Abstract
2,4,6-Trichlorophenol (2,4,6-TCP) is a hazardous pollutant. Several aerobic bacteria are known to degrade this compound. One of these, Ralstonia eutropha JMP134(pJP4), a well-known, versatile chloroaromatic compound degrader, is able to grow in 2,4,6-TCP by converting it to 2,6-dichlorohydroquinone, 6-chlorohydroxyquinol, 2-chloromaleylacetate, maleylacetate, and β-ketoadipate. Three enzyme activities encoded by tcp genes, 2,4,6-TCP monooxygenase (tcpA), 6-chlorohydroxyquinol 1,2-dioxygenase (tcpC), and maleylacetate reductase (tcpD), are involved in this catabolic pathway. Here we provide evidence that all these tcp genes are clustered in the R. eutropha JMP134(pJP4) chromosome, forming the putative catabolic operon tcpRXABCYD. We studied the presence of tcp-like gene sequences in several other 2,4,6-TCP-degrading bacterial strains and found two types of strains. One type includes strains belonging to the Ralstonia genus and possessing a set of tcp-like genes, which efficiently degrade 2,4,6-TCP and therefore grow in liquid cultures containing this chlorophenol as a sole carbon source. The other type includes strains belonging to the genera Pseudomonas, Sphingomonas, or Sphingopixis, which do not have tcp-like gene sequences and degrade this pollutant less efficiently and which therefore grow only as small colonies on plates with 2,4,6-TCP. Other than strain JMP134, none of the bacterial strains whose genomes have been sequenced possesses a full set of tcp-like gene sequences.
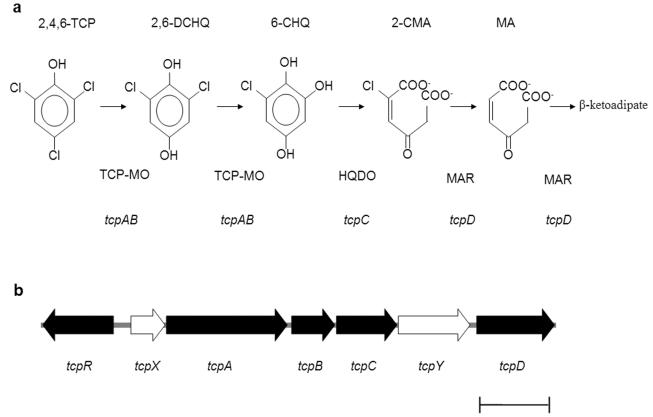
2,4,6-Trichlorophenol (2,4,6-TCP), one of the main components of the bleached Kraft pulp mill effluents, is widely used as a biocide and preservative (16, 23) and is considered a priority environmental pollutant worldwide (28). Aerobic bacteria have been reported to degrade this pollutant and in several cases to grow with it as the sole carbon source (6, 8, 19, 21). A catabolic pathway for 2,4,6-TCP (Fig. 1a), supported on biochemical evidence, has been proposed (22, 24, 31, 32). The pathway is initiated by the conversion of 2,4,6-TCP to 2,6-dichloro-p-hydroquinone (2,6-DCHQ) and then to 6-chlorohydroxyquinol (6-CHQ), both steps catalyzed by 2,4,6-TCP monooxygenase (TCP-MO). 6-CHQ is transformed to 2-chloromaleylacetate (2-CMA) by 6-chlorohydroxyquinol 1,2-dioxygenase (HQDO), and 2-CMA is converted to β-ketoadipate by maleylacetate reductase (MAR). However the genes involved in the 2,4,6-TCP catabolic pathway and their genetic organization are less well known. hadAB and hadC genes, encoding TCP-MO and HQDO, respectively, have been found in Ralstonia pickettii DTP0602 (14, 29). Recently, the tcpABC genes from R. eutropha JMP134(pJP4), a versatile, well-known chloroaromatic-compound-degrading strain (8, 11, 26), were shown to be encoding enzymes that convert 2,4,6-TCP to 2-CMA (22). However, no evidence about a gene encoding MAR activity was provided. Interestingly, tcpABC and hadABC are clustered (14, 22), which raises the possibility that 2,4,6-TCP degradation may be encoded in a catabolic operon in other bacterial strains that degrade this pollutant, including R. eutropha JMP134(pJP4).
FIG. 1.
Enzymes, intermediates, and genes involved in the degradation of 2,4,6-TCP in R. eutropha JMP134(pJP4). (a) Catabolic pathway for 2,4,6-TCP. (b) Genetic organization of tcp genes in R. eutropha JMP134(pJP4). MA, maleylacetate. tcpA (1,551 bp), tcpC (828 bp), and tcpD (1,065 bp): genes encoding TCP-MO, HQDO, and MAR, respectively. tcpR (969 bp); a putative LysR-type transcriptional activator regulatory gene. tcpB (594 bp), tcpX (465 bp), and tcpY (1,026 bp), ORFs of unknown function (see the text). Bar, 1 kb.
In this paper we report that all tcp genes, including the tcpD gene encoding MAR, are clustered in the chromosome of R. eutropha JMP134(pJP4) and organized in a catabolic operon, putatively regulated by a LysR-type transcriptional activator. In addition, we show that the presence of conserved tcp genes in other environmental strains is clearly linked to efficient growth in 2,4,6-TCP. In contrast, bacteria that grow poorly with this pollutant lack tcp-like genes.
MATERIALS AND METHODS
Bacterial strains, isolation of microorganisms, and growth conditions.
