Abstract
In this study, we determined the molecular mechanisms whereby forkhead transcription factor Foxo1, a key downstream signaling molecule of insulin-like growth factor 1 (IGF1)/insulin actions, regulates Runx2 activity and expression of the mouse osteocalcin gene 2 (Bglap2) in osteoblasts in vitro. We showed that Foxo1 inhibited Runx2-dependent transcriptional activity and osteocalcin mRNA expression and Bglap2 promoter activity in MC-4 preosteoblasts. Co-immunoprecipitation assay showed that Foxo1 physically interacted with Runx2 via its C-terminal region in osteoblasts or when co-expressed in COS-7 cells. Electrophoretic mobility shift assay demonstrated that Foxo1 suppressed Runx2 binding to its cognate site within the Bglap2 promoter. IGF1 and insulin prevented Foxo1 from inhibiting Runx2 activity by promoting Foxo1 phosphorylation and nuclear exclusion. In contrast, a neutralizing anti-IGF1 antibody decreased Runx2 activity and osteocalcin expression in osteoblasts. Chromatin immunoprecipitation assay revealed that IGF1 increased Runx2 interaction with a chromatin fragment of the proximal Bglap2 promoter in a PI3K/AKT-dependent manner. Conversely, knockdown of Foxo1 increased Runx2 interaction with the promoter. This study establishes that Foxo1 is a novel negative regulator of osteoblast-specific transcription factor Runx2 and modulates IGF1/insulin-dependent regulation of osteocalcin expression in osteoblasts.
Keywords: Bone, Insulin, Insulin-like Growth Factor (IGF), PI 3-Kinase, Promoters, FoxO1, Runx2, Osteoblast, Osteocalcin
Introduction
Foxo1 is a forkhead transcription factor that is defined by its amino forkhead DNA binding domain and carboxyl trans-activation domain. Foxo1 plays a pivotal role in mediating the effect of insulin or insulin-like growth factor 1 (IGF1)2 on the expression of genes involved in cell growth, differentiation, metabolism, and longevity (1–5). Insulin or IGF1 exerts its inhibitory effect on gene expression via a highly conserved sequence (TG/ATTTT/G), termed the insulin response element (IRE), in the promoter of genes that are negatively regulated by insulin/IGF1. In the absence of insulin/IGF1, Foxo1 resides in the nucleus and binds as a trans-activator to the IRE, enhancing promoter activity. In response to insulin, Foxo1 is phosphorylated at three highly conserved phosphorylation sites (Thr-24, Ser-256, and Ser-319) through the PI3K-dependent pathway, resulting in its nuclear exclusion and inhibition of target gene expression (1, 2, 6). Except for Foxo6 (7), all members of the Foxo superfamily undergo insulin/IGF1-dependent phosphorylation and nuclear exclusion. Failure to phosphorylate Foxo1 results in its permanent nuclear localization and constitutive trans-activation of gene expression. This phosphorylation-dependent Foxo1 translocation has been viewed as an acute mechanism for insulin or growth factors to inhibit gene expression, as insulin-induced Foxo1 phosphorylation is kinetically coupled to its subsequent translocation to the cytoplasm (1, 8).
Osteoblasts, the bone-forming cells, originate from multipotential mesenchymal cells. Osteoblast activity and function are regulated by a number of growth factors and hormones including IGF1, insulin, bone morphogenetic proteins (BMPs), basic fibroblast growth factor 2 (FGF-2), parathyroid hormone (PTH), tumor necrosis factor-α (TNF-α), and extracellular matrix signals (9–21). At the molecular level, osteoblast function is controlled by several key transcription factors including Runx2, osterix, and ATF4 (22–29). Runx2 is a runt domain-containing transcription factor that is characterized as a transcriptional activator of osteoblast differentiation and master gene for bone development (22–26). Runx2 expression and activity are controlled by a number of factors including IGF1, BMPs, FGF-2, PTH, TNF-α, and extracellular matrix signals (9, 12–14, 30, 31) as well as by nuclear factors via protein-protein interactions (29, 32–46).
Recent studies showed that osteocalcin, an osteoblast-specific product encoded by the Bglap2 (bone γ-carboxyglutamate protein) gene plays a critical role in regulating glucose metabolism (10, 11, 47). Using mice lacking Foxo1 selectively in osteoblasts, Rached et al. (48) recently showed that Foxo1 expressed in osteoblasts regulates glucose homeostasis through an osteocalcin-dependent mechanism. Specifically, osteoblast-conditional inactivation of Foxo1 increased β-cell proliferation and insulin secretion and sensitivity. Importantly, osteoblastic osteocalcin protein, which is active in the absence of δ-carboxylation, was found to be responsible for the metabolic actions of Foxo1 in regulating glucose homeostasis. Foxo1 decreases osteocalcin mRNA expression and increases osteocalcin carboxylation. Foxo1 achieves the latter by increasing the expression of Esp, a gene that encodes a protein that decreases osteocalcin function (i.e. increases its carboxylation). Therefore, Foxo1 negatively regulates both osteocalcin production/expression and function. However, the molecular mechanism whereby Foxo1 suppresses the Bglap2 gene is undefined. In this study, we hypothesized that Foxo1 inhibits Bglap2 gene expression and promoter activity, at least in part, via suppression of Runx2, a major transcriptional activator of the Bglap2 gene (25, 49). To address this hypothesis, we analyzed the molecular interplay between Runx2 and Foxo1 in Bglap2 gene expression in osteoblasts. We demonstrate that Foxo1 physically binds to and functionally antagonizes Runx2 from driving Bglap2 expression. Furthermore, we demonstrate that IGF1/insulin prevents Foxo1 from inhibiting Runx2, probably by promoting Foxo1 phosphorylation and nuclear exclusion in osteoblasts. This effect results in inhibition of Foxo1 action, which contributes to increased osteocalcin expression in osteoblasts and favors glucose homeostasis.
EXPERIMENTAL PROCEDURES
Reagents and Cell Lines
Tissue culture media and fetal bovine serum were obtained from HyClone (Logan, UT). Mouse MC3T3-E1 subclone 4 (MC-4) cells were described previously (31, 50) and maintained in AA (ascorbic acid)-free α-modified Eagle's medium (α-MEM), 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin and were not used beyond passage 15. COS-7 cells and rat UMR106–01 osteosarcoma cells were described (46, 51, 52). IGF1 was purchased from R&D Systems, Inc (Minnepolis, MN). Insulin and LY294002 were purchased from Sigma Aldrich.
DNA Constructs
Wild-type (wt) or mutated (mt) 6XRUNX2-LUC (also known as p6OSE2-luc (49)) or BGLAP2-LUC (also known as p657mOG2-luc (49)), which contains 2-bp substitution mutations in the OSE2 sites that abolishes Runx2 binding, 4XATF4-LUC (p4OSE1-luc (53, 54)), pCMV5/β-gal, pCMV/Runx2 expression plasmids (wt, amino acids 1–410, amino acids 1–330, amino acids 1–286, amino acids 1–258) containing cDNAs encoding either wt Runx2 or C-terminal deletions under CMV promoter control, and full-length GST-Runx2 and GST-Foxo1 fusion protein expression vectors were previously described (29, 46, 49, 53, 55, 56). HA-tagged pCMV/Runx2 expression plasmids containing cDNAs encoding wt or N-terminal deletions (ΔN97, ΔN242, and ΔN326) under CMV promoter control were previously described (57). To generate pCMV/Foxo1 expression plasmids expressing truncated forms of Foxo1 (amino acids 1–558, amino acids 1–456, amino acids 1–360, amino acids 1–258), a stop code (TAA, TAG, or TGA) that results in premature stop of Foxo1 protein at indicated amino acid residues was introduced into Foxo1 cDNA by PCR using pCMV/Foxo1 as a template. The pCMV/Foxo1(3A) expression plasmid expressing a mutant Foxo1 protein in which three insulin/AKT-dependent phosphorylation sites (Thr-24, Ser-256, and Ser-319) were mutated from Thr or Ser to Ala, was described previously (58). Adenovirus expressing Foxo1 under control of a CMV promoter (AdCMV/Foxo1) was constructed by subcloning full-length Foxo1 cDNA into pAdlox plasmid followed by CRE-mediated recombination as previously described (55, 56). Expression of Foxo1 protein was confirmed by Western blot analysis (data not shown). All sequences were verified by automatic DNA sequencing.
