res. 51, 2766–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simopoulos A. P. (2010) Exp. Biol. Med. 235, 785–795 [DOI] [PubMed] [Google Scholar]
- 47. McAdam B. F., Catella-Lawson F., Mardini I. A., Kapoor S., Lawson J. A., FitzGerald G. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 272–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu Y., Cheng Y., Fan J., Chen X. S., Klein-Szanto A., Fitzgerald G. A., Funk C. D. (2005) J. Clin. Invest. 115, 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu Y., Fan J., Hui Y., Rouzer C. A., Marnett L. J., Klein-Szanto A. J., FitzGerald G. A., Funk C. D. (2007) J. Biol. Chem. 282, 1498–1506 [DOI] [PubMed] [Google Scholar]
- 50. Muth-Selbach U. S., Tegeder I., Brune K., Geisslinger G. (1999) Anesthesiology 91, 231–239 [DOI] [PubMed] [Google Scholar]
- 51. Lapicque F., Vergne P., Jouzeau J. Y., Loeuille D., Gillet P., Vignon E., Thomas P., Velicitat P., Türck D., Guillaume C., Gaucher A., Bertin P., Netter P. (2000) Clin. Pharmacokinet. 39, 369–382 [DOI] [PubMed] [Google Scholar]
- 52. Burian M., Geisslinger G. (2005) Pharmacol. Ther. 107, 139–154 [DOI] [PubMed] [Google Scholar]
- 53. Grosser T., Yu Y., Fitzgerald G. A. (2010) Annu. Rev. Med. 61, 17–33 [DOI] [PubMed] [Google Scholar]
- 54. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.