The bacterial strains used in this study are listed in Table 1. Strains PZK and S37 were isolated by a 2,4,6-TCP enrichment of samples from the Biobío river, Laja, central Chile (3, 24). This sampling site was located 1,000 m downstream of the outlet of a chlorine-bleached Kraft mill effluent. Two soils were chosen to isolate additional strains degrading 2,4,6-TCP. One forest soil, from central Chile (sampling site located at 37°S, 73°W), has not been exposed to anthropogenic chloroorganic compounds. The other soil was collected from an industrial site in The Netherlands which contained about 5 ppm of chloroethenes. An enrichment procedure with each soil sample was performed using cultures prepared with a minimal saline medium, free of organic compounds (1), supplemented with 2,4,6-TCP. A 1-ml volume of an aqueous extraction from 3 g of soil was used to inoculate 50 ml of liquid minimal medium culture, supplemented with 0.5 mM 2,4,6-TCP, in a 250-ml Erlenmeyer flask. Growth and 2,4,6-TCP consumption were determined every 3 days, and a subculture of each positive sample was performed. After three subcultures, colonies were isolated on minimal medium agar plates supplemented with 0.5 mM 2,4,6-TCP. Minimal medium agar plates without 2,4,6-TCP were run to test whether these isolates grow on agar. The strains were maintained in minimal medium agar plates supplemented with 0.5 mM 2,4,6-TCP or liquid minimal medium cultures supplemented with 8 mM pyruvate. The strains isolated were classified on the basis of a 16S rRNA gene sequencing protocol (20). The PCR products were sequenced in an Applied Biosystems ABI 370 sequencing apparatus and analyzed using Sequencher software (Gene Code Corp.)and the FASTA 3 program, available at http://www2.ebi.ac.uk/fasta3/ (European Molecular Biology Laboratory).
TABLE 1.
Strains used in this work
| Strain | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| R. eutropha JMP134(pJP4) | pJP4, 2,4,6-TCP+, 2,4-D+, 3-CB+ | DSMZc |
| R. eutropha JMP222 | pJP4-cured JMP134 derivative, 2,4,6-TCP+, 2,4-D−, 3-CB−, Smr | H.-J. Knackmuss |
| R. eutropha MS1 | 2,4,6-TCP+, Apr, Kmr | This study |
| Ralstonia sp. strain PZK | 2,4,6-TCP+, Kmr | 24 |
| S. chilensis S37 | 2,4,6-TCPb | 3, 13 |
| S. paucimobilis MS2 | 2,4,6-TCPb | This study |
| P. tolaasii MS6 | 2,4,6-TCPb | This study |
| P. putida MS7 | 2,4,6-TCPb | This study |
2,4-D+, 3-CB+, 2,4,6-TCP+, able to grow with 2,4-D, 3-CB, or 2,4,6-TCP, respectively, as the sole carbon source in liquid medium.
Able to grow on agar plates supplemented with 2,4,6-TCP but not in liquid cultures as the sole carbon source. Apr, ampicillin resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen. Braunschweig, Germany.
Metabolic studies with 2,4,6-TCP.
Bacterial growth with 2,4,6-TCP was determined as an increase in the optical density at 660 nm (OD660) in liquid minimal medium cultures supplemented with 0.5 mM 2,4,6-TCP. 2,4,6-TCP degradation was determined using resting cells. Briefly, the strains were grown in liquid minimal medium cultures containing 8 mM pyruvate, supplemented with 0.5 mM 2,4,6-TCP, and incubated at 30°C with constant shaking (at 180 rpm). The cells were harvested and washed in minimal medium solution, and cell suspensions (OD660 = 1.0) were incubated with 0.5 mM 2,4,6-TCP for up to 72 h. Samples (1 ml) of cell-free supernatants were taken between 0 and 90 h and analyzed by UV spectroscopy in a diode array HP8452-A UV-visible spectrophotometer or by high-performance liquid chromatography (HPLC). Samples for HPLC (20 μl) were injected into a 126/166 System Gold Beckman liquid chromatograph equipped with a Waters Symmetry C18 4.6-μm-diameter column (Beckman Instruments, Fullerton, Calif.). A methanol-H2O (80:20) mixture containing 0.3% (by volume) phosphoric acid was used as the solvent at a flow rate of 1 ml min−1. The column effluent was monitored by measuring the absorbance at 210 nm. The retention volume for 2,4,6-TCP was 8.2 ml. Chloride release was determined by a previously described procedure based on the spectrophotometric detection of a colored product (3). Under our experimental conditions, this procedure gave a linear response in the 0.05 to 1 mM range, with relative errors below 5%. Controls without cells were routinely run to determine whether abiotic transformations had occurred. Incubations of these cell-free controls containing minimal medium plus 0.5 mM 2,4,6-TCP never showed chloride production or removal of this chlorophenol as determined by UV spectroscopy or HPLC.
PCR amplification of sequences involved in 2,4,6-TCP degradation.
PCR primer pairs were designed using previously published sequences corresponding to genes encoding TCP-MO, HQDO, and MAR (Table 2). Sequences were aligned using Clustal W software, and primer pairs were designed from the conserved regions using the Primer Select program (DNASTAR, Inc.). The expected product sizes were as follows: TCP-MO, 1,200 bp; HQDO, 636 bp; and MAR, 366 bp. PCR amplifications were carried out using total DNA, prepared by a standard procedure (4), as the template. To amplify the TCP-MO and HQDO sequences, the PCR conditions were 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 62°C for 45 s, and 72°C for 1.5 min, with a final step at 72°C for 10 min. To amplify MAR sequences, Taq polymerase was replaced by Pfx polymerase and the PCR amplification was performed under the same conditions as for amplification of TCP-MO and HQDO PCR products, except that the annealing temperature was set at 60°C and 28 cycles were run. All PCR amplifications were performed with a Perkin-Elmer GeneAmp PCR System 2400.