Transfection and Dual Luciferase Assay
Cells were plated on 35-mm dishes at a density of 5 × 104 cells/cm2. After 24 h, cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Each transfection contained 0.25 μg of the indicated reporter plasmids plus 0.01 μg of pRL-SV40, containing a cDNA for Renilla Reformis luciferase to control for transfection efficiency. Cells were harvested and assayed using the Dual Luciferase Assay kit (Promega, Madison, WI) on a ModuleTM Microplate Multimode Reader (Turner Biosystem, Sunnyvale, CA). For all transfection experiments, the amount of plasmid DNAs was balanced as necessary with β-galactosidase (β-gal) expression plasmid such that the total DNA was constant in each group. Experiments were performed in triplicates and repeated 3–4 times.
Adenoviral Infection
Adenoviral vectors for β-galactosidase (Ad-β-gal), Foxo1 (Ad-Foxo1), Foxo1 RNAi (Ad-Foxo1i), or control RNAi (USi) were described previously (29, 55, 56, 59). MC-4 cells were infected with adenovirus as described previously (29, 55, 56). Briefly, virus was added to cells in 1% FBS and incubated for 1 h at 37 °C. Dishes were rotated every 5 min for the first 15 min to ensure that all of the cells were exposed to virus. After 1 h, media were aspirated, and cultures were rinsed twice with serum-free medium, and then fresh media supplemented with 10% FBS were added to the dishes. The amount of adenovirus was balanced as necessary with a control adenovirus expressing β-galactosidase such that the total amount was constant in each group.
RNA Isolation, Reverse Transcription (RT), Quantitative RT-PCR, and Western Blot Analyses
RNA isolation, RT, and quantitative real-time RT-PCR were performed to measure the relative mRNA levels using SYBR Green kit (Bio-Rad) as previously described (46, 54, 60). Samples were normalized to Gapdh expression. The DNA sequences of mouse primers used for real-time PCR were as follows: Bglap2: 5′-TAG TGA ACA GAC TCC GGC GCT A-3′ (forward), 5′-TGT AGG CGG TCT TCA AGC CAT-3′ (reverse); Runx2: 5′-TAA AGT GAC AGT GGA CGG TCC C-3′ (forward), 5′-TGC GCC CTA AAT CAC TGA GG-3′ (reverse); Gapdh: 5′-CAG TGC CAG CCT CGT CCC GTA GA-3′ (forward), 5′-CTG CAA ATG GCA GCC CTG GTG AC-3′ (reverse). Western blot analysis was performed as previously described (46, 54). Antibodies used in this study were from the following sources: antibodies against Foxo1, Runx2, HA, anti-IGF1 neutralizing antibody, normal control IgG, and anti-rabbit or anti-mouse antibodies conjugated with horseradish peroxidase from Santa Cruz Biotechnology, Inc., and mouse monoclonal antibody against β-actin and M2 antibody from Sigma Aldrich.
Nuclear Extracts (NE) Preparation and Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared from rat UMR106–01 osteosarcoma cells, which express high level of Runx2 protein, and EMSA was performed as previously described (21, 31). GST-Runx2, GST-Foxo1, and GST proteins were purified using the Bulk GST Purification Module kit (GE Healthcare/Amersham Biosciences) as previously described (46). The DNA sequences of the oligonucleotides used for EMSA were as follows: OSE2 (Runx2 binding site) (49): 5-GAT CCG CTG CAA TCA CCA ACC ACA GCA-3. IRE (Insulin Response Element that contains a Foxo1 binding site) (55): 5-TGT AGT TTG TTT TGT TTT GTT GGC ATG-3. DNA oligonucleotide was labeled using a biotin 3′-end DNA labeling kit (cat: 89818, Pierce Biotechnology Inc.). 2 μg of nuclear extracts and 20 fmol biotin-labeled DNA probe were incubated in the presence and absence of indicated amounts of purified GST-Foxo1 or GST protein in 1x binding buffer for 30 min at room temperature. For supershift assay, 1 μg of IgG or anti-Runx2 antibody was first incubated with nuclear extracts prior to addition of DNA probe. Protein-DNA complexes were separated on 4% polyacrylamide gels in 0.5× TBE buffer, and transferred onto Biodyne B Nylon Membrane (cat: 77016, Pierce). The membrane was blocked in 1× blocking buffer, washed five times with 1× wash buffer, and visualized by a Chemiluminescent Nucleic Acid Detection Module (cat: 89880, Pierce).
Immunoprecipitation (IP) and Chromatin Immunoprecipitation (ChIP) Assays
Whole cell extracts and purified GST fusion proteins were used for immunoprecipitation using specific antibodies as previously described (46). ChIP assays were performed as described previously (21, 46). Briefly, the equivalent of 10 μg of DNA was used as starting material (input) in each ChIP reaction with 2 μg of the appropriate antibody (Runx2 or control rabbit IgG). Fractions of the purified ChIP DNA (5%) or inputs (0.02–0.05%) were used for PCR analysis. The reaction was performed with AmpliTaq Gold DNA Polymerase (Applied Biosystems) for 30 cycles of 15 s at 95 °C, 30 s at 60 °C, and 15 s at 72 °C. PCR primer pairs were generated to detect DNA segments located near the Runx2-binding site at −137/−131 (primers P1 and P2) and the Bglap2 cDNA region (primers P3 and P4) (46). The PCR products were separated on 3% agarose gels and visualized with ultraviolet light. All ChIP assays were repeated at least three times.
Statistical Analysis
Results were expressed as means ± standard deviation (S.D.). Students' t test was used to test for differences between two groups. Differences with a p < 0.05 was considered as statistically significant. All experiments were repeated a minimum of three times with triplicate samples.
