Abstract
Type 2 diabetes is a global problem, and current ineffective therapeutic strategies pave the way for novel treatments like small molecular activators targeting glucokinase (GCK). GCK activity is fundamental to beta cell and hepatocyte glucose metabolism, and heterozygous activating and inactivating GCK mutations cause hyperinsulinemic hypoglycemia (HH) and maturity onset diabetes of the young (MODY) respectively. Over 600 naturally occurring inactivating mutations have been reported, whereas only 13 activating mutations are documented to date. We report two novel GCK HH mutations (V389L and T103S) at residues where MODY mutations also occur (V389D and T103I). Using recombinant proteins with in vitro assays, we demonstrated that both HH mutants had a greater relative activity index than wild type (6.0 for V389L, 8.4 for T103S, and 1.0 for wild type). This was driven by an increased affinity for glucose (S0.5, 3.3 ± 0.1 and 3.5 ± 0.1 mm, respectively) versus wild type (7.5 ± 0.1 mm). Correspondingly, the V389D and T103I MODY mutants had markedly reduced relative activity indexes (<0.1). T103I had an altered affinity for glucose (S0.5, 24.9 ± 0.6 mm), whereas V389D also exhibited a reduced affinity for ATP and decreased catalysis rate (S0.5, 78.6 ± 4.5 mm; ATPKm, 1.5 ± 0.1 mm; Kcat, 10.3 ± 1.1s−1) compared with wild type (ATPKm, 0.4 ± <0.1; Kcat, 62.9 ± 1.2). Both Thr-103 mutants showed reduced inhibition by the endogenous hepatic inhibitor glucokinase regulatory protein. Molecular modeling demonstrated that Thr-103 maps to the allosteric activator site, whereas Val-389 is located remotely to this position and all other previously reported activating mutations, highlighting α-helix 11 as a novel region regulating GCK activity. Our data suggest that pharmacological manipulation of GCK activity at locations distal from the allosteric activator site is possible.
Keywords: Diabetes, Enzyme Kinetics, Enzyme Mutation, Enzyme Structure, Glucokinase, Glucose Metabolism, Enzyme Activation, Hyper/Hypoglycemia Genetics, Monogenic Disease, Point Mutation
Introduction
The global rise in the prevalence of type 2 diabetes (T2D)2 not only reflects worldwide changes in lifestyle, behavior, and diet but perhaps also suggests that our current therapeutic strategies against this disease are ineffective (1). This leaves an unmet medical need for treatments that are more effective at lowering glycemia with sustained efficacy (2–4). One drug target that has generated much interest over the last decade is glucokinase (GCK). Mutations within GCK have long been established as a cause of monogenic disorders of aberrant glucose homeostasis (5). This includes a spectrum of clinical phenotypes ranging from hyperinsulinemic hypoglycemia (HH) with heterozygous activating mutations (5–15) to maturity onset diabetes of the young (MODY) (5, 16, 17) and permanent neonatal diabetes mellitus (5, 18–22) with heterozygous and homozygous/compound heterozygous inactivating mutations, respectively. The association of common variation within this gene with both fasting plasma glucose (fpg) levels (23, 24) and T2D risk (25) also highlights the significance of this enzyme in maintaining normoglycemia. Additionally, the expression of GCK in both the pancreatic beta cell and liver hepatocyte (26, 27) provides the promise of a two-pronged attack on hyperglycemia with the opportunity of not only lowering the threshold for glucose-stimulated insulin secretion (GSIS) but also increasing hepatic glucose uptake, firmly establishing GCK as an important therapeutic target (2, 4). Several small molecular GCK activators (GKAs) have been patented and are in clinical trials (2–4, 28–39) with the results of one study showing a dose-dependent reduction in glycemia following administration to patients with T2D (30).
The elucidation of the GCK crystal structure has allowed us to study naturally occurring mutations in much greater molecular detail (40). Over 600 inactivating GCK mutations have been identified to date that are located throughout the enzyme, whereas only 13 naturally occurring activating substitutions are reported, the majority of which cluster at a discrete structural location deemed the allosteric activator site (5, 6, 12). Interestingly, this is also the site at which pharmacological GKAs bind (40). However, on greater scrutiny, the location of these activating mutations can be categorized in further detail. Group 1 includes residues that make direct contact with GKAs, group 2 encompasses residues that lie within the allosteric activator site but that do not contact the GKA, and group 3 includes residues greater than 15 Å from the allosteric activator site (40, 41). Importantly to date, only one GCK-HH mutation has been identified that falls within group 3: M197I (14, 40, 41).
GCK possesses certain biochemical properties that allow it to act as a glucose sensor in the pancreatic beta cell where its activity is coupled to GSIS. First, it has a low affinity for its substrate glucose (half-maximal activity at 7.5 mm (deemed the “S0.5 value”)) compared with other hexokinases, which become saturated well below physiological levels of this sugar. Additionally, lack of product inhibition by glucose 6-phosphate and cooperativity with respect to glucose (Hill number (nH), 1.7) despite functioning as a monomer mean that GCK is uniquely placed to fulfill its critical role in coupling fpg levels to insulin secretion (26, 27).
In this study, we report two novel GCK-HH mutations: V389L (c.1165C→G, p.Val-389 → Leu) and T103S (c.308C→G, p.Thr-103 → Ser). Importantly, these mutations occur at residues where GCK-MODY (formerly known as MODY-2) mutations V389D (c.1166T→A, p.Val-389 → Asp) and T103I (c.308C→T, p.Thr-103 → Ile), respectively, have also been reported (5), suggesting that these positions are of great regulatory importance (as has been shown with previous studies investigating contiguous mutations causing opposite phenotypes (6)). Therefore, we aimed to characterize all four naturally occurring mutations to further understand the mutational mechanisms behind these substitutions and learn more about these critical sites where the balance of enzyme activity can be “tipped” in opposite directions. We hypothesized that the structural location and physiochemical properties of the substituted residue would influence function and the corresponding clinical phenotype observed.
EXPERIMENTAL PROCEDURES
Patients
The male proband of family 1 is Caucasian and from the United Kingdom and was born by caesarean section at 38 weeks gestation weighing 5.4 kg. There are postnatal records of low capillary blood glucose levels, but no action was taken. He presented with seizures at 2 years of age and was diagnosed at 3.6 years with hyperinsulinemia (39.8 pm) and hypoglycemia (fpg, 2.9 mm). Following diagnosis, the proband was treated with diazoxide (5 mg/kg/day divided into three doses) and chlorothiazide (7 mg/kg/day divided into two doses). Within 3 days, diazoxide had to be increased to 7.5 and then 10 mg/kg/day to adequately control his blood glucose levels. The patient was then discharged from the hospital a week after his initial diagnosis with stable sugars. However, over the following month, the dose of diazoxide was increased further to 15 mg/kg/day. At the age of 5.2 years, the diazoxide dose per kg was reduced to 10 mg/kg/day but within a month had to be increased back up to 14 and then 15 mg/kg/day in response to periods of increased frequency of (typically morning) hypoglycemic episodes with symptoms (not seizures). Chlorothiazide was reduced to 3.5 mg/kg/day a fortnight after initial diagnosis, although it was quickly raised back up to 7 mg/kg/day a week later. At the age of 4.3 years, the chlorothiazide dose was gradually reduced and has now been discontinued. The proband now only suffers a handful of fasting hypoglycemic episodes per month (consumption of carbohydrate supplement restores normoglycemia promptly). Unfortunately, the proband has marked hypertrichosis as a side effect of the diazoxide treatment. The proband's father also carries the mutation. Following long periods without food, he feels “lightheaded” and “weak” and has capillary glucose levels of ∼2.9 mm.