TABLE 2.
PCR primers used in this work
| Primer | Sequence | Sourcea |
|---|---|---|
| MON-F | CGC AAT GTC TGG GTC GGC AA | tftD, hadA |
| MON-R | CAC GCG GCC GAT CTT CAC | tftD, hadA |
| HQ-F | ACC GCC ACC GGC CAC AAG TG | hadC |
| HQ-R | AGC GAT TCG CGC ACG CCG AAC ACA G | hadC |
| MAR-F | AAA TGA CGC CGA TCT ATG G | macA, tftE, tfdFI, tfdFII |
| MAR-R | AGC ACA TGG CAG AGC TTG TG | macA, tftE, tfdFI, tfdFII |
Southern analysis.
Southern analysis was performed with biotinylated DNA probes. The probe-labeling procedure was performed as described elsewhere (9), and hybridization was carried out under low-stringency conditions as recommended by the supplier. DNA was digested with ClaI or KpnI and electrophoresed in a 0.8% agarose gel. The results obtained with DNA digested with each of these restriction enzymes were essentially the same. To detect TCP-MO and MAR sequences, probes from the respective R. eutropha JMP222 PCR products were prepared. For HQDO detection, the probe was prepared from the R. eutropha MS1 PCR product.
DNA sequencing, sequence alignments, and sequence analysis.
PCR products obtained with primer pairs for TCP-MO, HQDO, and MAR were cloned in pGem-T vector and sequenced using primer pairs M13 and rM13 (Gibco-BRL, Rockville, Md.). Partial sequences were compared using Clustal W from the MegAlign program of DNASTAR, Inc. The R. eutropha JMP134(pJP4) genome sequence is available at http://www.jgi.doe.gov. Contig analysis was performed using the WU-Blast 2.0 program from the ProWeb project (http://www.proweb.org/tools/wu-blast.html). The computational resource of the National Center for Biotechnology Information was used through the BLASTX and TBLASTN software facilities.
RESULTS AND DISCUSSION
All the tcp genes involved in 2,4,6-TCP degradation are clustered in the chromosome of R. eutropha JMP134(pJP4).
A previous study had shown that conversion of 2,4,6-TCP to 2-CMA in R. eutropha JMP134(pJP4) was encoded by the tcpABC genes, which were located in a 3.2-kb DNA fragment (22). A gene encoding the conversion of 2-CMA to β-ketoadipate was not reported in that work, and a gene encoding MAR in R. pickettii DTP0602 has not been reported, either. Alternatively, it is possible that a MAR activity such as that encoded by macA (GenBank accession no. AF130250) in the closely related strain R. eutropha 335 may be involved in 2,4,6-TCP degradation by R. eutropha JMP134(pJP4), and its cured derivative JMP222 (24). This prompted us to look for additional putative tcp sequences in R. eutropha JMP134(pJP4). This was performed in the annotated, 18X coverage, 7.4-Mb draft genome sequence of this bacterium (http://genome.jgi-psf.org/draft_microbes/raleu/raleu.home.html). The presence of one copy of a cluster containing the tcpABC sequences was evident (Fig. 1b). The sequence of tcpABC perfectly matched that reported previously (22). Interestingly, an open reading frame (ORF), tcpD, with significant identity to other MAR-encoding genes (see below) was located downstream of and close to the tcpABC genes, suggesting a functional relation among these tcp genes. The R. eutropha JMP134(pJP4) genome also carries a macA-like gene (whose product has 64% amino acid identity to the tcpD gene product, and 60% amino acid identity to the macA product from R. eutropha 335), but its genetic context (ORFs encoding functions putatively involved in sugar transport) is not related to chloroaromatic catabolism. Two other ORFs were found in the tcp DNA region (Fig. 1b). One of them, designed tcpX, encoded a product with a 54% amino acid identity to the tftC gene product from the 2,4,5-TCP degradation pathway in Burkholderia cepacia AC1100 (15). The other ORF, designed tcpY, encoded a product with no significant identity to known proteins. More significantly, upstream of the tcpXABCYD gene sequences, and located divergently, a seventh ORF was found whose product showed homology to LysR-type transcriptional activators. This ORF, named tcpR, encodes a product with a higher identity (35% amino acid identity) to the pcpR gene product involved in pentachlorophenol (PCP) degradation by Sphingobium chlorophenolicum ATCC 39723 (7).
The functional relationship of the tcpABCD (and probably tcpXABCYD) genes is supported by several observations. Except for tcpR, all these sequences are transcribed in the same direction, with tcpC and tcpY genes having a 22-bp overlap in their codogenic regions. ORFs that flank these tcp genes are not related to chloroaromatic catabolism because they encode proteins putatively involved in amino acid metabolism and transport (data not shown). All these tcp sequences have the same GC content (65%) and codon usage as the R. eutropha JMP134(pJP4) chromosome (data not shown). These observations allowed us to propose that all tcp genes required for conversion of 2,4,6-TCP to β-ketoadipate in R. eutropha JMP134(pJP4) are in single copy and are clustered forming part of a catabolic operon, putatively regulated by a LysR-type transcriptional regulator. This is the first report of such genetic organization in trichlorophenol catabolism. An operon-like organization has been reported for the tft genes involved in the degradation of 2,4,5-TCP in B. cepacia AC1100, but no putative regulatory genes were found in association with it (10). In addition, the genes involved in 2,4,5-TCP catabolism, tftEFGH and tftCD (two copies), are located in multiple replicons (15). The presence of a single copy of each of the tcp genes located in one locus in the chromosome of R. eutropha JMP134(pJP4) may explain the stability of this catabolic property (8, 24), which contrasts with the instability of the tft-encoded catabolic phenotype in B. cepacia AC1100 (15). The genetic organization of the pcp genes for PCP degradation by S. chlorophenolicum ATCC 39723 has been recently reported (7). The pcp genes are found in four locations in the genome. Available evidence does not seem to indicate that pcp genes are regulated by only one transcriptional activator (7), as is suggested by the tcp gene organization in R. eutropha JMP134(pJP4).