RESULTS
Foxo1 Decreases Bglap2 Expression and Promoter Activity and Runx2-dependent Transcriptional Activity
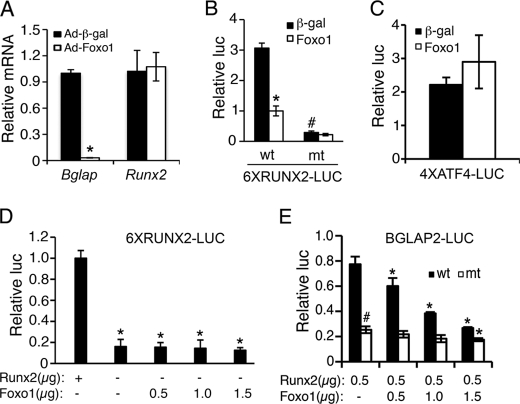
To determine the effect of Foxo1 on Bglap2 expression in osteoblasts, MC3T3-E1 subclone 4 (MC-4) preosteoblasts were infected with equal amount of adenovirus expression vectors for Foxo1 (Ad-Foxo1) or β-galactosidase (Ad-β-gal), followed by RNA preparation and quantitative real-time RT/PCR for Bglap2 and Runx2 mRNAs, which were normalized to Gapdh mRNA. Consistent with the result from a recent study (48), adenoviral overexpression of Foxo1 dramatically reduced the level of Bglap2 mRNA in the MC-4 cells (Fig. 1A). Surprisingly, Foxo1 did not alter the mRNA level of Runx2, a major upstream transcriptional activator of the Bglap2 (25, 26, 49, 61). A similar result was observed in rat UMR106–01 osteoblast-like cells (data not shown). Therefore, we next evaluated whether Foxo1 modulated Runx2 activity. To this end, COS-7 cells were co-transfected with 6XRUNX2-LUC, previously known as p6OSE2-luc, a reporter plasmid containing 6 copies of OSE2 (osteoblast-specific element 2, a well-defined Runx2-response element (49)) upstream of a minimal 34 bp Bglap2 promoter (19, 25, 31), pRL-SV40 (for normalization), and pCMV/Runx2 with and without expression vector for Foxo1. For these studies, we used COS-7 cells because they contain undetectable levels of endogenous Runx2 and Foxo1 proteins (20) and data not shown). As shown in Fig. 1B, Foxo1 significantly reduced the Runx2-dependent transcriptional activity. The repression was totally lost with mutations in the Runx2 DNA binding core sequence from AACCACA to AAGAACA in the reporter plasmid, which abolishes Runx2 DNA binding (29). Foxo1 inhibition of Runx2 is specific because Foxo1 did not suppress ATF4 (activating transcription factor 4)-dependent OSE1 (osteoblast-specific element 1, an ATF4-response element from the Bglap2 promoter (49)) activity (Fig. 1C). Further supporting the specificity of this regulation, Foxo1 had no effect on basal promoter activity in the absence of Runx2 (Fig. 1D). As shown in Fig. 1E, Foxo1 also inhibited the 657-bp Bglap2 promoter activity, which was dependent upon the presence of the intact Runx2 DNA binding sites in the promoter. The result with the 657-bp Bglap2 promoter is of particular significance because this promoter fragment contains sufficient information to direct osteoblast-specific expression in transgenic mice (62). Notably, high concentration of Foxo1(1.5 μg) slightly, but significantly, inhibited the luciferase activity driven by the 657-bp Bgalp2 promoter in which the Runx2 DNA binding sites were mutated, indicating that a small part of Foxo1 inhibition of the promoter activity is Runx2-independent. Collectively, these results demonstrate that Foxo1 suppresses Bglap2 gene expression, at least in part, through Runx2 inhibition.
FIGURE 1.
Foxo1 inhibits osteocalcin expression and Bglap2 promoter activity and Runx2-dependent transcriptional activity. A, MC3T3E-1 cells were infected with and without adenovirus expressing Foxo1. Adenovirus expressing β-galactosidase was used as control. Cells were then differentiated in AA-containing α-MEM media for 5 days, followed by quantitative real-time RT/PCR for Bglap2 and Runx2 mRNAs, which were normalized to Gapdh mRNA. *, p < 0.05, versus Ad-β-gal. B and C, COS-7 cells were transiently transfected with 6XRUNX2-LUC, 6XRUNX2mt-LUC (B) in which the Runx2-binding sequence was mutated, or 4XATF4-LUC (C), pRL-SV40 (for normalization), and Runx2 expression plasmid with and without Foxo1 expression plasmid. β-Galactosidase expression plasmid was used as control. After 48 h, cells were harvested for dual luciferase assay. Firefly luciferase was normalized to Rotylenchulus reniformis luciferase to control the transfection efficiency. *, p < 0.05, versus 0 μg Foxo1; #, p < 0.05, versus wt. D, COS-7 cells were transfected with 6XRUNX2-LUC and pRL-SV40 and increasing amounts of Foxo1 plasmid with and without Runx2 expression plasmid. *, p < 0.05, versus Runx2. E, COS-7 cells were transfected with BGLAP2-LUC or BGLAP2mt-LUC in which two previously defined Runx2-binding sites were mutated (29), pRL-SV40, and FLAG-Runx2 plasmid with and without increasing amounts of Foxo1 expression plasmid. *, p < 0.05, versus 0 μg Foxo1; #, p < 0.05, versus wt.
Foxo1 Physically Interacts with Runx2
To define the molecular mechanism through which Foxo1 inhibits Runx2, we next determined whether Foxo1 interacted with Runx2 by performing immunoprecipitation (IP) assays using nuclear extracts from the rat UMR106–01 osteoblastic cells. As shown in Fig. 2A, Runx2 protein was present in an anti-Foxo1 antibody immunoprecipitate (lanes 1 and 2). Reciprocal IP assay showed that an anti-Runx2 antibody, but not control IgG, immunoprecipitated the Foxo1 protein (lanes 3 and 4). As expected, ATF4 protein, a known Runx2-interacting factor (29), was present in the anti-Runx2 antibody immunoprecipitate (lanes 5 and 6). In contrast, an NFATc1 antibody failed to immunoprecipitate Runx2 (lanes 7 and 8). As shown in Fig. 2B, Foxo1 and Runx2 can also be co-immunoprecipitated from COS-7 cells exogenously expressing both factors. To determine if Foxo1 directly interacted with Runx2 in the absence of other nuclear proteins, we conducted IP assays using purified GST-Foxo1 and GST-Runx2 and GST proteins. As shown in Fig. 2C, GST-Runx2 protein was immunoprecipitated by an anti-Foxo1 antibody (lanes 1 and 2) and, vice versa, GST-Foxo1 was immunoprecipitated by an anti-Runx2 antibody (lanes 3 and 4). In contrast, GST protein was not immunoprecipitated by antibodies against Runx2 or Foxo1 or normal IgG (lanes 5–8), thus demonstrating a direct interaction between Foxo1 and Runx2.
FIGURE 2.
Foxo1 physically interacts with Runx2. A and B, IP assays using nuclear extracts from UMR106-01 osteoblast-like cells (A) or from COS-7 cells overexpressing Foxo1 and Runx2 (B). Extracts were immunoprecipitated with control rabbit IgG or indicated antibodies, followed by Western blot analysis using respective antibodies. C, IP assays using GST fusion proteins. Mixture of purified GST-Foxo1, GST-Runx2 or GST proteins was immunoprecipitated by control IgG (lanes 1, 3, 5, and 7), Foxo1 antibody (lanes 2 and 6) or Runx2 antibody (lanes 4 and 8), followed by Western blot for Runx2 (lanes 1 and 2), Foxo1 (lanes 3 and 4) or GST (lanes 5–8).