The female proband of family 2 is part Caucasian, part Metis and from Canada. She was born at 38 weeks gestation weighing 3.2 kg. There was no history of neonatal hypoglycemia, but in retrospect, she had an episode of symptomatic hypoglycemia at 15 years after a long hike. She presented at 22 years with frequent episodes of fatigue, weakness, and unexplained sweating, which could be prevented by frequent snacks in addition to her regular meals (body mass index of 22 kg/m2), and was subsequently diagnosed with hyperinsulinemia (36.1 pm) and hypoglycemia (2.8 mm). Triglyceride levels were in the normal range (1.38 mm). Investigations including endoscopic ultrasound, computed tomography, and MRI of abdomen and an exploratory laparotomy were negative for an insulioma. While the proband was in the hospital, her 27-year-old sister was found to have hypoglycemia, and subsequent investigations confirmed hyperinsulinemia (143.1 pm) with hypoglycemia (2.5 mm). She was born at 42 weeks gestation weighing 4.1 kg. With both sisters having symptomatic hypoglycemia when glucose readings are lower than 3 mm, they are both currently managed with frequent small meals, 50–75 mg of diazoxide twice daily, and 12.5–25 mg of hydrochlorothiazide once a day. Screening of the family revealed that the proband inherited the mutation from her 56-year-old father who also has two affected male siblings, and her 78-year-old paternal grandmother who was diagnosed with T2D at age 75. The father's fasting glucose values are between 3.1 and 4.0 mm with corresponding insulin levels of 51.4 and 56.3 pm. Although he does not have symptomatic fasting hypoglycemia, he does have occasional hypoglycemic symptoms postexercise. All subjects gave informed consent.
Screening of GCK Gene and Identification of Mutations
Genomic DNA was extracted from peripheral leukocytes. The 10 exons of the pancreatic beta cell isoform of the GCK gene were amplified by PCR (primer sequences are available upon request). Unidirectional sequencing was performed using universal M13 primers and Big Dye Terminator Cycler Sequencing kit v3.1 (Applied Biosystems) according to the manufacturer's instructions. Reactions were analyzed on an ABI 3730 capillary sequencer (Applied Biosystems), and sequences were compared with the reference sequence (NM_000162) using Mutation Surveyor v3.24 (SoftGenetics). Mutation testing was undertaken in family members to establish co-segregation and in 400 normal chromosomes to establish mutation frequency.
Protein Extraction
Two preparations of each mutant glutathione S-transferase-tagged human pancreatic glucokinase were prepared as described previously following heterologous expression in Escherichia coli (42). Extracted enzyme purities were determined using the Agilent 230 Protein kit (Agilent Technologies UK Ltd.) with a cutoff for purity of ≥95%. Protein concentration was calculated using the Bio-Rad Bradford reagent assay (BioRad Laboratories, Ltd.). Recombinant FLAG-tagged human glucokinase regulatory protein (GKRP; gene name, GCKR) was also prepared using previously described protocols (43, 44).
Glucose-dependent Kinetic Assays
GCK activity was determined spectrophotometrically using glucose-6-phosphate dehydrogenase-linked assays as described previously (42). In these assays, GCK activity was observed at 30 °C, pH 7.4 over a 0–100 mm glucose concentration range with ATP in excess. As the mutants V389D, V389R, and V389P had greatly reduced affinity for glucose versus wild-type enzyme, the glucose S0.5 was instead determined from assays using 0–600 mm glucose.
Briefly, the reaction mixture was prepared on ice and contained 7.5 mm MgCl2, 187.5 mm KCl, 125 mm HEPES buffer, 1.25 mm NADP+, 6.25 mm ATP, 2.5 mm DTT, 0.13% BSA, and 66 units of glucose-6-phosphate dehydrogenase. The activity of 10 milliunits of GCK was observed over the following glucose concentration range for 5 min: 100.00, 50.00, 25.00, 12.50, 6.25, 3.13, 1.56, 0.78, 0.39, 0.20, 0.10, and 0.00 mm. One milliunit of GCK was defined as that which converted 1 nmol of substrate/min.
ATP-dependent Assays
These assays were conducted as for the glucose-dependent assays, although GCK activity was instead measured over a 0–5 mm ATP concentration range with glucose in excess. The altered reaction mixture contained 18.75 mm glucose and no ATP. Ten milliunits of GCK were assayed in the presence of 5.00, 4.00, 3.00, 2.50, 2.00, 1.50, 1.00, 0.75, 0.50, 0.25, 0.10, and 0.00 mm ATP.
Glucose S0.5, nH, and turnover number (Kcat) values were calculated using the Hill equation, whereas the affinity for ATP (ATPKm) was derived from the Michaelis-Menten equation. All data fits were performed using Kaleidagraph v3.5 (Synergy Software).
GKRP Competitive Inhibition Assays
GKRP-mediated inhibition of GCK activity was determined spectrophotometrically using glucose-6-phosphate dehydrogenase-linked assays based on published protocols (43, 44). The assays were carried out at 5 mm glucose, 37 °C, pH 7.1.
The number of GKRP units used per assay was 0.5, 1.0, and 1.5 with 1 GKRP unit inhibiting 10 milliunits of GCK by 50% under standard assay conditions. The reaction mixture contained 25 mm HEPES buffer, 0.5 mm NADP+, 1 mm ATP, 2 mm MgCl2, 25 mm KCl, 1 mm DTT, 3.8 μm BSA, and 4 units of glucose-6-phosphate dehydrogenase. The reaction rate was recorded over 5 min as before. Data were presented as “percentage of GCK activity,” which was normalized to that obtained with no GKRP, and IC50 values (half-maximal inhibitory concentration) were also calculated from these data.
GKA-mediated Activation Assays
These assays were performed as described previously in the presence of 10 milliunits of GCK and saturating GKA (0–60 μm) and glucose (0–100 mm) concentrations (45). The reaction mixture was the same as for the glucose-dependent assays but also contained RO0281675 (resuspended in DMSO) at the following concentrations: 60.00, 27.00, 9.00, 3.00, 1.00, 0.30, and 0.00 μm. Data were expressed as “-fold activation,” which described the ratio Kcat/S0.5 determined in the presence and absence of saturating levels of RO0281675 (60 μm). The GKA half-maximal effect concentration (EC50) was also calculated and described the drug concentration that gave GCK activity 50% between base-line and the maximal responses.
Calculation of Relative Activity Index (RAI) and GSIS Threshold
RAI values allow the direct comparison of mutants with wild-type enzyme and take into account all GCK intrinsic kinetic parameters (S0.5, ATPKm, nH, and Kcat). The relationship between the RAI for a heterozygous mutant and the GSIS threshold can also be calculated mathematically. Details of all equations used have been published previously (7, 46, 47).
Structural Analysis
Crystal structures for the closed and super-open conformations of GCK (40) were obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank. Molecular modeling was carried out using PyMOL v0.99 (Schrödinger, LLC).
RESULTS
Novel GCK Mutations in Two Families with HH
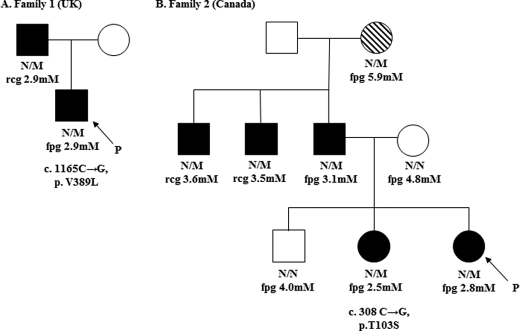
Fig. 1, A and B, show the pedigrees for the HH probands identified in the UK and Canada, respectively. Direct sequencing of GCK in the proband of family 1 identified a novel heterozygous mutation, V389L (c.1165C→G, p.Val-389 → Leu). Family studies established that the mutation was inherited from the proband's affected father.
FIGURE 1.
Pedigrees for HH families 1 and 2 (A and B, respectively). Males are shown as squares, and females are shown as circles. Filled symbols show individuals with hypoglycemia, open symbols show normoglycemic individuals, and hatched symbols show individuals with T2D. Fpg or random capillary glucose (rcg) levels and mutation status for each tested individual are shown under the symbols (N/M denotes family members heterozygous for mutation, and N/N denotes those who do not carry the mutation). P denotes the probands.