2,4,6-TCP degradation in other environmental isolates.
tcp genes are responsible for 2,4,6-TCP degradation in R. eutropha JMP134(pJP4), and the closely related had genes (see below) play the same role in R. pickettii DTP0602. Therefore, we hypothesized that tcp-like gene sequences would be involved in trichlorophenol degradation in other 2,4,6-TCP-degrading bacterial isolates. We used an enrichment procedure to isolate new bacterial strains that degrade this pollutant. Three different bacteria were isolated from the unpolluted Chilean soil, and one was isolated from the Dutch soil contaminated with chloroethenes. Sequencing of its rRNA genes revealed that the strains from the Chilean uncontaminated soil were Pseudomonas tolaasii MS6 (98.4% similarity to the corresponding type strain), P. putida MS7 (99.6% similarity), and S. paucimobilis MS2 (99.2% similarity). The bacterial strain isolated from the Dutch soil was R. eutropha MS1 (98.2% similarity). These four bacteria were able to grow on agar plates containing 0.5 mM 2,4,6-TCP, but only R. eutropha MS1 was also able to grow in liquid cultures supplemented with up to 2 mM 2,4,6-TCP. None of these strains produced colonies on agar plates prepared in minimal saline medium without 2,4,6-TCP. Strain MS1 was unable to grow with several other chlorinated compounds (0.5 to 1.0 mM): PCP, 2,4,5-TCP, 2,3-, and 2,4-dichlorophenol, 4-chlorophenol, 3,5-dichlorobenzoate, 3-chlorobenzoate (3-CB), or 2,4-dichlorophenoxyacetic acid (2,4-D).
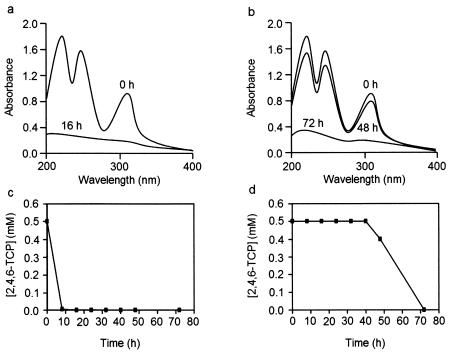
Degradation of 2,4,6-TCP in these new isolates was studied and compared with that in strains previously shown to degrade this pollutant. The latter group included R. eutropha JMP134(pJP4) and JMP222, Ralstonia sp. strain PZK (24), and strain S37 (3), recently reclassified as Sphingopixis chilensis (13). Since some of the new isolates did not grow well in liquid cultures containing 2,4,6-TCP, this study was performed with resting bacterial cell suspensions (OD660 = 1.0). UV spectroscopy analysis of the supernatants of resting cells pregrown with pyruvate and incubated with 2,4,6-TCP showed that strain MS1 degraded 2,4,6-TCP as efficiently as did R. eutropha JMP134(pJP4). R. eutropha JMP222, and Ralstonia sp. strain PZK (Fig. 2a). In contrast, strains S. paucimobilis MS2, S. chilensis S37, P. tolaasii MS6, and P. putida MS7 degraded 2,4,6-TCP, but less efficiently (Fig. 2b). This behavior is in agreement with that reported for S. chilensis S37 (3, 13). Degradation of 2,4,6-TCP was confirmed by HPLC analysis. Cell suspensions from strains JMP134, JMP222, MS1, and PZK degraded this chlorophenol faster (Fig. 2c) than did suspensions from strains MS2, MS6, MS7, and S37 (Fig. 2d). The latter group exhibited a clear adaptation phase (lasting >24 h) during degradation of this chlorophenol. In these incubations of resting cells, UV spectroscopy or HPLC analysis did not allow the detection of intermediates. It has been reported that intermediates in 2,4,6-TCP catabolism by R. eutropha JMP134 are accumulated in minor amounts (22, 24) and that CHQs are unstable compounds (22, 24, 32). Determinations with dense suspensions (OD660 = 5.0) of resting cells from all these strains showed that in all cases 90 to 100% of the available chlorine from 2,4,6-TCP (molar stoichiometry, 3:1) was released as chloride. However, strains MS2, MS6, MS7, and S37 released 50% of chloride four- to fivefold slower than did strains JMP134(pJP4), JMP222, MS1, and PZK. The first group of strains took more than 24 h to complete chloride release whereas the second group required only 12 to 16 h of incubation for quantitative release of chloride.
FIG. 2.
Comparison of the degradation of 2,4,6-TCP by different types of 2,4,6-TCP-degrading bacteria. Resting cells (OD660 = 1; pyruvate grown and 2,4,6-TCP-induced) of R. eutropha MS1 (a and c), and P. tolaasii MS6 (b and d), chosen as representatives of each bacterial type, were incubated in the presence of 0.5 mM 2,4,6-TCP. Samples were taken at intervals, and the UV spectra of the cell-free fluid were obtained (a and b). Representative profiles from several independent determinations are shown. In addition, the concentration of 2,4,6-TCP in the culture fluid was determined by HPLC (c and d). Each point is the average of two replicates; standard deviations were too low to be depicted. Time points (hours) are indicated.