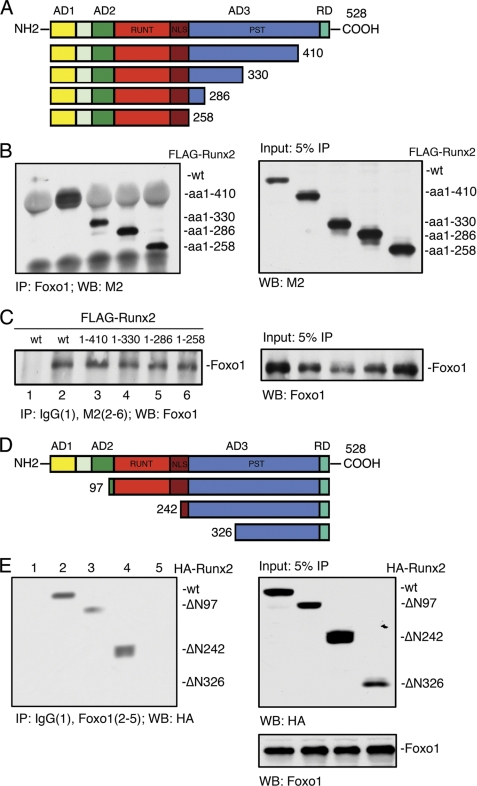
Deletion Analysis of the Runx2 cDNA
To identify the Foxo1-binding domain within the Runx2 molecule, COS-7 cells were co-transfected with the Foxo1 expression plasmid and wt FLAG-Runx2 or various FLAG-Runx2 C-terminal deletion mutant expression vectors. Forty hours later, nuclear extracts were prepared for IP using an anti-Foxo1 antibody, followed by Western blot analysis using a M2 antibody. As shown in Fig. 3B, deletion of Runx2 from amino acid 528 (wt) to amino acid 258 did not abolish the Foxo1 binding. This result was confirmed by the reciprocal IP using a M2 antibody, followed by Western blot for Foxo1 (Fig. 3C). As shown in Fig. 3E, the deletion of the N-terminal 242 amino acid of Runx2 did not reduce its ability to bind to Foxo1. However, further deletion from amino acid 242 to amino acid 326 completely abrogated the Foxo1-Runx2 interaction. Collectively, these results suggest that the amino acid 242–258 region of Runx2 is critical for interaction with Foxo1.
FIGURE 3.
Deletion analysis of the Runx2 cDNA. A, schematic showing the domain structure of Runx2 and the C-terminal deletion mutants. Runx2 contains three transcriptional activation domains (AD1, 2, 3), a transcriptional repression domain (RD) containing a VWRPY motif at C-terminal, a RUNT domain responsible for DNA binding, a nuclear localization sequence (NLS), and a large C-terminal PST domain which is rich in serine/threonine/tyrosine residues. B and C, whole cell extracts from COS-7 cells cotransfected with expression vectors for Foxo1 and wt FLAG-Runx2 or various FLAG-Runx2 C-terminal deletion mutants (amino acid 1–410, amino acid 1–330, amino acid 1–286, and amino acid 1–258) were immunoprecipitated with Foxo1 antibody, followed by Western blot using M2 antibody (B). In reciprocal IP, the same extracts were immunoprecipitated with control IgG (lane 1) or M2 antibody (lanes 2–6), followed by Western blot using Foxo1 antibody (C). D, schematic showing the domain structure of Runx2 and its N-terminal deletion mutants. E, whole cell extracts from COS-7 cells cotransfected with expression vectors for Foxo1 and wt HA-Runx2 or various HA-Runx2 N-terminal deletion mutants (ΔN97, ΔN242, and ΔN326) were immunoprecipitated with control IgG (lane 1) or Foxo1 antibody (lanes 2–5), followed by Western blot using HA antibody.
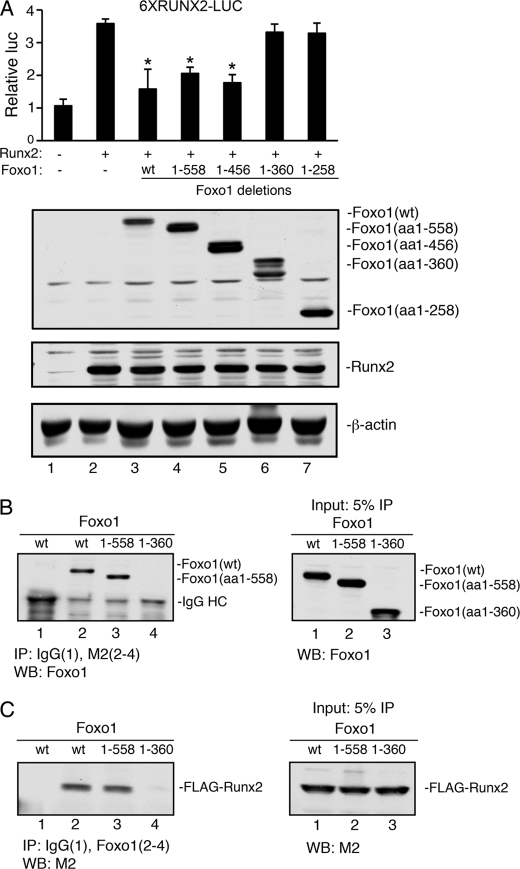
Deletion Analysis of the Foxo1 cDNA
Several C-terminal Foxo1 deletion mutant expression vectors were generated and tested for their ability to suppress Runx2 activity in COS-7 cells. As shown in Fig. 4A (top), the deletion of Foxo1 cDNA from amino acids 653 (wt) to 456 did not alter its inhibition of Runx2 activity. However, further deletion from amino acids 456 to 360 completely abolished its Runx2 inhibitory activity. As shown in Fig. 4A (bottom), Western blot analysis showed that both wt and mutant Foxo1 proteins were expressed at comparable levels. Likewise, co-expression of wt or mutant Foxo1 expression vectors did not markedly alter the level of Runx2 protein. Consistent with the results from the functional study, IP assays revealed that the deletion from amino acids 456 to 360 of Foxo1 also abolished the Foxo1-Runx2 interaction (Fig. 4, B and C). These results suggest that the C-terminal region (amino acids 360–456) is essential for Foxo1 to bind to and inhibit Runx2 activity.
FIGURE 4.
Deletion analysis of the Foxo1 cDNA. A, COS-7 cells were transiently transfected with 6XRUNX2-LUC and pRL-SV40 with and without Runx2 plasmid as well as with and without wt or various deletion mutant Foxo1 plasmids (amino acids 1–558, amino acids 1–456, amino acids 1–360, and amino acids 1–258), followed by dual luciferase assay (top) and Western blot (bottom). *, p < 0.05, versus 0 μg Foxo1. B and C, whole cell extracts from COS-7 cells overexpressing FLAG-Runx2 and wt or two C-terminal deletion Foxo1 mutants (amino acids 1–558 and amino acids 1–360) were immunoprecipitated with control IgG (lane 1) or M2 antibody (lanes 2–4), followed by Western blot using an anti-Foxo1 antibody (B). In reciprocal IP, the same extracts were immunoprecipitated with control IgG (lane 1) or Foxo1 antibody (lanes 2–4), followed by Western blot using a M2 antibody (C).
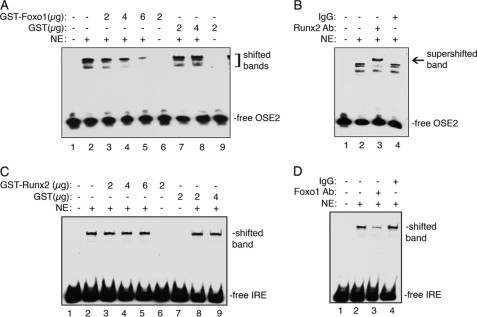
Foxo1 Inhibits Runx2 Binding to Its Cognate Site (OSE2) within the Bglap2 Promoter
To study the mechanism whereby Foxo1 inhibits Runx2 as demonstrated above, we next determined whether Foxo1 affected Runx2 DNA binding activity by performing EMSA using the wild-type (wt) OSE2 (Osteoblast-Specific Element 2) oligo, a well-established Runx2 binding element from the Bglap2 promoter (49), as probe and 2 μg of nuclear extracts (NE) from UMR106-01 cells in the presence and absence of increasing amounts of purified GST-Foxo1 protein. As shown in Fig. 5A, while GST-Foxo1 protein itself did not bind to the OSE2 oligo (lane 6), it dose-dependently inhibited the binding of Runx2 to the OSE2 oligo (lanes 3–5). This inhibition was specific because the GST protein neither altered the Runx2 DNA binding activity (lanes 7 and 8) nor bound to the OSE2 DNA itself (lane 9). In contrast, purified GST-Runx2 or GST did not inhibit the binding of Foxo1 to its cognate site (Fig. 5C). The existence of Runx2 or Foxo1 in the shifted DNA-protein complexes was confirmed by antibody analyses that either supershifted the complex (anti-Runx2 antibody, Fig. 5B) or reduced the complex (anti-Foxo1 antibody, Fig. 5D), in contrast to the control IgG.
