Abstract
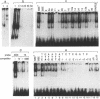
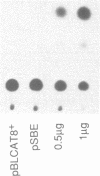
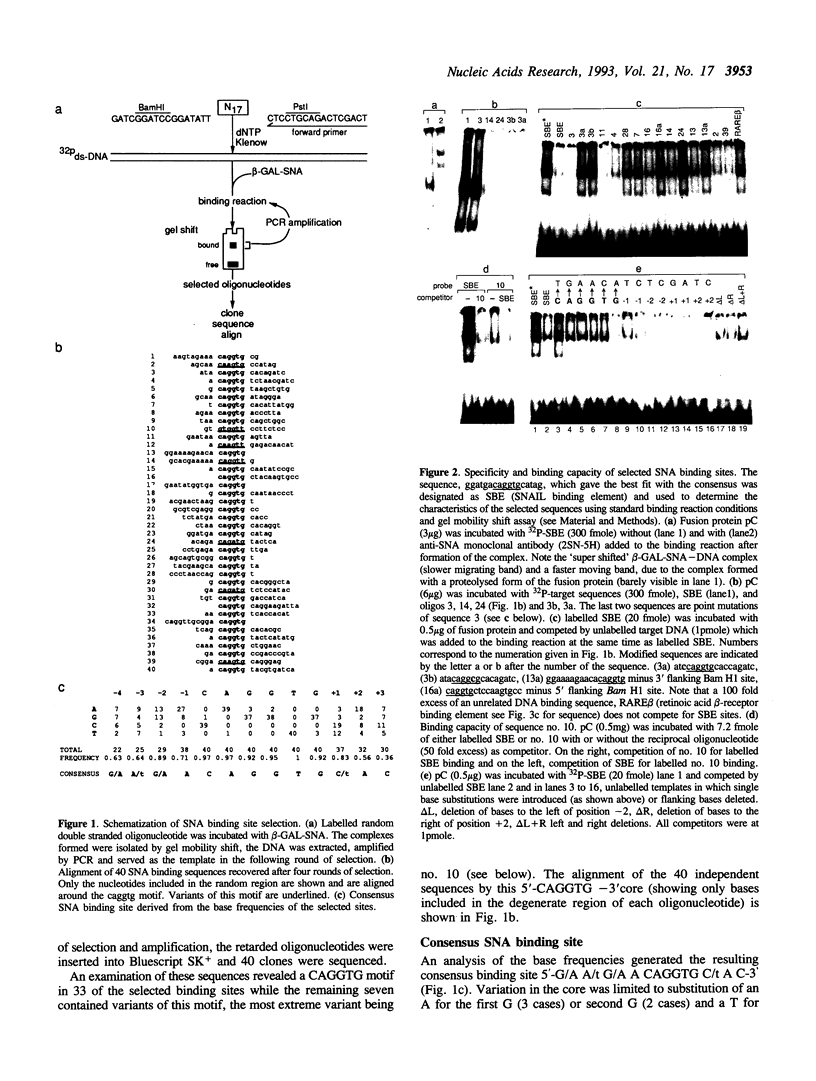
The Drosophila gene snail (sna) which encodes a zinc finger protein is essential for dorsal-ventral pattern formation in the developing embryo. We have defined a repertoire of SNAIL (SNA) binding sites using recombinant SNA proteins to select specific binding sequences from a pool of random sequence nucleotides. The bound sequences which were selected by multiple rounds of gel retardation and amplification by the polymerase chain reaction (PCR) were subsequently cloned and sequenced. The consensus sequence, 5'G/A A/t G/A A CAGGTG C/t A C 3', with a highly conserved core of 6 bases, CAGGTG, shares no significant homology with known binding sequences of other Drosophila zinc finger proteins. However, the CAGGTG core is identical to the core motif of aHLH (helix-loop-helix) binding sites. The strongest SNA binding is obtained with sequences containing this core motif whereas reduced binding is seen for sequences with canonical CANNTG HLH motifs. Interestingly, SNA binding is detected in the promoter region of the snail gene. Transient expression in co-transfection experiments using a SNA binding element (SBE) linked to a heterologous promoter indicates that SNA has the ability to function as a transcription activator.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberga A., Boulay J. L., Kempe E., Dennefeld C., Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991 Apr;111(4):983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- Bocquel M. T., Kumar V., Stricker C., Chambon P., Gronemeyer H. The contribution of the N- and C-terminal regions of steroid receptors to activation of transcription is both receptor and cell-specific. Nucleic Acids Res. 1989 Apr 11;17(7):2581–2595. doi: 10.1093/nar/17.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay J. L., Dennefeld C., Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. 1987 Nov 26-Dec 2Nature. 330(6146):395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Caudy M., Vässin H., Brand M., Tuma R., Jan L. Y., Jan Y. N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988 Dec 23;55(6):1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Diamond M. I., Miner J. N., Yoshinaga S. K., Yamamoto K. R. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990 Sep 14;249(4974):1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Grau Y., Carteret C., Simpson P. Mutations and Chromosomal Rearrangements Affecting the Expression of Snail, a Gene Involved in Embryonic Patterning in DROSOPHILA MELANOGASTER. Genetics. 1984 Oct;108(2):347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Issemann I., Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988 Jan 11;16(1):369–369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y., Gehring W. J. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987 Sep 11;50(6):963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Bier E., Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992 Sep;6(9):1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Jiang J., Rushlow C. A., Zhou Q., Small S., Levine M. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 1992 Aug;11(8):3147–3154. doi: 10.1002/j.1460-2075.1992.tb05387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y., Nambu J. R., Lieberman P. M., Crews S. T. Dorsal-ventral patterning in Drosophila: DNA binding of snail protein to the single-minded gene. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3414–3418. doi: 10.1073/pnas.89.8.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Hitpass L., Ryffel G. U., Heitlinger E., Cato A. C. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 1988 Jan 25;16(2):647–663. doi: 10.1093/nar/16.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Ip Y. T., Levine M., Arora K. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science. 1991 Oct 4;254(5028):118–122. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Saffman E. E., Kornfeld K., Hogness D. S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989 Jun 16;57(6):1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Locker J., Buzard G. A dictionary of transcription control sequences. DNA Seq. 1990;1(1):3–11. doi: 10.3109/10425179009041342. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Beal G., Papas T. S. Defining target sequences of DNA-binding proteins by random selection and PCR: determination of the GCN4 binding sequence repertoire. DNA Cell Biol. 1990 Dec;9(10):783–788. doi: 10.1089/dna.1990.9.783. [DOI] [PubMed] [Google Scholar]
- Meyer M. E., Gronemeyer H., Turcotte B., Bocquel M. T., Tasset D., Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989 May 5;57(3):433–442. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Franks R. G., Hu S., Crews S. T. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. 1990 Oct 5;63(1):63–75. doi: 10.1016/0092-8674(90)90288-p. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Brandl C. J., Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989 Jul;9(7):2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Courey A. J. The same dorsal binding site mediates both activation and repression in a context-dependent manner. EMBO J. 1992 May;11(5):1837–1842. doi: 10.1002/j.1460-2075.1992.tb05235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payre F., Vincent A. Genomic targets of the serendipity beta and delta zinc finger proteins and their respective DNA recognition sites. EMBO J. 1991 Sep;10(9):2533–2541. doi: 10.1002/j.1460-2075.1991.tb07793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y., Vaessin H., Jan L. Y., Jan Y. N. Neuroectoderm in Drosophila embryos is dependent on the mesoderm for positioning but not for formation. Genes Dev. 1991 Sep;5(9):1577–1588. doi: 10.1101/gad.5.9.1577. [DOI] [PubMed] [Google Scholar]
- Serfling E. Autoregulation--a common property of eukaryotic transcription factors? Trends Genet. 1989 May;5(5):131–133. doi: 10.1016/0168-9525(89)90049-8. [DOI] [PubMed] [Google Scholar]
- Stanojević D., Hoey T., Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. 1989 Sep 28;341(6240):331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Thisse C., Perrin-Schmitt F., Stoetzel C., Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991 Jun 28;65(7):1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- Treisman J., Desplan C. The products of the Drosophila gap genes hunchback and Krüppel bind to the hunchback promoters. Nature. 1989 Sep 28;341(6240):335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]