The proband from family 2 was also heterozygous for a novel mutation, T103S (c.308C→G, p.Thr-103 → Ser). Family studies established that the mutation was also present in the affected sister, father, two paternal uncles, and paternal grandmother, with all but the latter exhibiting fasting hypoglycemia. The paternal grandmother was diagnosed with T2D at age 75 years, although notably she is of Metis descent (a First Nation Canadian population at high risk of developing diabetes (48)).
Neither variant was identified in >400 normal chromosomes. Mutations in other known HH genes (ABCC8, KCNJ11, and HNF4A) were also excluded.
Kinetic Analyses of Recombinant Human Wild-type and Mutant GCKs
Table 1 shows the kinetic characteristics of wild-type, V389L, V389D, T103S, and T103I GCKs as determined using in vitro glucose-6-phosphate dehydrogenase-linked assays. Both the V389L and T103S HH mutations have an increased RAI when compared with wild type (6.0 and 8.4, respectively, versus 1.0). This was largely driven by an increased affinity for glucose demonstrated by reduced S0.5 values (3.5 ± 0.1 and 3.3 ± 0.1 mm, respectively, versus 7.5 ± 0.1 mm). The ATPKm, nH, and Kcat values for these HH mutations were similar to those obtained with wild-type enzyme. Conversely, the MODY mutations V389D and T103I had greatly reduced RAI values (both <0.1 versus 1.0 for wild type). Again, this effect was largely driven by a decreased affinity for glucose (S0.5, 78.6 ± 4.5 mm for V389D and 24.9 ± 0.6 mm for T103I). However, with the V389D mutant, there was also a reduced affinity for the second substrate ATP when compared with wild type (ATPKm, 1.5 ± 0.1 versus 0.4 ± <0.1 mm) and turnover number (Kcat, 10.3 ± 1.1 versus 62.9 ± 1.2 s−1). It was also of note that the yield of V389D recombinant protein was greatly reduced compared with the other enzymes (only 0.9 ± 0.3 mg).
TABLE 1.
Kinetic data for wild-type and four naturally occurring mutant GCKs
Data are shown as means ± S.E. for n = 18 experiments and are representative of two preparations of wild-type and each mutant protein.
| GCK | S0.5 | nH | ATPKm | Kcat | RAI | GSIS threshold | Yield | Activity/mg |
|---|---|---|---|---|---|---|---|---|
| mm | mm | s−1 | mm | mg | ||||
| Wild type | 7.5 ± 0.1 | 1.6 ± <0.1 | 0.4 ± <0.1 | 62.9 ± 1.2 | 1.0 | 5.0 | 17.7 ± 0.7 | 6.6 ± 0.5 |
| V389L | 3.5 ± 0.1 | 1.6 ± <0.1 | 0.5 ± <0.1 | 67.8 ± 1.6 | 6.0 | 2.9 | 20.3 ± 1.8 | 11.3 ± 0.8 |
| V389D | 78.6 ± 4.5 | 1.5 ± <0.1 | 1.5 ± 0.1 | 10.3 ± 1.1 | <0.1 | 7.1 | 0.9 ± 0.3 | 0.16 ± 0.1 |
| T103S | 3.3 ± 0.1 | 1.6 ± <0.1 | 0.4 ± <0.1 | 59.2 ± 1.7 | 8.4 | 2.9 | 9.5 ± <0.1 | 11.4 ± 2.2 |
| T103I | 24.9 ± 0.6 | 1.7 ± <0.1 | 0.6 ± <0.1 | 68.1 ± 0.1 | <0.1 | 6.7 | 12.5 ± 0.5 | 11.2 ± 0.5 |
Table 2 shows the results of kinetic assays characterizing a range of amino acid substitutions designed to further probe the physiochemical requirements of positions 389 and 103. There is a clear preference for hydrophobic residues at position 389 with hydrophilic substitutions having a deleterious effect on protein function. Substitutions at residue 103 result in a spectrum of effects on enzyme function with activating, near-normal, and inactivating kinetics.
TABLE 2.
Kinetic data for wild type and a range of amino acid substitutions at positions 103 and 389
Data are shown as mean ± S.E. for n = 6 experiments and are representative of two preparations of wild-type protein and one preparation of each mutant protein.
| GCK | Hydro- | S0.5 | nH | ATPKm | Kcat | RAI | GSIS threshold |
|---|---|---|---|---|---|---|---|
| mm | mm | s−1 | mm | ||||
| Wild type | Phobe | 7.5 ± 0.1 | 1.6 ± <0.1 | 0.4 ± <0.1 | 62.9 ± 1.2 | 1.0 | 5.0 |
| V389L | Phobe | 3.5 ± 0.1 | 1.6 ± <0.1 | 0.5 ± <0.1 | 67.8 ± 1.6 | 6.0 | 2.9 |
| V389C | Phobe | 8.3 ± 0.3 | 1.5 ± <0.1 | 0.5 ± <0.1 | 58.5 ± 2.4 | 0.8 | 5.0 |
| V389S | Phobe | 23.9 ± 2.5 | 1.3 ± <0.1 | 0.7 ± <0.1 | 39.9 ± 2.1 | <0.1 | 6.5 |
| V389P | Phobe | 39.4 ± 0.6 | 1.8 ± <0.1 | 0.6 ± <0.1 | 31.3 ± 0.3 | <0.1 | 6.8 |
| V389D | Phile | 78.6 ± 4.5 | 1.5 ± <0.1 | 1.5 ± 0.1 | 10.3 ± 1.1 | <0.1 | 7.1 |
| V389R | Phile | 68.8 ± 0.8 | 1.6 ± <0.1 | 0.5 ± <0.1 | 42.3 ± 0.4 | <0.1 | 6.8 |
| T103S | Phobe | 3.3 ± 0.1 | 1.6 ± <0.1 | 0.4 ± <0.1 | 59.2 ± 1.7 | 8.4 | 2.9 |
| T103P | Phobe | 6.0 ± 0.1 | 1.9 ± <0.1 | 0.6 ± <0.1 | 51.9 ± 0.7 | 1.2 | 4.7 |
| T103Y | Phobe | 9.1 ± 1.0 | 1.3 ± 0.1 | 0.4 ± <0.1 | 39.0 ± 0.9 | 0.9 | 5.5 |
| T103C | Phobe | 11.2 ± 0.2 | 1.5 ± <0.1 | 0.5 ± <0.1 | 52.3 ± 1.4 | 0.3 | 5.7 |
| T103I | Phobe | 24.9 ± 0.6 | 1.7 ± <0.1 | 0.6 ± <0.1 | 68.1 ± 0.1 | <0.1 | 6.7 |
| T103N | Phile | 5.8 ± 0.1 | 1.6 ± <0.1 | 0.5 ± <0.1 | 51.5 ± 1.1 | 1.2 | 4.5 |
| T103K | Phile | 7.5 ± 0.3 | 1.3 ± <0.1 | 0.4 ± <0.1 | 25.6 ± 0.7 | 0.9 | 5.6 |
In Silico Calculation of GSIS Thresholds
Table 1 shows the calculated GSIS thresholds for patients with the four individual mutations. Both HH mutations have a predicted GSIS threshold of 2.9 mm, which accurately mirrors the fpg levels of 2.9 and 2.8 mm observed in the V389L and T103S probands, respectively. Calculated GSIS thresholds of 7.1 and 6.7 mm for the V389D and T103I mutations, respectively, are in line with the expected fpg levels in patients with MODY mutations (∼5.5–8.0 mm) (5). The severely deleterious effect of the V389D mutation on enzyme function is not reflected in the predicted fpg level due to compensation by the wild-type allele (49).