FIGURE 5.
Foxo1 inhibits Runx2 binding to the OSE2 DNA. A, EMSA. Labeled OSE2 DNA probe was incubated with 2 μg of nuclear extracts (NE) from UMR106-01 cells in the presence and absence of indicated amounts of purified GST-Foxo1 or GST proteins. B, super gel shift assay. Labeled OSE2 DNA probe was incubated with 2 μg of nuclear extracts from UMR106-01 cells in the presence of Runx2 antibody (lane 3) or control IgG (lane 4). C, EMSA. Labeled IRE DNA probe was incubated with 2 μg of NE from UMR106-01 cells in the presence and absence of indicated amounts of purified GST-Runx2 or GST proteins. C, super gel shift assay. Labeled IRE DNA probe was incubated with 2 μg nuclear extracts from UMR106-01 cells in the presence of Foxo1 antibody (lane 3) or control IgG (lane 4).
IGF1 Prevents Foxo1 from Inhibiting Runx2 in Osteoblasts
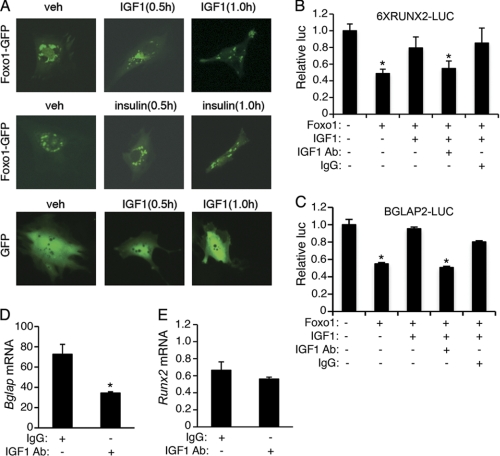
IGF1 or insulin phosphorylates Foxo1 through the PI3K-dependent pathway, which results in Foxo1 nuclear exclusion in many cell types (1, 2, 6). To determine whether this also occurs in osteoblasts, MC-4 cells were electroporated with expression plasmids for Foxo1-GFP or GFP proteins as previously described (63). Thirty hours later, cells were treated with and without 10 ng/ml mouse recombinant IGF1 or 100 nm insulin for the indicated times. As shown in Fig. 6A, top and middle, GFP-Foxo1 protein was rapidly translocated from the nucleus to the cytoplasm 0.5–1 h after IGF1 or insulin addition to the MC-4 cells, which contain a high level of endogenous Runx2 (12, 20). Conversely, IGF1 treatment did not alter subcellular distribution of the GFP protein in the same cells (Fig. 6A, bottom). A similar result was observed in insulin-treated MC-4 cells (data not shown). Based on these observations, we reasoned that IGF1 or insulin should abolish or reduce Foxo1 inhibition of Runx2 by phosphorylation and subsequent translocation of Foxo1 to the cytoplasm from the nucleus where Runx2 is located and activates Bglap2 transcription. To test this, MC-4 cells were co-transfected with 6XRUNX2-LUC (Fig. 6B) or BGLAP2-LUC (Fig. 6C) and pRL-SV40 with and without expression vector for Foxo1. Twenty hours later, cells were treated with and without 10 ng/ml IGF1 as well as with and without 0.5 μg/ml of IGF1 neutralizing antibody or control IgG for 6 h, followed by dual luciferase assay. Importantly, Foxo1-mediated inhibition of Runx2 activity was completely reversed by the addition of IGF1 (Fig. 6B) or insulin (Fig. 7E). Furthermore, the effect of IGF1 was completely blocked by the addition of a specific IGF1 neutralizing antibody, but not by the control IgG. A similar result was obtained when the 657-bp BGLAP2-LUC was used in MC-4 cells (Fig. 6C). The IGF1 neutralizing antibody significantly diminished the level of Bglap2 but not Runx2 mRNAs in the MC-4 cells (Fig. 6D and E), which suggests that endogenous IGF1 signaling, probably via an autocrine mechanism, plays a critical role for maintaining the Bglap2 mRNA expression in osteoblasts. Taken together, these results suggest that IGF1/insulin favors Bglap2 expression probably by preventing Foxo1 from inhibiting Runx2 in osteoblasts.
FIGURE 6.
Effects of IGF1 and IGF1 neutralizing antibody on Foxo1 inhibition of Runx2 activity and Bglap expression in MC-4 cells. A, MC-4 cells were transfected with Foxo1-GFP or GFP expression plasmids and treated with and without IGF1 (10 ng/ml) or insulin (100 nm) for the indicated times. Images were captured by using a Olympus 1X70 fluorescent microscope (Olympus 1X70; Olympus America, Inc., Melville, NY) attached a digital camera (original magnification, ×100). B and C, MC-4 cells were transfected with 6XRUNX2-LUC (B) or BGLAP2-LUC (C) and pRL-SV40 with and without Foxo1 expression plasmid and treated with and without 10 ng/ml IGF1 as well as with and without 0.5 μg/ml IGF1 neutralizing antibody for 24 h, followed by dual luciferase assay. *, p < 0.05, versus 0 μg Foxo1. D and E, MC-4 cells were treated with and without 0.5 μg/ml IGF1 neutralizing antibody for 24 h, followed by quantitative real-time RT/PCR for Bglap2 and Runx2 mRNAs, which were normalized to Gapdh mRNA. *, p < 0.05, versus 0 μg IGF1.
FIGURE 7.
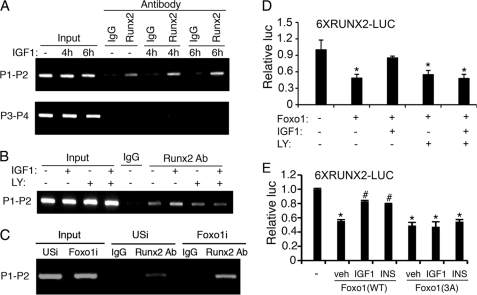
IGF1 increases Runx2 interactions with the Bglap2 promoter in a PI3K/AKT-dependent manner. A, ChIP assay of the Bglap2 promoter in MC-4 cells. Cells were incubated with and without 10 ng/ml IGF1 for the indicated times, followed by ChIP assay using a Runx2 antibody or a control IgG. B, PI3K/AKT inhibition blocks IGF1-induced interactions of Runx2 with the Bglap2 promoter. MC-4 cells were pre-treated with and without 10 μg/ml LY294002 for 2 h and further incubated with and without 10 ng/ml IGF1 and 10 μg/ml LY294002 for another 2 h, followed by ChIP assay using a Runx2 antibody or a control IgG. C, RNAi knockdown of Foxo1 increases Runx2 interaction with the Bglap2 promoter in MC-4 cells. Cells were infected with equal amount of adenoviral vectors for Foxo1 RNAi (Foxo1i) or control RNAi (USi). 48 h later, cells were harvested for ChIP assay using a Runx2 antibody or a control IgG. D, LY294002 inhibition of IGF1 activation of Runx2 activity. MC-4 cells were transfected with 6XRUNX2 and pRL-SV40 with and without Foxo1 expression plasmid and treated with and without 10 ng/ml IGF1 as well as with and without 10 μg/ml LY294002 for 12 h, followed by dual luciferase assay. *, p < 0.05, versus veh. E, insulin/IGF1 fail to prevent Foxo1(3A) from inhibiting Runx2. MC-4 cells were transfected with 6XRUNX2-LUC and pRL-SV40 with and without Foxo1(wt) and Foxo1(3A) and treated with and without 10 ng/ml IGF1 or insulin (100 nm), followed by dual luciferase assay. *, p < 0.05, versus 0 μg Foxo1; #, p < 0.05, versus veh.