Despite the limited number of bacteria available for analysis, it is clear that all 2,4,6-TCP-degrading strains isolated from polluted sites, R. eutropha JMP134(pJP4) and JMP222 (11), R. pickettii DTP1062 (19), Ralstonia sp. strain PZK (24), R. eutropha MS1 (this work), Alcaligenes eutrophus TCP (2), strain GP1 (21), and other strains (5), belong to the Ralstonia genus, although some of them were initially classified in other genus-species combinations (5). All these strains efficiently degrade 2,4,6-TCP and therefore grow in liquid cultures with this TCP. Another interesting aspect, previously reported (24), is that these bacterial strains showed a very narrow growth substrate range and that the TCP-degrading ability is apparently encoded in the chromosome. In contrast, the 2,4,6-TCP-degrading strains that do not belong to the Ralstonia genus degrade this chlorophenol less efficiently and do not grow in liquid cultures (3, 13; this work). A recent report also shows that several non-Ralstonia strains belonging to a consortium degrading 2,4,6-TCP were unable to grow efficiently with this pollutant (18). A similar situation has been observed for strains degrading PCP, but in this case the growth-efficient bacteria belong to the sphingomonad group (12, 30).
In summary, efficient degrading strains grow with 2,4,6-TCP in solid and liquid cultures and their resting cells fully degrade this chlorophenol in short (less than 12-16 h) incubations. In contrast, the less efficient strains only grow on solid medium, and their resting cells degrade 2,4,6-TCP in longer incubations.
Detection of tcp-like gene sequences in strains that degrade 2,4,6-TCP.
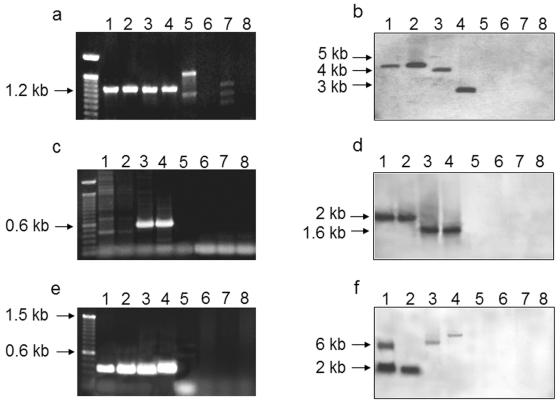
To detect the presence of tcp genes in 2,4,6-TCP-degrading bacterial isolates, PCR amplification and Southern analysis were performed. For PCR detection, primer pairs were designed that targeted conserved regions found after an alignment of published sequences involved in TCP metabolism (Table 2). tcp gene sequences were excluded from these alignments to minimize bias toward R. eutropha JMP134(pJP4) genes. When the primer pair MON-F and MON-R was used, PCR products putatively corresponding to TCP-MO-like sequences were found in strains R. eutropha JMP134(pJP4), JMP222, and MS1 and Ralstonia sp. strain PZK (Fig. 3a). Each of the four 1,200-bp PCR products was partially sequenced (about 600 bp). In the case of the PCR product from R. eutropha JMP134(pJP4), the sequence matched tcpA perfectly. The same situation was observed with R. eutropha JMP222. The sequence analysis of tcpA revealed 93% amino acid identity to the hadA gene product of R. pickettii DTP1062. The corresponding sequence analysis indicated that the PCR product of R. eutropha MS1 has 86 and 99% amino acid identity to the tcpA and hadA gene products, respectively. For Ralstonia sp. strain PZK, the amino acid identity to the tcpA and hadA gene products was 88 and 90%, respectively. No PCR products of the correct size were obtained with DNA from P. tolaasii MS6, P. putida MS7, S. paucimobilis MS2, and S. chilensis S37 (Fig. 3a). Bands of unexpected sizes were occasionally observed (Fig. 3a, lanes 5 and 7; Fig. 3c, lanes 1 and 2). This material corresponds to nonspecific PCR products, as shown by sequence analysis. In some cases, these bands were also observed in PCR amplification performed with only one primer of each pair.
FIG. 3.
Detection of gene sequences involved in 2,4,6-TCP metabolism by PCR and Southern analysis. (a, c, and e) Gel images showing PCR products obtained with primer pairs for (a) TCP-MO (tcpA). (c) HQDO (tcpC), and (e) MAR (tcpD). (b, d, and f) Southern blot using the strain JMP222 tcpA probe (b), the strain MS1 tcpC probe (d), and the strain JMP222 tcpD probe (f). Total DNA from R. eutropha JMP134(pJP4) (lanes 1) R. eutropha JMP222 (lanes 2), R. eutropha MS1 (lanes 3), Ralstonia sp. strain PZK (lanes 4), S. chilensis S37 (lanes 5), S. paucimobilis MS2 (lanes 6), P. tolaasii MS6 (lanes 7), and P. putida MS7 (lanes 8) was digested with Clal. The left lane in panels a, c, and e shows 1-kb (a) or 100-bp (c and e) DNA standards. Arrows indicate fragment sizes for orientation.
The presence or absence of tcpA-like sequences was also explored by Southern analysis, using as a probe the cloned PCR product obtained from R. eutropha JMP222. As shown in Fig. 3b, the strains that were positive for PCR amplification were also positive with the JMP222 probe, supporting the presence of a TCP-MO-like sequence. The JMP222 probe hybridized with a ClaI (Fig. 3b) or KpnI (data not shown) fragment of the same size (4,375 bp, determined from the strain JMP134 genome sequence) in DNA from strains JMP134(pJP4) and JMP222 (the pJP4-cured derivative of strain JMP134), indicating the same chromosomal location for the tcpA gene. The strains that were negative for PCR products using the primer pairs based on TCP-MO were also negative in Southern analysis using the tcpA-like JMP222 probe (Fig. 3b).