Response to Physiological Hepatic Inhibitor GKRP
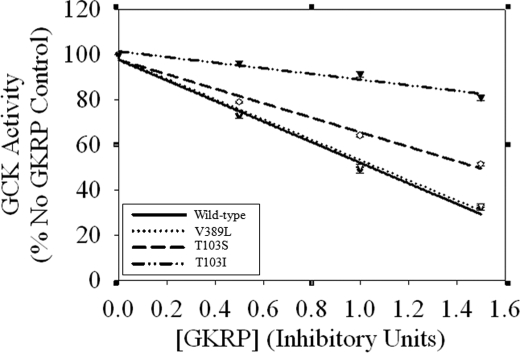
Fig. 2 shows the results of inhibition assays where 10 milliunits of GCK (wild type, V389L, T103S, and T103I) were incubated with 0.5, 1.0, and 1.5 units of GKRP. V389D was not included in these experiments as its activity per mg was so low that to see measurable inhibition by regulatory protein inappropriately large quantities of the enzyme would be needed. As expected, the IC50 value for wild type was 1.1 ± 0.1 units of GKRP; the HH mutant V389L yielded identical results. GKRP was less able to inhibit mutant T103S and T103I activity with IC50 values of 1.5 ± <0.1 and 4.7 ± 0.5 GKRP units, respectively.
FIGURE 2.
Wild-type, V389L, T103S, and T103I GCKs incubated with 0.5, 1.0, and 1.5 units of GKRP. GCK activity was normalized to a control in which the reaction conditions were identical, but GKRP was absent. IC50 values were calculated using these data and defined as the half-maximal inhibitory concentration of GKRP in each experiment. Data are shown as means ± S.E. for n = 6 experiments and are representative of two preparations of wild-type and each mutant protein.
Activation by Small Molecular GKA RO0281675
Table 3 shows -fold activation of each enzyme in terms of Kcat/S0.5 in the presence of 60 μm GKA. The MODY mutation T103I showed the greatest -fold increase in activity, roughly 6 times that of the wild-type enzyme (90.9 ± 4.0 versus 14.4 ± 0.3, respectively). The V389D MODY mutation also demonstrated a high -fold increase in activity of 33.9 ± 2.1. Unsurprisingly, both HH mutations were activated to a lesser degree with -fold increases in activity of 5.7 ± 0.5 for T103S and 7.4 ± 1.3 for V389L.
TABLE 3.
Data obtained from assaying wild-type and the mutant GCKs in presence of 60 μm RO0281675
-Fold activation was calculated by dividing Kcat by the S0.5 value at this drug concentration, and EC50 is the concentration of drug that gives a GCK activity 50% between base-line and the maximal responses. Data are shown as mean ± S.E. for n = 6 experiments and are representative of two preparations of wild-type and each mutant protein.
| GCK | -Fold activation (Kcat/S0.5) | EC50 |
|---|---|---|
| μm | ||
| Wild type | 14.4 ± 0.3 | 5.8 ± 1.9 |
| V389L | 7.4 ± 1.3 | 3.9 ± 0.1 |
| V389D | 33.9 ± 2.1 | 34.6 ± <0.1 |
| T103S | 5.7 ± 0.5 | 2.6 ± 1.3 |
| T103I | 90.9 ± 4.0 | 12.8 ± 4.2 |
Structural Modeling: Thr-103 but Not Val-389 Maps to Allosteric Activator Site
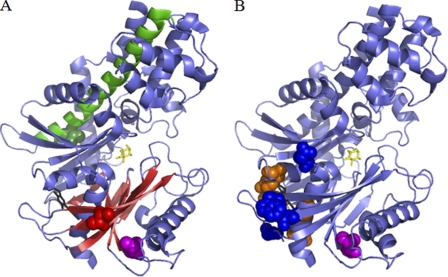
Fig. 3A shows the location of residues Thr-103 and Val-389 within the closed conformation of human GCK (40). Thr-103 lies within the allosteric activator site, specifically within β-strand 4 of the small domain, a region where the majority of other activating mutations are reported (Fig. 3B). However, this residue does not form direct contacts with bound GKAs, and so T103S can be described as a group 2 activating mutation (41). Surprisingly, residue Val-389 is located far from the allosteric activator site in α-helix 11 of the large domain. Not only is this distal from the site of GKA binding, but it is also far from the only other group 3 activating mutation, M197I (14, 41), highlighting a novel regulatory region within the enzyme.
FIGURE 3.
A shows the positions of residues Thr-103 (red) and Val-389 (green) within the crystal structure of closed human GCK (40). Thr-103 maps to the allosteric activator site close to where GKAs (black) bind, specifically within β-strand 4 of the small domain β-sheet (red). Val-389 (green) is both distal from this site and residue Met-197 (magenta) (14) and instead maps to α-helix 11 (green) within the large domain. B shows the position of all other naturally occurring activating mutations reported. Group 1 mutations (those in the allosteric activator site making direct contact with GKA) are shown in orange, group 2 mutations (those in the allosteric activator site but not contacting GKA) are in blue, and group 3 mutations (residues greater than 15 Å from the allosteric activator site) are in magenta (41). The binding sites of the GKA “compound A” (black) and glucose (yellow) are also shown in both figures (40).
DISCUSSION
The current unmet medical need for more effective T2D treatments capable of sustainably lowering glycemia to desirable levels means the quest for novel therapeutic targets is both a timely and important one (2–4). The fundamental role of GCK in glucose homeostasis (specifically within the beta cell and liver) (26, 27) makes it an interesting and well established drug target. Our functional characterization of two novel GCK-HH activating mutations and the corresponding MODY mutations at the same amino acid residues provides further insight into important regulatory regions within GCK, more specifically sites where the functional balance can sway in either direction.
Intrinsic kinetic parameters for wild-type and the four mutant GCKs measured via spectrophotometric in vitro assays revealed that the increased activity index of both HH mutations (V389L and T103S) was largely driven by an increased affinity for glucose (Table 1), a result observed with previously reported naturally occurring GCK-HH mutations (5, 7, 9–11, 13–15). The reduced activity index of the GCK-MODY mutant T103I followed this theme; however, the substitution of an aspartate for a valine at residue 389 (V389D) had disastrous consequences with regard to enzyme function with affinity for the two substrates glucose and ATP decreased and a substantially reduced overall rate of catalysis. This deleterious effect on GCK function was also mirrored by the protein yield in both recombinant V389D protein preparations. This suggests that not only does V389D directly alter GCK activity, but protein stability as a whole may be reduced. The result of these kinetic characterizations provides a potential explanation for the pathogenesis behind these four mutations as they correspond to both an elevated or diminished capacity to bind and turn over glucose (V389L/T103S and V389D/T103I, respectively).
As GCK activity is fundamental to glucose metabolism in both the beta cell and hepatocyte (26, 27), it is subject to a number of different transcriptional (50) and post-translational regulatory mechanisms (5, 51–58). Therefore, the pathogenic mechanism behind a GCK mutation may not just lie at the level of altered enzyme kinetics. This has been shown with mutations that co-segregate with hyperglycemia in MODY families but whose kinetics are near-normal or even mildly activating. Two such examples are V62M (5, 45, 59) and G72R (5, 59, 60). Kinetic analysis revealed both mutations to be mildly activating and to have only marginal thermolability versus wild-type enzyme and the previously reported instability mutant E300K (5, 47, 61, 62). Further investigation revealed that both mutants exhibited defective binding of the endogenous hepatic inhibitor GKRP and a small molecular GKA (5, 34, 45, 59). As well as its direct inhibitory role, GKRP is thought to paradoxically bind and maintain a “reserve pool” of GCK in the hepatocyte nucleus that may be released rapidly upon postprandial increases in glucose (63). Additionally, small molecular GKAs are thought to bind at the same allosteric site as an as yet unidentified endogenous GCK regulator within the beta cell (34). Therefore, it was postulated that loss of these interactions upon GCK mutation could have physiologically pathogenic consequences, a result that was confirmed in Min6 cells where both the V62M and G72R mutants showed reduced catalytic and protein stability compared with wild-type enzyme (59). In vitro assays studying the interaction of GCK mutants with both GKRP and GKAs can therefore provide insight into whether the pathogenic mechanism behind a particular mutation lies at levels in addition to directly altered kinetics. Studies performed by Heredia et al. (64) investigated the regulation of a number of naturally occurring GCK activating mutants by GKRP. Their data showed differences in GKRP binding affinity between the mutants that was proposed to reflect the mutational mechanism. For example, mutations that enhanced the interaction between glucose and the apoenzyme had the lowest GKRP affinity (as these substitutions stabilized a more compact structure in which the GKRP-binding site was altered), whereas those that enhanced the glucose-bound GCK conformational change demonstrated only slightly reduced GKRP affinity versus wild-type enzyme (64).