IGF1 Increases Runx2 Interaction with the Bglap2 Promoter in a PI3K/AKT-dependent Manner in the MC-4 Cells
To determine whether Runx2 associates with the endogenous Bglap2 promoter in vivo, we performed ChIP assays using MC-4 cells with and without IGF1 treatment for the indicated times. Consistent with our previous observation (46), Runx2 specifically interacted with a chromatin fragment of the proximal Bglap2 promoter that contains the Runx2-binding site (primers P1/P2) (Fig. 7A). Importantly, this interaction was markedly stimulated by IGF1 treatment. In contrast, Runx2 antibody failed to immunoprecipitate a 3′ chromatin fragment from the transcribed region of the Bglap2 that contains no Runx2-binding sites (primers P3/P4) (46) (Fig. 7A, bottom). Further, the IGF1-induced increase in Runx2 binding to the Bglap2 promoter was abolished by treatment with LY294002, a specific inhibitor of the PI3K/AKT pathway (Fig. 7B). We next determine the effect of Foxo1 knockdown on Runx2 interaction with the Bglap2 promoter in MC-4 cells. Adenoviral overexpression of a mouse Foxo1 RNAi (Foxo1i) dramatically reduced the level of Foxo1 protein in MC-4 cells (supplemental Fig. S3). As shown in Fig. 7C, knockdown of Foxo1 markedly increased Runx2 binding to the Bglap2 promoter. As shown in Fig. 7D, the PI3K/AKT inhibition abrogated IGF1-induced reversal of Foxo1 inhibition of Runx2 activity. Foxo1 is phosphorylated at three highly conserved phosphorylation sites (Thr-24, Ser-256, and Ser-319) through the PI3K/AKT-dependent pathway, resulting in its nuclear exclusion and inhibition of target gene expression (1, 2, 6). To further study the role of the PI3K/AKT pathway in Foxo1 modulation of Runx2 activity, we compared the effects of wt Foxo1 and a mutant Foxo1(3A), in which the PI3K/AKT-dependent phosphorylation sites were mutated from either Thr or Ser to Ala, on Runx2 activity in the presence and absence of IGF1 or insulin. Significantly, in contrast to result from the wt Foxo1 group, neither IGF1 nor insulin prevented the mutant Foxo1(3A) from inhibiting Runx2 in the MC-4 cells (Fig. 7E).
DISCUSSION
This study identifies Foxo1 as a negative regulator of the bone transcription factor Runx2 in osteoblasts. Foxo1 physically interacts with Runx2 and inhibits Runx2 activity and decreases expression of the Bglap2 gene encoding osteocalcin Most importantly, we demonstrated that both IGF1 and insulin, which phosphorylate and export Foxo1 from the nucleus to the cytoplasm, prevent Foxo1 from inhibiting Runx2 and increase Runx2 activity, thereby favoring osteocalcin expression, an osteoblast-secreted hormone that plays a critical role in regulating glucose metabolism via its actions in β-cells in the pancreas.
Results from the present study established that Foxo1 indirectly down-regulates osteocalcin expression, at least in part, by inhibiting Runx2, a major upstream transcriptional activator of the Bglap2 gene in osteoblasts. Foxo1 interaction with Runx2 in osteoblasts, or when coexpressed in COS-7 cells, requires the presence of the Foxo1 C-terminal 360–456 amino acid region (96 amino acids) and the 242–258 amino acid (17 amino acids) region of Runx2. Using highly purified GST-fusion proteins in pull-down assays, we were able to demonstrate a direct physical interaction between Foxo1 and Runx2 proteins in vitro. Additionally, EMSA clearly revealed that purified GST-Foxo1, but not GST, dose-dependently inhibited Runx2 binding to its cognate site within the Bglap2 promoter. Therefore, our results demonstrate a novel molecular mechanism through which Foxo1 modulates Runx2 activity and thereby Bglap2 expression in osteoblasts. It should be noted that Foxo1 also decreases osteocalcin function by altering the carboxylation state. It is the uncarboxylated form of circulating osteocalcin that plays a critical role in regulating glucose homeostasis (i.e. enhanced insulin secretion by β-cells, insulin sensitivity and energy expenditure) (47). Foxo1 increases the expression of Esp (48), a gene that encodes a protein called protein tyrosine phosphatase (OST-PTP), which decreases the function of the osteocalcin protein by increasing its carboxylation through an as-yet-unknown mechanism (47). Therefore, Foxo1 regulates both osteocalcin expression and function.
A recent study showed that Foxo1 interacted with the Bglap2 promoter and the first intron region as demonstrated by ChIP assay in primary osteoblast cultures (48). However, potential Foxo1 DNA binding site(s) in the Bglap2 promoter were not identified and characterized in this study. Our study clearly established that Foxo1 inhibits Bglap2 expression, at least in part, via Runx2, a well-established transcriptional activator of the Bglap2 gene, and intact Runx2 DNA binding sites in the Bglap2 promoter.
Insulin signaling in osteoblasts was recently shown to be critical for postnatal bone acquisition and remodeling (10, 11). Fulzele et al. (10) recently showed that the insulin receptor (IR) is expressed in osteoblasts and that osteoblast-specific deletion of the IR severely impairs osteoblast differentiation. Mice lacking the IR in osteoblasts have reduced bone mass with increased adiposity and insulin resistance. Importantly, this study further revealed that the metabolic dysregulation in these mice was caused by reduced osteocalcin expression/function. Our results from the present study suggest that insulin favors osteocalcin expression by preventing Foxo1 from inhibiting Runx2, a key activator of the Bglap2 gene. This notion is strongly supported by: (i) the well-established role of IGF1/insulin signaling via the PI3K/AKT pathway that phosphorylates and exports Foxo1 from the nucleus to the cytoplasm, where it binds to 14-3-3 proteins; (ii) Foxo1 inhibition of Runx2 activity and Bglap2 expression was completely prevented by IGF1 treatment in osteoblasts, which expresses both the IR and IGF1 receptors; (iii) IGF1 neutralizing antibody reduced Runx2 activity and Bglap2 expression in osteoblasts; and (iv) IGF1 increased Runx2 interaction with the Bglap2 promoter in osteoblasts, which was abolished by PI3K/AKT inhibition. Interestingly, IR deficiency in osteoblasts caused increased expression of Twist2, a known Runx2 inhibitor (45). However, the molecular mechanism whereby insulin signaling modulates the expression of Twist2 is still unknown. Additionally, IGF1 and probably insulin were shown to activate Runx2 activity via the Erk1/2 MAPK pathway (30), a major signaling route in osteoblasts (14, 19). Erk1/2 phosphorylation sites in Runx2 were recently identified and characterized (57). This phosphorylation is critical for Runx2 activity and osteoblast differentiation as well as bone formation (57, 64). Collectively, these studies suggest that IGF1/insulin signaling favors Runx2 activity and Bglap2 expression via at least three distinct molecular mechanisms. Therefore, our findings from this study add a new layer to the molecular mechanisms through which IGF1/insulun signaling in osteoblasts modulates glucose homeostasis as well as bone metabolism.