The presence of HQDO-encoding gene sequences was also investigated using the primer pair HQ-F and HQ-R (Table 2). PCR products of the expected size (636 bp) were observed only with DNA from R. eutropha MS1 and Ralstonia sp. strain PZK (Fig. 3c). These two PCR products were partially sequenced and showed 65 and 87% amino acid identity (in the case of strain MS1) and 71 and 81% amino acid identity (in the case of Ralstonia sp. strain PZK) to TcpC and HadC, respectively. Using the R. eutropha MS1 tcpC-like PCR product as a probe, a Southern analysis was performed. In this case, in addition to the positive hybridization with strains MS1 and PZK, same-size bands (2,150 bp as determined from the strain JMP134 genome sequence) for strains JMP134(pJP4) and JMP222 were also evident (Fig. 3d). The strains that gave no PCR product with the primer pairs for HQDO were also negative for hybridization with the tcpC-like probe from strain MS1 (Fig. 3d).
MAR-encoding gene sequences were also searched for in the DNA from all these 2,4,6-TCP-degrading strains. In this case, a PCR primer pair based on the conserved regions deduced from the alignment of four different MAR-encoding sequences, including the tfdFI and tfdFII genes from pJP4 plasmid (Table 2), was used. Positive amplification of a fragment of the expected size (366 bp) was found with the four Ralstonia strains (Fig. 3e). These PCR products were partially sequenced and showed high identity to macA (GenBank accession no. AF130250). The amino acid identities were 64, 77, and 64% for R. eutropha JMP222, R. eutropha MS1, and Ralstonia sp. strain PZK, respectively. The sequence found for strains JMP134(pJP4) and JMP222 perfectly matched the tcpD gene sequence. Southern analysis using the PCR product obtained from strain JMP222 as a probe confirmed the results obtained by PCR (Fig. 3f). A positive signal was found for the four Ralstonia strains, and absence of signals was detected with the strains that were negative for PCR amplification (Fig. 3e). Weaker signals for DNA from strains PZK and MS-1 than for DNA from strains JMP222 and JMP134(pJP4) were also observed with DNA digested with KpnI (data not shown). Since the same amount of digested DNA was loaded in each well, we supposed that the MAR-encoding sequences from strains MS-1 and PZK are less similar to that of strain JMP222 than TCP-MO- or HQDO-encoding sequences. The bands observed with DNA from strain JMP134(pJP4) corresponded to the TfdFI (6.8 kb) and TfdFII (2.6 kb) encoded in pJP4 and to TcpD (2.0 kb).
The results described in this section clearly suggest that Ralstonia strains are able to grow efficiently with 2,4,6-TCP due to the presence of conserved tcp-like sequences. In contrast, Sphingomonas, Sphingopixis, and Pseudomonas strains, which do not have tcp-like sequences, degrade 2,4,6-TCP less efficiently. Several possibilities may explain this difference. The less efficient strains may possess the same catabolic pathway but the pathway may be encoded by genes whose similarity to the tcp genes is low and therefore undetectable by the molecular tools used in this work. Alternatively, a different pathway (and therefore different genes) may be used in the less efficient degraders. The first possibility has been detected for 2,4-dichlorophenoxyacetic acid (2,4-D) degradation. Kamagata et al. (17) demonstrated that 2,4-D-degrading bacteria isolated from pristine oligotrophic environments were slowly degrading bacteria and that their catabolic genes differed from those of 2,4-D degraders typically isolated from contaminated environments (17). Moreover, most of the efficiently 2,4-D-degrading bacteria were members of the β subdivision of Proteobacteria, and the slowly degrading strains belonged to α and γ subdivisions. Our results also show an interesting correlation: efficient microorganisms degrading 2,4,6-TCP, like Ralstonia strains, belong to the β subdivision, and less efficient 2,4,6-TCP degraders, like sphingomonads and pseudomonads, belong to the α and γ subdivisions of Proteobacteria, respectively.
tcp-like genes in sequenced bacterial genomes.
The presence of tcp-like sequences was also investigated in bacteria whose genome sequences are available for analysis. The genome sequences of more than 30 bacterial strains with or without known or putative catabolic abilities toward aromatic compounds were analyzed. We found tcp homologues in 13 strains (Table 3), but the complete set of tcp genes was not present in any of them. In the bacterial genomes with positive matches, the higher amino acid identities were around 60 to 65%. The tcpA-like gene sequence was less frequently found, whereas positive matches for tcpD were found in all genomes except for that of Deinococcus radiodurans. In two genomes, Burkholderia cepacia and Ralstonia solanacearum, more than one match was found for genes putatively encoding MAR. Two MAR-encoding chromosomal genes have also been reported for R. eutropha (this work) and S. chlorophenolicum (7). None of the strains listed in Table 3 has been reported to utilize 2,4,6-TCP efficiently in the way that R. eutropha strains do, including the related R. metallidurans (formerly R. eutropha) and R. solanacearum strains. The absence of tcp genes, especially the tcpA gene, encoding the first two steps in the 2,4,6-TCP degradation pathway, is clearly related to this catabolic limitation and is in agreement with the observations about the less efficient 2,4,6-TCP-degrading environmental bacterial strains reported above. Additional evidence to support this point was obtained with two strains whose genomes have been fully sequenced (Table 3): P. putida KT2440, a well-known aromatic compounds degrader, and Agrobacterium tumefaciens C58, a phytopathogen. Suspensions (OD660 = 1.0) of resting cells previously grown in 8 mM pyruvate and induced with 0.5 mM 2,4,6-TCP degrade this chlorophenol only after 48 h of incubation (data not shown); i.e., they behaved identically to the other, less efficient strains reported in this work.