The results of the GKRP inhibition assays performed in this study (Fig. 2) show that GKRP was able to inhibit wild-type GCK and the V389L mutant equally. However, GKRP-mediated inhibition of both Thr-103 mutants was less effective compared with the wild-type enzyme with the binding to mutant T103I affected the most. This is consistent with data suggesting that the interaction of GKRP with GCK occurs at leucine-asparagine motifs within the hinge region of the enzyme (65), a site that maps next to the allosteric activator site (within which Thr-103 is found). Additionally, these data suggest that the T103S activating mutation may directly increase glucose binding to the apoenzyme, whereas V389L may enhance the conformational change from the inactive to active structure once glucose is bound (64). As GKRP is specific to the liver and not expressed within the beta cell (43, 66, 67), it is difficult to postulate the precise consequence of a reduced GKRP binding affinity on overall GCK activity, particularly as the ability to bind this inhibitor contributes to but is not a determinant of clinical phenotype (5, 14, 45, 59, 60, 64). As GKRP binds T103S less effectively, it could be assumed that there is less inhibition of GCK activity in the liver, thus leading to an increase in hepatocyte glycolytic flux (43, 68), which would contribute to the hypoglycemia of this HH patient. Equally, the greatly decreased GKRP affinity observed with T103I may mean that there are markedly reduced hepatic nuclear reserves of this mutant enzyme, consequently hindering postprandial glucose clearance by the liver (63), contributing to the hyperglycemia in this MODY patient.
Table 3 shows the results of the GKA RO0281675-mediated stimulation of wild-type and the four GCK mutants. Interestingly, all four mutants responded to the drug, suggesting that not even the particularly deleterious V389D substitution resulted in an enzyme that could not be converted into the active conformation. The MODY mutants V389D and T103I showed the greatest -fold increase in activity. This is perhaps a reflection of the reduced activity of the MODY mutants prior to stimulation, a hypothesis supported by both HH mutants in this study that showed the smallest -fold increases in activity and also by the previous findings of Grimsby and Grippo (10, 69) who demonstrated that the V455M GCK activating mutant was resistant to GKA-mediated stimulation. The much greater response of the inactivating T103I mutant versus that of V389D may be explained by the physiochemical properties of these substituted residues. Both threonine and isoleucine are hydrophobic, meaning it may be easier to “force” GCK into the closed active structure using small molecular activators, whereas the V389D mutant has a hydrophilic residue (aspartate) being pushed into a hydrophobic pocket normally occupied by valine. This is also reflected in V389D having the highest EC50 value of the four mutants.
Molecular modeling of these mutations within the GCK crystal structure (40) allows the mechanisms behind their pathogenesis to be further probed. Kamata et al. (40) proposed a three-GCK conformation model that explained both the sigmoidal binding curve and cooperativity with respect to glucose. The transition between functional states requires a global conformational change and the breaking and reforming of numerous intramolecular bonds (40), both of which are easily affected by the physiochemical properties of substituted residues at sites critical to function.
Val-389 is situated near the C-terminal end of α-helix 11. It is surrounded by hydrophobic residues Leu-386, Ile-390, Met-393, Met-41, Tyr-234, Trp-257, and Ala-232. The observation that certain amino acid substitutions at residue 389 may increase GCK activity identifies this structural position and α-helix 11 as a novel site regulating GCK function, one that is distal from both the allosteric activator site and the only other reported naturally occurring group 3 activating mutation, M197I (14, 40, 41). Residue 389 lies within a solvent-excluded region of the molecule, a location suited by hydrophobic valine and the even more hydrophobic leucine but not by hydrophilic aspartate. Table 2 shows the kinetic parameters measured for a series of mutations at position 389. These data showed that all hydrophilic substitutions drastically reduced GCK activity, whereas all hydrophobic replacements except serine gave activating or near-normal kinetics (except the substitution of proline (P), which may break this α-helix). This finding is similar to that observed with the other group 3 activating mutant, M197I. This residue is also located within a hydrophobic pocket where the introduction of hydrophilic residues is deleterious to GCK function (14).
The mutational mechanisms behind the substitutions at residue 103 are much harder to explain than those at residue 389. Thr-103 is a surface residue; however, it is unclear how a conservative mutation such as the replacement of a threonine with a serine would affect GCK activity. Mutating the threonine to an aliphatic isoleucine puts a non-polar hydrophobic residue in a solvent-exposed environment, which may account for the inactivating kinetics of T103I. A series of amino acid substitutions were carried out at this position to see how GCK activity was altered. Table 2 shows these mutants to demonstrate a spectrum of kinetic properties. T103S was the only activating mutation observed. The mutants T103N, T103K, T103P, and T103Y all demonstrate near-normal kinetics, meaning that either (i) these mutations would not be pathogenic or (ii) their mutational mechanism would not lie at the level of altered glucose catalysis (59). Importantly, only one of these mutations (T103N) has been identified in a GCK-MODY patient (70). Finally, the mutants T103I and T103C had a reduced activity index, reflecting the potential intolerance of an aliphatic side chain in this solvent-exposed region.
In summary, the newly identified importance of α-helix 11 suggests that further activating mutations are likely to exist and that they may not necessarily map to the allosteric activator site. Our results build upon earlier work by Pal and Miller (41) who preemptively identified further activating mutations by genetic screening as well as that by Sayed et al.(14) who characterized the first activating mutation outside of the allosteric activator site. This study highlights the importance of the continued study of naturally occurring GCK activating mutations as detailed kinetic analysis at these residues may enable the fine tuning of GKAs for the treatment of diabetes.
Acknowledgment
We are grateful to Dr. Mars Skae for the clinical details on the UK patient.
This work was supported in part in Oxford by the Medical Research Council (MRC).
- T2D
- type 2 diabetes
- GCK
- glucokinase
- HH
- hyperinsulinemic hypoglycemia
- MODY
- maturity onset diabetes of the young
- fpg
- fasting plasma glucose
- GSIS
- glucose-stimulated insulin secretion
- GKA
- small molecular glucokinase activator
- S0.5
- glucose concentration needed for half-maximal GCK activity
- nH
- Hill number (measure of cooperativity)
- ATPKm
- concentration of ATP needed for half-maximal enzyme activity
- Kcat
- turnover number
- EC50
- GKA half-maximal effect concentration
- RAI
- relative activity index
- GKRP
- glucokinase regulatory protein.