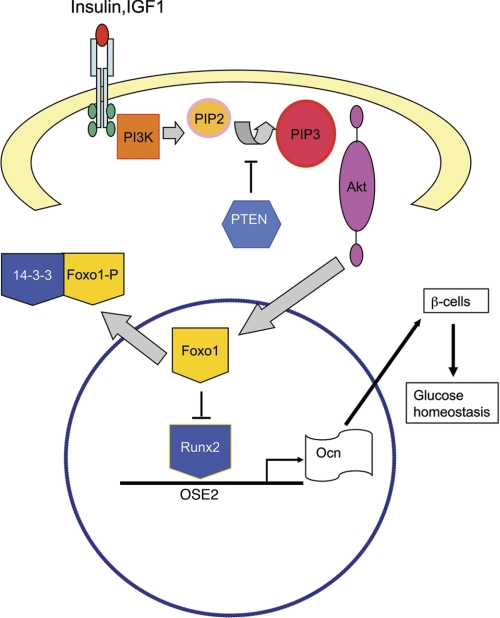
Based on the findings from this and other studies, we propose the following molecular model for Foxo1 modulation of osteocalcin expression in osteoblasts and the effect of IGF1/insulin signaling on this function of Foxo1 (Fig. 8). Binding of IGF1/insulin to their receptors, which are expressed in osteoblasts (10, 11), activates the PI3K/AKT pathway, which phosphorylates and translocates Foxo1 from the nucleus to the cytoplasm, where it binds to 14-3-3 chaperone proteins (65). This nuclear exclusion prevents Foxo1 from binding to and inhibiting Runx2, which results in enhanced Bglap2 expression. In the meantime, insulin and probably IGF1 down-regulate the expression of Twist2 (10), a known Runx2 suppressor, and thereby increase Runx2 activity (not shown), which further up-regulates osteocalcin expression. Finally, the uncarboxylated form of osteocalcin, via the circulation, regulates glucose homeostasis (i.e. insulin secretion and production and sensitivity) (47). It should be noted that Foxo1 can also bind independently to its target genes (not shown). In summary, we, for the first time to our knowledge, demonstrate that Foxo1 is a novel negative regulator of Runx2 and mediates IGF1/insulin actions in regulating osteocalcin expression in osteoblasts.
FIGURE 8.
Molecular model of Foxo1 and IGF1/insulin regulation of osteocalcin expression. In osteoblasts, in the absence of IGF1/insulin signaling, Foxo1 binds to Runx2 and inhibits Runx2 DNA binding to the OSE2 sites of the Bglap2 promoter, resulting in a reduction of osteocalcin (Ocn) expression. In the presence of IGF1/insulin signaling, Foxo1 is phosphorylated via the PI3K/AKT pathway and translocated from the nucleus to the cytoplasm where it binds to the 14-3-3 protein, thus eliminating the inhibition of Runx2. Runx2 serves as a transcriptional activator that binds to the Bglap2 promoter and increases osteocalcin expression, which favors glucose metabolism.
Supplementary Material
Acknowledgments
We thank Dr. Deborah L. Galson of the University of Pittsburgh for critical reading of this manuscript, Dr. Marc Montminy of the Salk Institute for Biological Studies for kindly providing us with adenoviral vectors for Foxo1 shRNA (Foxo1i) and control shRNA (USi) (59). We thank Dr. Min Luo of the University of Pittsburgh for technical assistance and Dr. Xiaoyan Zhang of Peking University for assistance in preparation of the Working Model figure (Fig. 8).
This work was supported, in whole or in part, by National Institutes of Health Grants DK072230, AR059647, and DK066301 and by Department of Defense Grant W81XWH-07-1-0160 and by Chinese Ministry of Science and Technology Grant 2009CB918902.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- IGF1
- insulin-like growth factor 1
- IRE
- insulin response element
- AA
- ascorbic acid
- NE
- nuclear extracts
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1. Accili D., Arden K. C. (2004) Cell 117, 421–426 [DOI] [PubMed] [Google Scholar]
- 2. Barthel A., Schmoll D., Unterman T. G. (2005) Trends Endocrinol. Metab. 16, 183–189 [DOI] [PubMed] [Google Scholar]
- 3. Lee S. S., Kennedy S., Tolonen A. C., Ruvkun G. (2003) Science 300, 644–647 [DOI] [PubMed] [Google Scholar]
- 4. Hwangbo D. S., Gershman B., Tu M. P., Palmer M., Tatar M. (2004) Nature 429, 562–566 [DOI] [PubMed] [Google Scholar]
- 5. Dong X. C., Copps K. D., Guo S., Li Y., Kollipara R., DePinho R. A., White M. F. (2008) Cell Metab. 8, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arden K. C. (2004) Mol. Cell 14, 416–418 [DOI] [PubMed] [Google Scholar]
- 7. van der Heide L. P., Jacobs F. M., Burbach J. P., Hoekman M. F., Smidt M. P. (2005) Biochem. J. 391, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Der Heide L. P., Hoekman M. F., Smidt M. P. (2004) Biochem. J. 380, 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang S., Wei D., Wang D., Phimphilai M., Krebsbach P. H., Franceschi R. T. (2003) J. Bone Miner Res. 18, 705–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fulzele K., Riddle R. C., DiGirolamo D. J., Cao X., Wan C., Chen D., Faugere M. C., Aja S., Hussain M. A., Bruning J. C., Clemens T. L. (2010) Cell 142, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferron M., Wei J., Yoshizawa T., Del Fattore A., DePinho R. A., Teti A., Ducy P., Karsenty G. (2010) Cell 142, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiao G., Jiang D., Gopalakrishnan R., Franceschi R. T. (2002) J. Biol. Chem. 277, 36181–36187 [DOI] [PubMed] [Google Scholar]
- 13. Gilbert L., He X., Farmer P., Rubin J., Drissi H., van Wijnen A. J., Lian J. B., Stein G. S., Nanes M. S. (2002) J. Biol. Chem. 277, 2695–2701 [DOI] [PubMed] [Google Scholar]
- 14. Xiao G., Gopalakrishnan R., Jiang D., Reith E., Benson M. D., Franceschi R. T. (2002) J. Bone Miner Res. 17, 101–110 [DOI] [PubMed] [Google Scholar]
- 15. Zhang X., Sobue T., Hurley M. M. (2002) Biochem. Biophys. Res. Commun. 290, 526–531 [DOI] [PubMed] [Google Scholar]
- 16. Hurley M. M., Tetradis S., Huang Y. F., Hock J., Kream B. E., Raisz L. G., Sabbieti M. G. (1999) J. Bone Miner Res. 14, 776–783 [DOI] [PubMed] [Google Scholar]
- 17. Montero A., Okada Y., Tomita M., Ito M., Tsurukami H., Nakamura T., Doetschman T., Coffin J. D., Hurley M. M. (2000) J. Clin. Invest. 105, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yakar S., Rosen C. J., Beamer W. G., Ackert-Bicknell C. L., Wu Y., Liu J. L., Ooi G. T., Setser J., Frystyk J., Boisclair Y. R., LeRoith D. (2002) J. Clin. Invest. 110, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao G., Jiang D., Thomas P., Benson M. D., Guan K., Karsenty G., Franceschi R. T. (2000) J. Biol. Chem. 275, 4453–4459 [DOI] [PubMed] [Google Scholar]
- 20. Xiao G., Wang D., Benson M. D., Karsenty G., Franceschi R. T. (1998) J. Biol. Chem. 273, 32988–32994 [DOI] [PubMed] [Google Scholar]
- 21. Yu S., Franceschi R. T., Luo M., Fan J., Jiang D., Cao H., Kwon T. G., Lai Y., Zhang J., Patrene K., Hankenson K., Roodman G. D., Xiao G. (2009) PLoS One 4, e7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 23. Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 24. Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 25. Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 26. Banerjee C., McCabe L. R., Choi J. Y., Hiebert S. W., Stein J. L., Stein G. S., Lian J. B. (1997) J. Cell. Biochem. 66, 1–8 [DOI] [PubMed] [Google Scholar]