TABLE 3.
Presence of tcp-like genes in sequenced bacterial genomesa
| Strain | Amino acid identity (%) tob:
|
|||
|---|---|---|---|---|
| tcpA | tcpB | tcpC | tcpD | |
| Agrobacterium tumefaciens C58 | 60 (114/190) | 50 (127/256) | 54 (192/354) | |
| Burkholderia cepacia J2315 | 25 (119/461) | 60 (174/287) | 63 (222/351) | |
| 58 (209/357) | ||||
| Burkholderia fungorum LB400 | 60 (118/196) | 32 (86/261) | 51 (182/352) | |
| Novosphingobium aromaticivorans SMCCf199 | 32 (86/263) | 27 (103/380) | ||
| Deinococcus radiodurans R1 | 25 (119/463) | 30 (47/153) | ||
| Pseudomonas fluorescens Pf0-1 | 32 (84/262) | 27 (106/380) | ||
| Pseudomonas syringae B728a | 27 (103/373) | |||
| Pseudomonas aeruginosa PAO-1 | 26 (125/470) | 34 (89/260) | (90/268) | |
| 27 (131/482) | ||||
| Pseudomonas putida KT2440 | 34 (91/263) | (88/289) | ||
| Ralstonia metallidurans CH34 | 45 (87/191) | 41 (116/281) | 56 (200/357) | |
| Ralstonia solanacearum GMI1000 | 50 (98/190) | (83/256) | ||
| (84/295) | ||||
| Shewanella oneidensis MR-1 | (89/288) | |||
| Sulfolobus solfataricus P2 | 23 (106/456) | 28 (69/238) | ||
Genome sequence were obtained from The Institute for Genomic Research, The Sanger Institute, and the Department of Energy—Joint Genome Institute public data banks. The Tcp protein lengths in R. eutropha JMP134(pJP4) are as follows: TcpA, 517 amino acids TcpB, 197 amino acids, TcpC, 276 amino acids; TcpD, 355 amino acids.
The number of amino acids used in the comparison and the total number of amino acids in each protein sequence are shown in parentheses. No percent similarity for MAR sequences is indicated in cases when only part of the protein exhibits homology.
Comparisons of the tcp sequences with those of other genes encoding polychlorophenol catabolism were carried out. The tcpA sequences from R. eutropha MS1, R. eutropha JMP134(pJP4), and Ralstonia sp. strain PZK clustered apart from the tftD sequence, which codes for a similar enzyme activity but one involved in 2,4,5-TCP degradation in B. cepacia AC1100 (15). On the other hand, tcpD sequences from R. eutropha JMP134(pJP4) and MS1, but not strain PZK, are clustered. The tcpD-like product from Ralstonia sp. strain PZK clustered with that of other polychlorophenol degraders, e.g., B. cepacia AC1100 and S. chlorophenolicum ATCC 39723. It is interesting that the chromosomal tcpD gene in strain JMP134(pJP4) clustered apart from the two pJP4-located MAR genes, which are involved in metabolism of 3-CB and 2,4-D (25, 27). No significant identities were found between the tcp and pcp genes. Despite the related chemical structure of PCP and 2,4,6-TCP and the similar catabolic pathways, bacteria that grow efficiently with one of them do not perform as well with the other. This behavior may be a consequence of a different evolutionary origin of pcp and tcp genes that might explain the lack of significant identities among these genes and the differences in gene organization between the tcp and pcp genes indicated above.
Acknowledgments
This work was supported by FONDECYT (grants 8990004 and 2980041), FONDAP-FONDECYT (grant 1501-0001, program 7), and Universidad de Concepción (grant 200.036.020-1.0). M.A.S is a FUNDACION ANDES undergraduate fellow.
We acknowledge the Biodegradation Group, Division of Microbiology, GBF, Braunschweig, Germany, for help with some initial experiments. We also acknowledge the Millennium Institute for Fundamental and Applied Biology and the Center of Genomics and Bioinformatics, Pontificia Universidad Católica de Chile, for their help.