REFERENCES
- 1. Zimmet P., Alberti K. G., Shaw J. (2001) Nature 414, 782–787 [DOI] [PubMed] [Google Scholar]
- 2. Coghlan M., Leighton B. (2008) Expert Opin. Investig. Drugs 17, 145–167 [DOI] [PubMed] [Google Scholar]
- 3. Matschinsky F. M. (2009) Nat. Rev. Drug Discov. 8, 399–416 [DOI] [PubMed] [Google Scholar]
- 4. Matschinsky F. M., Porte D. (2010) F1000 Med. Rep. 2, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osbak K. K., Colclough K., Saint-Martin C., Beer N. L., Bellanné-Chantelot C., Ellard S., Gloyn A. L. (2009) Hum. Mutat. 30, 1512–1526 [DOI] [PubMed] [Google Scholar]
- 6. Barbetti F., Cobo-Vuilleumier N., Dionisi-Vici C., Toni S., Ciampalini P., Massa O., Rodriguez-Bada P., Colombo C., Lenzi L., Garcia-Gimeno M. A., Bermudez-Silva F. J., Rodriguez de Fonseca F., Banin P., Aledo J. C., Baixeras E., Sanz P., Cuesta-Muñoz A. L. (2009) Mol. Endocrinol. 23, 1983–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christesen H. B., Jacobsen B. B., Odili S., Buettger C., Cuesta-Munoz A., Hansen T., Brusgaard K., Massa O., Magnuson M. A., Shiota C., Matschinsky F. M., Barbetti F. (2002) Diabetes 51, 1240–1246 [DOI] [PubMed] [Google Scholar]
- 8. Christesen H. B., Tribble N. D., Molven A., Siddiqui J., Sandal T., Brusgaard K., Ellard S., Njølstad P. R., Alm J., Brock Jacobsen B., Hussain K., Gloyn A. L. (2008) Eur. J. Endocrinol. 159, 27–34 [DOI] [PubMed] [Google Scholar]
- 9. Cuesta-Muñoz A. L., Huopio H., Otonkoski T., Gomez-Zumaquero J. M., Näntö-Salonen K., Rahier J., López-Enriquez S., García-Gimeno M. A., Sanz P., Soriguer F. C., Laakso M. (2004) Diabetes 53, 2164–2168 [DOI] [PubMed] [Google Scholar]
- 10. Glaser B., Kesavan P., Heyman M., Davis E., Cuesta A., Buchs A., Stanley C. A., Thornton P. S., Permutt M. A., Matschinsky F. M., Herold K. C. (1998) N. Engl. J. Med. 338, 226–230 [DOI] [PubMed] [Google Scholar]
- 11. Gloyn A. L., Noordam K., Willemsen M. A., Ellard S., Lam W. W., Campbell I. W., Midgley P., Shiota C., Buettger C., Magnuson M. A., Matschinsky F. M., Hattersley A. T. (2003) Diabetes 52, 2433–2440 [DOI] [PubMed] [Google Scholar]
- 12. Kassem S., Bhandari S., Rodríguez-Bada P., Motaghedi R., Heyman M., García-Gimeno M. A., Cobo-Vuilleumier N., Sanz P., Maclaren N. K., Rahier J., Glaser B., Cuesta-Muñoz A. L. (2010) N. Engl. J. Med. 362, 1348–1350 [DOI] [PubMed] [Google Scholar]
- 13. Meissner T., Marquard J., Cobo-Vuilleumier N., Maringa M., Rodríguez-Bada P., García-Gimeno M. A., Baixeras E., Weber J., Olek K., Sanz P., Mayatepek E., Cuesta-Muñoz A. L. (2009) Horm. Metab. Res. 41, 320–326 [DOI] [PubMed] [Google Scholar]
- 14. Sayed S., Langdon D. R., Odili S., Chen P., Buettger C., Schiffman A. B., Suchi M., Taub R., Grimsby J., Matschinsky F. M., Stanley C. A. (2009) Diabetes 58, 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wabitsch M., Lahr G., Van de Bunt M., Marchant C., Lindner M., von Puttkamer J., Fenneberg A., Debatin K. M., Klein R., Ellard S., Clark A., Gloyn A. L. (2007) Diabet. Med. 24, 1393–1399 [DOI] [PubMed] [Google Scholar]
- 16. Froguel P., Vaxillaire M., Sun F., Velho G., Zouali H., Butel M. O., Lesage S., Vionnet N., Clément K., Fougerousse F., Tanizawa Y., Weissenbach J., Beckmann J. S., Lathrop G. M., Passa P. H., Permutt M. A., Cohen D. (1992) Nature 356, 162–164 [DOI] [PubMed] [Google Scholar]
- 17. Hattersley A. T., Turner R. C., Permutt M. A., Patel P., Tanizawa Y., Chiu K. C., O'Rahilly S., Watkins P. J., Wainscoat J. S. (1992) Lancet 339, 1307–1310 [DOI] [PubMed] [Google Scholar]
- 18. Njølstad P. R., Sagen J. V., Bjørkhaug L., Odili S., Shehadeh N., Bakry D., Sarici S. U., Alpay F., Molnes J., Molven A., Søvik O., Matschinsky F. M. (2003) Diabetes 52, 2854–2860 [DOI] [PubMed] [Google Scholar]
- 19. Njølstad P. R., Søvik O., Cuesta-Muñoz A., Bjørkhaug L., Massa O., Barbetti F., Undlien D. E., Shiota C., Magnuson M. A., Molven A., Matschinsky F. M., Bell G. I. (2001) N. Engl. J. Med. 344, 1588–1592 [DOI] [PubMed] [Google Scholar]
- 20. Porter J. R., Shaw N. J., Barrett T. G., Hattersley A. T., Ellard S., Gloyn A. L. (2005) J. Pediatr. 146, 131–133 [DOI] [PubMed] [Google Scholar]
- 21. Rubio-Cabezas O., Díaz González F., Aragonés A., Argente J., Campos-Barros A. (2008) Pediatr. Diabetes 9, 245–249 [DOI] [PubMed] [Google Scholar]
- 22. Turkkahraman D., Bircan I., Tribble N. D., Akçurin S., Ellard S., Gloyn A. L. (2008) J. Pediatr. 153, 122–126 [DOI] [PubMed] [Google Scholar]
- 23. Weedon M. N., Clark V. J., Qian Y., Ben-Shlomo Y., Timpson N., Ebrahim S., Lawlor D. A., Pembrey M. E., Ring S., Wilkin T. J., Voss L. D., Jeffery A. N., Metcalf B., Ferrucci L., Corsi A. M., Murray A., Melzer D., Knight B., Shields B., Smith G. D., Hattersley A. T., Di Rienzo A., Frayling T. M. (2006) Am. J. Hum. Genet. 79, 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weedon M. N., Frayling T. M., Shields B., Knight B., Turner T., Metcalf B. S., Voss L., Wilkin T. J., McCarthy A., Ben-Shlomo Y., Davey Smith G., Ring S., Jones R., Golding J., Byberg L., Mann V., Axelsson T., Syvänen A. C., Leon D., Hattersley A. T. (2005) Diabetes 54, 576–581 [DOI] [PubMed] [Google Scholar]