- 27. Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H. C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T. M., Hanauer A., Karsenty G. (2004) Cell 117, 387–398 [DOI] [PubMed] [Google Scholar]
- 28. Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 29. Xiao G., Jiang D., Ge C., Zhao Z., Lai Y., Boules H., Phimphilai M., Yang X., Karsenty G., Franceschi R. T. (2005) J. Biol. Chem. 280, 30689–30696 [DOI] [PubMed] [Google Scholar]
- 30. Qiao M., Shapiro P., Kumar R., Passaniti A. (2004) J. Biol. Chem. 279, 42709–42718 [DOI] [PubMed] [Google Scholar]
- 31. Xiao G., Cui Y., Ducy P., Karsenty G., Franceschi R. T. (1997) Mol. Endocrinol. 11, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 32. Kundu M., Javed A., Jeon J. P., Horner A., Shum L., Eckhaus M., Muenke M., Lian J. B., Yang Y., Nuckolls G. H., Stein G. S., Liu P. P. (2002) Nat. Genet. 32, 639–644 [DOI] [PubMed] [Google Scholar]
- 33. Miller J., Horner A., Stacy T., Lowrey C., Lian J. B., Stein G., Nuckolls G. H., Speck N. A. (2002) Nat. Genet. 32, 645–649 [DOI] [PubMed] [Google Scholar]
- 34. Yoshida C. A., Furuichi T., Fujita T., Fukuyama R., Kanatani N., Kobayashi S., Satake M., Takada K., Komori T. (2002) Nat. Genet. 32, 633–638 [DOI] [PubMed] [Google Scholar]
- 35. Javed A., Guo B., Hiebert S., Choi J. Y., Green J., Zhao S. C., Osborne M. A., Stifani S., Stein J. L., Lian J. B., van Wijnen A. J., Stein G. S. (2000) J. Cell Sci. 113, 2221–2231 [DOI] [PubMed] [Google Scholar]
- 36. McLarren K. W., Theriault F. M., Stifani S. (2001) J. Biol. Chem. 276, 1578–1584 [DOI] [PubMed] [Google Scholar]
- 37. Selvamurugan N., Chou W. Y., Pearman A. T., Pulumati M. R., Partridge N. C. (1998) J. Biol. Chem. 273, 10647–10657 [DOI] [PubMed] [Google Scholar]
- 38. D'Alonzo R. C., Kowalski A. J., Denhardt D. T., Nickols G. A., Partridge N. C. (2002) J. Biol. Chem. 277, 24788–24798 [DOI] [PubMed] [Google Scholar]
- 39. Hess J., Porte D., Munz C., Angel P. (2001) J. Biol. Chem. 276, 20029–20038 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y. W., Yasui N., Ito K., Huang G., Fujii M., Hanai J., Nogami H., Ochi T., Miyazono K., Ito Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10549–10554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas D. M., Carty S. A., Piscopo D. M., Lee J. S., Wang W. F., Forrester W. C., Hinds P. W. (2001) Mol. Cell 8, 303–316 [DOI] [PubMed] [Google Scholar]
- 42. Kim S., Koga T., Isobe M., Kern B. E., Yokochi T., Chin Y. E., Karsenty G., Taniguchi T., Takayanagi H. (2003) Genes Dev. 17, 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aslam F., McCabe L., Frenkel B., van Wijnen A. J., Stein G. S., Lian J. B., Stein J. L. (1999) Endocrinology 140, 63–70 [DOI] [PubMed] [Google Scholar]
- 44. Kahler R. A., Westendorf J. J. (2003) J. Biol. Chem. 278, 11937–11944 [DOI] [PubMed] [Google Scholar]
- 45. Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D. M., Olson E. N., Justice M. J., Karsenty G. (2004) Dev. Cell 6, 423–435 [DOI] [PubMed] [Google Scholar]
- 46. Yu S., Jiang Y., Galson D. L., Luo M., Lai Y., Lu Y., Ouyang H. J., Zhang J., Xiao G. (2008) J. Biol. Chem. 283, 5542–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoshizawa T., Hinoi E., Jung D. Y., Kajimura D., Ferron M., Seo J., Graff J. M., Kim J. K., Karsenty G. (2009) J. Clin. Invest. 119, 2807–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rached M. T., Kode A., Silva B. C., Jung D. Y., Gray S., Ong H., Paik J. H., DePinho R. A., Kim J. K., Karsenty G., Kousteni S. (2010) J. Clin. Invest. 120, 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ducy P., Karsenty G. (1995) Mol. Cell. Biol. 15, 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang D., Christensen K., Chawla K., Xiao G., Krebsbach P. H., Franceschi R. T. (1999) J. Bone Miner Res. 14, 893–903 [DOI] [PubMed] [Google Scholar]
- 51. Benson M. D., Cui Y., Aubin J. E., Franceschi R. T. (1998) J. Bone Mineral Res. 12, Suppl 1, S277 (abstract F206) [Google Scholar]
- 52. Benson M. D., Bargeon J. L., Xiao G., Thomas P. E., Kim A., Cui Y., Franceschi R. T. (2000) J. Biol. Chem. 275, 13907–13917 [DOI] [PubMed] [Google Scholar]
- 53. Jiang D., Franceschi R. T., Boules H., Xiao G. (2004) J. Biol. Chem. 279, 5329–5337 [DOI] [PubMed] [Google Scholar]
- 54. Yu S., Franceschi R. T., Luo M., Zhang X., Jiang D., Lai Y., Jiang Y., Zhang J., Xiao G. (2008) Endocrinology 149, 1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kamagate A., Qu S., Perdomo G., Su D., Kim D. H., Slusher S., Meseck M., Dong H. H. (2008) J. Clin. Invest. 118, 2347–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu Z., Jiao P., Huang X., Feng B., Feng Y., Yang S., Hwang P., Du J., Nie Y., Xiao G., Xu H. (2010) J. Clin. Invest. 120, 3901–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ge C., Xiao G., Jiang D., Yang Q., Hatch N. E., Roca H., Franceschi R. T. (2009) J. Biol. Chem. 284, 32533–32543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Asada S., Daitoku H., Matsuzaki H., Saito T., Sudo T., Mukai H., Iwashita S., Kako K., Kishi T., Kasuya Y., Fukamizu A. (2007) Cell Signal. 19, 519–527 [DOI] [PubMed] [Google Scholar]
- 59. Liu Y., Dentin R., Chen D., Hedrick S., Ravnskjaer K., Schenk S., Milne J., Meyers D. J., Cole P., Yates J., 3rd, Olefsky J., Guarente L., Montminy M. (2008) Nature 456, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cao H., Yu S., Yao Z., Galson D. L., Jiang Y., Zhang X., Fan J., Lu B., Guan Y., Luo M., Lai Y., Zhu Y., Kurihara N., Patrene K., Roodman G. D., Xiao G. (2010) J. Clin. Invest. 120, 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Banerjee C., Hiebert S. W., Stein J. L., Lian J. B., Stein G. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4968–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Frendo J. L., Xiao G., Fuchs S., Franceschi R. T., Karsenty G., Ducy P. (1998) J. Biol. Chem. 273, 30509–30516 [DOI] [PubMed] [Google Scholar]
- 63. Zhang X., Yu S., Galson D. L., Luo M., Fan J., Zhang J., Guan Y., Xiao G. (2008) J. Cell. Biochem. 105, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ge C., Xiao G., Jiang D., Franceschi R. T. (2007) J. Cell Biol. 176, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.