REFERENCES
- 1.Adriaens, P., H. P. Kohler, D. Kohler-Staub, and D. D. Focht. 1989. Bacterial dehalogenation of chlorobenzoates and coculture biodegradation of 4,4′-dichlorobiphenyl. Appl. Environ. Microbiol. 55:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni, V., G. Baggi, M. Colombo, L. Cavalca, M. Zangrossi, and S. Bernasconi. 1998. Degradation of 2,4,6-trichlorophenol by a specialized organism and by indigenous soil microflora: bioaugmentation and self-remediability for soil restoration. Lett. Appl. Microbiol. 27:86-92. [DOI] [PubMed] [Google Scholar]
- 3.Aranda, C., F. Godoy, B. González, J. Homo, and M. Martínez. 1999. Effects of glucose and phenylalanine upon 2,4,6-trichlorophenol degradation by Pseudomonas paucimobilis S37 cells in non-growth state. Microbios 100:73-82. [PubMed] [Google Scholar]
- 4.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1992. Short protocols in molecular biology: a compendium of methods from, current protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bock, C., R. M. Kroppenstedt, U. Schmidt, and H. Diekmann. 1996. Degradation of prochloraz and 2,4,6-trichlorophenol by environmental bacterial strains. Appl. Microbiol. Biotechnol. 45:257-262. [DOI] [PubMed] [Google Scholar]
- 6.Briglia, M., F. A. Rainey, E. Stackebrandt, G. Schraa, and M. S. Salkinoja-Salonen. 1996. Rhodococcus percolatus sp. nov., a bacterium degrading 2,4,6-trichlorophenol. Int. J. Syst. Bacteriol. 46:23-30. [DOI] [PubMed] [Google Scholar]
- 7.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. [DOI] [PubMed] [Google Scholar]
- 9.Clément, P., D. H. Pieper, and B. González. 2001. Molecular characterization of a deletion/duplication rearrangement in tfd genes from Ralstonia eutropha JMP134(pJP4) that improves growth on 3-chlorobenzoic acid but abolishes growth on 2,4-dichlorophenoxyacetic acid. Microbiology 147:2141-2148. [DOI] [PubMed] [Google Scholar]
- 10.Daubaras, D. L., C. D. Hershberger, K. Kitano, and A. M. Chakrabarty. 1995. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl. Environ. Microbiol. 62:1279-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ederer, M. M., R. L. Crawford, R. P. Herwig, and C. S. Orser. 1997. PCP degradation is mediated by closely related strains of the genus Sphingomonas. Mol. Ecol. 6:39-49. [DOI] [PubMed] [Google Scholar]
- 13.Godoy, F. A., M. Bunster, V. Matus, C. Aranda, B. González, and M. Martinez. 2003. Poly-β-hydroxyalkanoates consumption during degradation of 2,4,6-trichlorophenol by Sphingopixis chilensis S37. Lett. Appl. Microbiol. 36:315-320. [DOI] [PubMed] [Google Scholar]
- 14.Hatta, T., O. Nakano, N. Imai, N. Takizawa, and H. Kiyohara. 1999. Cloning and sequence analysis of hydroxyquinol 1,2-dioxygenase gene in 2,4,6-trichlorophenol-degrading Ralstonia pickettii DTP0602 and characterization of its product. J. Biosci. Bioeng. 87:267-272. [DOI] [PubMed] [Google Scholar]
- 15.Hübner, A., C. E. Danganan, L. Xun, A. M. Chakrabarty, and W. Hendrickson. 1998. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC1100: characterization of the tftC and tftD genes and location of the tft operons on multiple replicons. Appl. Environ. Microbiol. 64:2086-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh, V. B., H. M. Chang, T. W. Joyce, and T. K. Kirk. 1985. Dechlorination of chloro-organics by a white-rot fungus. TAPPI J. 68:98-102. [Google Scholar]
- 17.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kharoune, L., K. Kharoune, and J. M. Lebeault. 2002. Aerobic degradation of 2,4,6-trichlorophenol by a microbial consortium—selection and characterization of microbial consortium. Appl. Microbiol. Biotechnol. 59:112-117. [DOI] [PubMed] [Google Scholar]
- 19.Kiyohara, H., T. Hatta, Y. Ogawa, T. Kakuda, H. Yokohama, and N. Takizawa. 1992. Isolation of Pseudomonas pickettii strains that degrade 2,4,6-trichlorophenol and their dechlorination of chlorophenols. Appl. Environ. Microbiol. 58:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing. In Nucleic acid techniques in bacterial systematics, p. 115-175. In E. Stackebrandt and M. Goodfellows (ed.), John Wiley & Sons, Ltd., Chicheter, United Kingdom.
- 21.Li, D.-Y., J. Eberspächer, B. Wagner, J. Kuntzer, and F. Lingens. 1991. Degradation of 2,4,6-trichlorophenol by Azotobacter sp. strain GP1. Appl. Environ. Microbiol. 57:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louie, T. M., C. M. Webster, and L. Xun. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J. Bacteriol. 184:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAllister, K. A., H. Lee, and J. T. Trevors. 1996. Microbial degradation of pentachlorophenol. Biodegradation 7:1-40. [Google Scholar]
- 24.Padilla, L., V. Matus, P. Zenteno, and B. González. 2000. Degradation of 2,4,6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J. Basic. Microbiol. 40:243-249. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Pantoja, D., L. Guzmán, M. Manzano, D. H. Pieper, and B. González. 2000. Role of tfdCIDIEIFI and tfDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 66:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieper, D. H., W. Reineke, K. H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 27.Plumeier, I., D. Pérez-Pantoja, S. Heim, B. González, and D. H. Pieper. 2002. The importance of different tfd genes during the degradation of chloroaromatics by Ralstonia eutropha JMP134. J. Bacteriol. 184:4054-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sittig, M. 1981. Handbook of toxic and hazardous chemicals. Noyes Publications, Park Ridge, N.J.
- 29.Takizawa, N., H. Yokohama, K. Yanagihara, T. Hatta, and H. Kiyohara. 1995. A locus of Pseudomonas pickettii DTP0602, had, that encodes 2,4,6-trichlorophenol 4-dechlorinase with hydroxylase activity, and hydroxylation of various chlorophenols by the enzyme. J. Ferment. Bioeng. 80:318-326. [Google Scholar]
- 30.Tiirola, M. A., H. Wang, L. Paulin, and M. S. Kulomaa. 2002. Evidence for natural horizontal transfer of the pcpB gene in the evolution of polychlorophenol-degrading sphingomonads. Appl. Environ. Microbiol. 68:4495-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieser, M., B. Wagner, J. Eberspächer, and F. Lingens. 1997. Purification and characterization of 2,4,6-trichlorophenol 4-monooxygenase, a dehalogenating enzyme from Azotobacter sp. strain GP1. J. Bacteriol. 179:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaborina, O., M. Latus, J. Eberspächer, L. A. Golovleva, and F. Lingens. 1995. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobacter sp. strain GP1. J. Bacteriol. 177:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]