- 25. Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., Lindgren C. M., Mägi R., Morris A. P., Randall J., Johnson T., Elliott P., Rybin D., Thorleifsson G., Steinthorsdottir V., Henneman P., Grallert H., Dehghan A., Hottenga J. J., Franklin C. S., Navarro P., Song K., Goel A., Perry J. R., Egan J. M., Lajunen T., Grarup N., Sparsø T., Doney A., Voight B. F., Stringham H. M., Li M., Kanoni S., Shrader P., Cavalcanti-Proença C., Kumari M., Qi L., Timpson N. J., Gieger C., Zabena C., Rocheleau G., Ingelsson E., An P., O'Connell J., Luan J., Elliott A., McCarroll S. A., Payne F., Roccasecca R. M., Pattou F., Sethupathy P., Ardlie K., Ariyurek Y., Balkau B., Barter P., Beilby J. P., Ben-Shlomo Y., Benediktsson R., Bennett A. J., Bergmann S., Bochud M., Boerwinkle E., Bonnefond A., Bonnycastle L. L., Borch-Johnsen K., Böttcher Y., Brunner E., Bumpstead S. J., Charpentier G., Chen Y. D., Chines P., Clarke R., Coin L. J., Cooper M. N., Cornelis M., Crawford G., Crisponi L., Day I. N., de Geus E. J., Delplanque J., Dina C., Erdos M. R., Fedson A. C., Fischer-Rosinsky A., Forouhi N. G., Fox C. S., Frants R., Franzosi M. G., Galan P., Goodarzi M. O., Graessler J., Groves C. J., Grundy S., Gwilliam R., Gyllensten U., Hadjadj S., Hallmans G., Hammond N., Han X., Hartikainen A. L., Hassanali N., Hayward C., Heath S. C., Hercberg S., Herder C., Hicks A. A., Hillman D. R., Hingorani A. D., Hofman A., Hui J., Hung J., Isomaa B., Johnson P. R., Jørgensen T., Jula A., Kaakinen M., Kaprio J., Kesaniemi Y. A., Kivimaki M., Knight B., Koskinen S., Kovacs P., Kyvik K. O., Lathrop G. M., Lawlor D. A., Le Bacquer O., Lecoeur C., Li Y., Lyssenko V., Mahley R., Mangino M., Manning A. K., Martínez-Larrad M. T., McAteer J. B., McCulloch L. J., McPherson R., Meisinger C., Melzer D., Meyre D., Mitchell B. D., Morken M. A., Mukherjee S., Naitza S., Narisu N., Neville M. J., Oostra B. A., Orrù M., Pakyz R., Palmer C. N., Paolisso G., Pattaro C., Pearson D., Peden J. F., Pedersen N. L., Perola M., Pfeiffer A. F., Pichler I., Polasek O., Posthuma D., Potter S. C., Pouta A., Province M. A., Psaty B. M., Rathmann W., Rayner N. W., Rice K., Ripatti S., Rivadeneira F., Roden M., Rolandsson O., Sandbaek A., Sandhu M., Sanna S., Sayer A. A., Scheet P., Scott L. J., Seedorf U., Sharp S. J., Shields B., Sigurethsson G., Sijbrands E. J., Silveira A., Simpson L., Singleton A., Smith N. L., Sovio U., Swift A., Syddall H., Syvänen A. C., Tanaka T., Thorand B., Tichet J., Tönjes A., Tuomi T., Uitterlinden A. G., van Dijk K. W., van Hoek M., Varma D., Visvikis-Siest S., Vitart V., Vogelzangs N., Waeber G., Wagner P. J., Walley A., Walters G. B., Ward K. L., Watkins H., Weedon M. N., Wild S. H., Willemsen G., Witteman J. C., Yarnell J. W., Zeggini E., Zelenika D., Zethelius B., Zhai G., Zhao J. H., Zillikens M. C., Borecki I. B., Loos R. J., Meneton P., Magnusson P. K., Nathan D. M., Williams G. H., Hattersley A. T., Silander K., Salomaa V., Smith G. D., Bornstein S. R., Schwarz P., Spranger J., Karpe F., Shuldiner A. R., Cooper C., Dedoussis G. V., Serrano-Ríos M., Morris A. D., Lind L., Palmer L. J., Hu F. B., Franks P. W., Ebrahim S., Marmot M., Kao W. H., Pankow J. S., Sampson M. J., Kuusisto J., Laakso M., Hansen T., Pedersen O., Pramstaller P. P., Wichmann H. E., Illig T., Rudan I., Wright A. F., Stumvoll M., Campbell H., Wilson J. F., Bergman R. N., Buchanan T. A., Collins F. S., Mohlke K. L, Tuomilehto J., Valle T. T., Altshuler D., Rotter J. I., Siscovick D. S., Penninx B. W., Boomsma D. I., Deloukas P., Spector T. D., Frayling T. M., Ferrucci L., Kong A., Thorsteinsdottir U., Stefansson K., van Duijn C. M., Aulchenko Y. S., Cao A., Scuteri A., Schlessinger D., Uda M., Ruokonen A., Jarvelin M. R., Waterworth D. M., Vollenweider P., Peltonen L., Mooser V., Abecasis G. R., Wareham N. J., Sladek R., Froguel P., Watanabe R. M., Meigs J. B., Groop L., Boehnke M., McCarthy M. I., Florez J. C., Barroso I. (2010) Nat. Genet. 42, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matschinsky F. M. (1990) Diabetes 39, 647–652 [DOI] [PubMed] [Google Scholar]
- 27. Matschinsky F. M. (2002) Diabetes 51, Suppl. 3, S394–S404 [DOI] [PubMed] [Google Scholar]
- 28. McKerrecher D., Johnstone C., Caulkett P. W. R., James R., Jones C. D., Hargreaves R. B., Hayter B. R., Currie G. S., Allen J. V., Gaskin H. (April 7, 2004) European Patent (EP) 1404335, World Intellectual Property Organization Patent (WO) 03000267
- 29. Array BioPharma Inc (2009) Glucokinase Activator ARRY-403: Phase I Single Ascending Dose Top-Line Results, Array BioPharma Inc., Boulder, CO [Google Scholar]
- 30. Bonadonna R. C., Heise T., Arbet-Engels C., Kapitza C., Avogaro A., Grimsby J., Zhi J., Grippo J. F., Balena R. (2010) J. Clin. Endocrinol. Metab. 95, 5028–5036 [DOI] [PubMed] [Google Scholar]
- 31. Bonadonna R. C., Kapitza C., Heinse T., Avogaro A., Boldrin M., Grimsby J., Mulligan M. E., Arbet-Engels C., Balena R. (2008) Diabetologia 51, Suppl. 1, S371 (Abstr. 927) [Google Scholar]
- 32. Brocklehurst K. J., Payne V. A., Davies R. A., Carroll D., Vertigan H. L., Wightman H. J., Aiston S., Waddell I. D., Leighton B., Coghlan M. P., Agius L. (2004) Diabetes 53, 535–541 [DOI] [PubMed] [Google Scholar]
- 33. Castelhano A. L., Dong H., Fyfe M. C., Gardner L. S., Kamikozawa Y., Kurabayashi S., Nawano M., Ohashi R., Procter M. J., Qiu L., Rasamison C. M., Schofield K. L., Shah V. K., Ueta K., Williams G. M., Witter D., Yasuda K. (2005) Bioorg. Med. Chem. Lett. 15, 1501–1504 [DOI] [PubMed] [Google Scholar]
- 34. Grimsby J., Sarabu R., Corbett W. L., Haynes N. E., Bizzarro F. T., Coffey J. W., Guertin K. R., Hilliard D. W., Kester R. F., Mahaney P. E., Marcus L., Qi L., Spence C. L., Tengi J., Magnuson M. A., Chu C. A., Dvorozniak M. T., Matschinsky F. M., Grippo J. F. (2003) Science 301, 370–373 [DOI] [PubMed] [Google Scholar]
- 35. McKerrecher D., Allen J. V., Bowker S. S., Boyd S., Caulkett P. W., Currie G. S., Davies C. D., Fenwick M. L., Gaskin H., Grange E., Hargreaves R. B., Hayter B. R., James R., Johnson K. M., Johnstone C., Jones C. D., Lackie S., Rayner J. W., Walker R. P. (2005) Bioorg. Med. Chem. Lett. 15, 2103–2106 [DOI] [PubMed] [Google Scholar]
- 36. McKerrecher D., Allen J. V., Caulkett P. W., Donald C. S., Fenwick M. L., Grange E., Johnson K. M., Johnstone C., Jones C. D., Pike K. G., Rayner J. W., Walker R. P. (2006) Bioorg. Med. Chem. Lett. 16, 2705–2709 [DOI] [PubMed] [Google Scholar]
- 37. Holland G. W., Sarabu R., Kester R. F., Haynes N. E., Bizzarro F. T., Corbett W. L., Grippo J. F., Mahaney P. E. (October 6, 2004) European Patent (EP) 1169312, World Intellectual Property Organization Patent (WO) 0058293
- 38. Zhai S., Mulligan M. E., Grimsby J., Arbet-Engels C., Boldrin M., Balena R., Zhi J. (2008) Diabetologia 51, Suppl. 1, S372, Abstr. 928 [Google Scholar]
- 39. Zhi J., Zhai S., Mulligan M. E., Grimsby J., Arbet-Engels C., Boldrin M., Balena R. (2008) Diabetologia 51, Suppl. 1, S23 (Abstr. 42) [Google Scholar]
- 40. Kamata K., Mitsuya M., Nishimura T., Eiki J., Nagata Y. (2004) Structure 12, 429–438 [DOI] [PubMed] [Google Scholar]
- 41. Pal P., Miller B. G. (2009) Biochemistry 48, 814–816 [DOI] [PubMed] [Google Scholar]
- 42. Liang Y., Kesavan P., Wang L. Q., Niswender K., Tanizawa Y., Permutt M. A., Magnuson M. A., Matschinsky F. M. (1995) Biochem. J. 309, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beer N. L., Tribble N. D., McCulloch L. J., Roos C., Johnson P. R., Orho-Melander M., Gloyn A. L. (2009) Hum. Mol. Genet. 18, 4081–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brocklehurst K. J., Davies R. A., Agius L. (2004) Biochem. J. 378, 693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gloyn A. L., Odili S., Zelent D., Buettger C., Castleden H. A., Steele A. M., Stride A., Shiota C., Magnuson M. A., Lorini R., d'Annunzio G., Stanley C. A., Kwagh J., van Schaftingen E., Veiga-da-Cunha M., Barbetti F., Dunten P., Han Y., Grimsby J., Taub R., Ellard S., Hattersley A. T., Matschinsky F. M. (2005) J. Biol. Chem. 280, 14105–14113 [DOI] [PubMed] [Google Scholar]
- 46. Davis E. A., Cuesta-Muñoz A., Raoul M., Buettger C., Sweet I., Moates M., Magnuson M. A., Matschinsky F. M. (1999) Diabetologia 42, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 47. Gloyn A. L., Odili S., Buettger C., Njolstad P., Shiota C., Magnuson M. A., Matschinsky F. M. (2004) in Glucokinase and Glycaemic Disease: from Basics to Novel Therapeutics (Matschinsky F. M., Magnuson M. A. eds) pp. 92–109, Karger, Basel [Google Scholar]
- 48. Bruce S. (2000) Am. J. Hum. Biol. 12, 542–551 [DOI] [PubMed] [Google Scholar]
- 49. Liang Y., Najafi H., Smith R. M., Zimmerman E. C., Magnuson M. A., Tal M., Matschinsky F. M. (1992) Diabetes 41, 792–806 [DOI] [PubMed] [Google Scholar]
- 50. Lynedjian P. (2004) in Glucokinase and Glycaemic Disease: from Basics to Novel Therapeutics (Matschinsky F. M., Magnuson M. A. eds) pp. 156–166, Karger, Basel [Google Scholar]
- 51. Alvarez E., Roncero I., Chowen J. A., Vázquez P., Blázquez E. (2002) J. Neurochem. 80, 45–53 [DOI] [PubMed] [Google Scholar]
- 52. Baltrusch S., Lenzen S., Okar D. A., Lange A. J., Tiedge M. (2001) J. Biol. Chem. 276, 43915–43923 [DOI] [PubMed] [Google Scholar]
- 53. Bjørkhaug L., Molnes J., Søvik O., Njølstad P. R., Flatmark T. (2007) J. Biol. Chem. 282, 22757–22764 [DOI] [PubMed] [Google Scholar]
- 54. Danial N. N., Gramm C. F., Scorrano L., Zhang C. Y., Krauss S., Ranger A. M., Datta S. R., Greenberg M. E., Licklider L. J., Lowell B. B., Gygi S. P., Korsmeyer S. J. (2003) Nature 424, 952–956 [DOI] [PubMed] [Google Scholar]
- 55. Danial N. N., Walensky L. D., Zhang C. Y., Choi C. S., Fisher J. K., Molina A. J., Datta S. R., Pitter K. L., Bird G. H., Wikstrom J. D., Deeney J. T., Robertson K., Morash J., Kulkarni A., Neschen S., Kim S., Greenberg M. E., Corkey B. E., Shirihai O. S., Shulman G. I., Lowell B. B., Korsmeyer S. J. (2008) Nat. Med. 14, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Massa L., Baltrusch S., Okar D. A., Lange A. J., Lenzen S., Tiedge M. (2004) Diabetes 53, 1020–1029 [DOI] [PubMed] [Google Scholar]
- 57. Shiota C., Coffey J., Grimsby J., Grippo J. F., Magnuson M. A. (1999) J. Biol. Chem. 274, 37125–37130 [DOI] [PubMed] [Google Scholar]
- 58. Van Schaftingen E., Veiga da Cunha M. (2004) in Glucokinase and Glycaemic Disease: from Basics to Novel Therapeutics (Matschinsky F. M., Magnuson M. A. eds) pp. 193–207, Karger, Basel [Google Scholar]
- 59. Arden C., Trainer A., de la Iglesia N., Scougall K. T., Gloyn A. L., Lange A. J., Shaw J. A., Matschinsky F. M., Agius L. (2007) Diabetes 56, 1773–1782 [DOI] [PubMed] [Google Scholar]
- 60. Sagen J. V., Odili S., Bjørkhaug L., Zelent D., Buettger C., Kwagh J., Stanley C., Dahl-Jørgensen K., de Beaufort C., Bell G. I., Han Y., Grimsby J., Taub R., Molven A., Søvik O., Njølstad P. R., Matschinsky F. M. (2006) Diabetes 55, 1713–1722 [DOI] [PubMed] [Google Scholar]
- 61. Burke C. V., Buettger C. W., Davis E. A., McClane S. J., Matschinsky F. M., Raper S. E. (1999) Biochem. J. 342, 345–352 [PMC free article] [PubMed] [Google Scholar]
- 62. Miller S. P., Anand G. R., Karschnia E. J., Bell G. I., LaPorte D. C., Lange A. J. (1999) Diabetes 48, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 63. Slosberg E. D., Desai U. J., Fanelli B., St Denny I., Connelly S., Kaleko M., Boettcher B. R., Caplan S. L. (2001) Diabetes 50, 1813–1820 [DOI] [PubMed] [Google Scholar]
- 64. Heredia V. V., Carlson T. J., Garcia E., Sun S. (2006) J. Biol. Chem. 281, 40201–40207 [DOI] [PubMed] [Google Scholar]
- 65. Baltrusch S., Francini F., Lenzen S., Tiedge M. (2005) Diabetes 54, 2829–2837 [DOI] [PubMed] [Google Scholar]
- 66. Grimsby J., Coffey J. W., Dvorozniak M. T., Magram J., Li G., Matschinsky F. M., Shiota C., Kaur S., Magnuson M. A., Grippo J. F. (2000) J. Biol. Chem. 275, 7826–7831 [DOI] [PubMed] [Google Scholar]
- 67. Zawalich W. S., Rognstad R., Pagliara A. S., Matschinsky F. M. (1977) J. Biol. Chem. 252, 8519–8523 [PubMed] [Google Scholar]
- 68. O'Doherty R. M., Lehman D. L., Télémaque-Potts S., Newgard C. B. (1999) Diabetes 48, 2022–2027 [DOI] [PubMed] [Google Scholar]
- 69. Grimsby J., Grippo J. F. (2004) in Glucokinase and Glycaemic Disease: from Basics to Novel Therapeutics (Matschinsky F. M., Magnuson M. A. eds) pp. 360–378, Karger, Basel [Google Scholar]
- 70. Massa O., Meschi F., Cuesta-Munoz A., Caumo A., Cerutti F., Toni S., Cherubini V., Guazzarotti L., Sulli N., Matschinsky F. M., Lorini R., Iafusco D., Barbetti F. (2001) Diabetologia 44, 898–905 [DOI] [PubMed] [Google Scholar]