Abstract
In the title compound, C7H6O5, the three hydroxy groups on the ring are oriented in the same direction. There are two intramolecular O—H⋯O hydrogen bonds in the ring. In the crystal, there are several intermolecular O—H⋯O hydrogen bonds and a short contact of 2.7150 (18) Å between the O atoms of the para-OH groups of adjacent molecules.
Related literature
For the biological activity of the title compound, see: Gomes et al. (2003 ▶); Priscilla & Prince (2009 ▶); Lu et al. (2010 ▶). For the structure of gallic acid monohydrate, see: Okabe et al. (2001 ▶); Jiang et al. (2000 ▶); Billes et al. (2007 ▶).
Experimental
Crystal data
C7H6O5
M r = 170.12
Monoclinic,
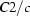
a = 25.629 (2) Å
b = 4.9211 (4) Å
c = 11.2217 (9) Å
β = 106.251 (1)°
V = 1358.77 (19) Å3
Z = 8
Mo Kα radiation
μ = 0.15 mm−1
T = 293 K
0.30 × 0.19 × 0.11 mm
Data collection
Bruker APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2003 ▶) T min = 0.916, T max = 1.000
7171 measured reflections
1254 independent reflections
1172 reflections with I > 2s(I)
R int = 0.022
Refinement
R[F 2 > 2σ(F 2)] = 0.032
wR(F 2) = 0.091
S = 1.06
1254 reflections
121 parameters
4 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.16 e Å−3
Δρmin = −0.21 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 2003 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶) and Mercury (Macrae et al., 2008) ▶; software used to prepare material for publication: SHELXL97 and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811007471/fj2397sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811007471/fj2397Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1 | 0.82 (1) | 2.19 (2) | 2.6625 (14) | 117 (1) |
| O3—H3⋯O2 | 0.82 (1) | 2.35 (2) | 2.7464 (14) | 110 (1) |
| O1—H1⋯O5i | 0.84 (1) | 1.89 (2) | 2.7324 (13) | 176 (2) |
| O3—H3⋯O3ii | 0.82 (1) | 2.04 (2) | 2.8167 (9) | 157 (2) |
| O4—H4⋯O5iii | 0.85 (2) | 1.81 (2) | 2.6570 (13) | 175 (2) |
Symmetry codes: (i) 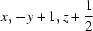 ; (ii)
; (ii) 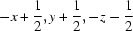 ; (iii)
; (iii) 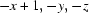 .
.
Acknowledgments
This work was supported by the Thailand Research Fund through the Royal Golden Jubilee PhD Program under grant No. PHD/0259/2549, the Prince of Songkla University under grant No. PHA520036S and the National Research University Project of Thailand’s Office of the Higher Education Commission.
supplementary crystallographic information
Comment
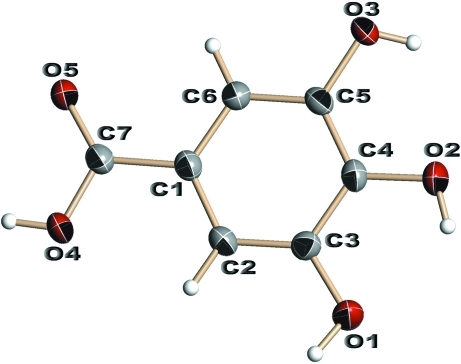
Gallic acid, 3,4,5-trihydroxybenzoic acid, has been reported to have various biological activities such as antioxidant, antimutagenic, anticarcinogenic, antihyperglycemic and cardioprotective effects (Gomes et al., 2003; Priscilla & Prince, 2009; Lu et al., 2010). It has been shown that the activity of polyphenolic compounds, including gallic acid, is dependent on their structural characteristics (Gomes et al., 2003). Thus, the investigation of its crystal structure is important for a better understanding of its biological functions. Recently, different crystal structures of gallic acid monohydrate have been reported (Jiang et al., 2000; Okabe et al., 2001; Billes et al., 2007). Here, for the first time, the crystal structure of anhydrous gallic acid (I) was determined. The molecular structure of I is planar [Fig.1]. All the H atoms of the three hydroxy groups are oriented in the same direction.
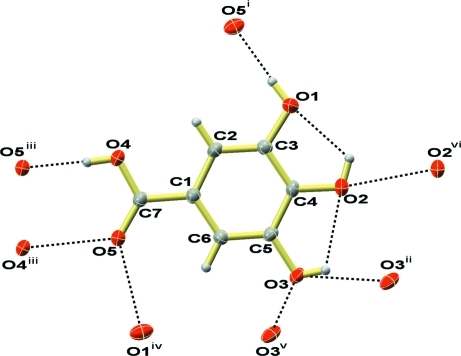
The intra-hydrogen bonds are found between these hydroxy groups, O2···O1 = 2.6625 (14) and O3···O2 = 2.7464 (14)Å [Table.1]. This agrees with the report of Okabe et al. (2001). However, this orientation is inconsistent with those described by Billes et al. (2007) and Jiang et al. (2000), in which one H atom of the hydroxy group is oriented in the opposite direction to the others. The dissimilarity between I and the gallic acid monohydrate structure reported by Okabe is the different orientation of their carboxyl groups in relation to the direction of the three hydroxy groups.
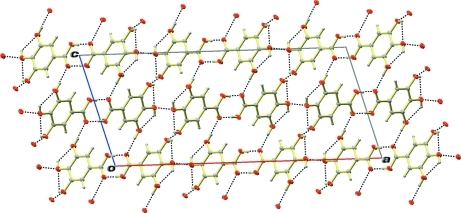
The inter-hydrogen bonds in the crystal packing of I are found between oxygen atoms,O3···O3ii [2.8167 (9) Å, symmetry code (ii): 1/2 - x, y + 1/2 - z - 1/2], O1···O5i [2.7324 (13) Å, symmetry code (i): x, -y + 1, z + 1/2] and O4···O5iii [2.6570 (13) Å, symmetry code (iii): -x + 1, -y, -z] [Table 1]. Moreover, the short contact between the oxygen of the hydroxy groups of the adjacent molecule is observed, O2···O2vi [2.7150 (18) Å, symmetry code (vi): 1/2 - x, 2.5 - y, -z]. All intra- and intermolecular interactions including short contacts are depicted in Fig. 2 and the packing interactions as plotted down the b axis are shown in Fig. 3.
Experimental
Gallic acid monohydrate was obtained from Fluka Chemie GmbH (Buchs, Switzerland). The anhydrous gallic acid crystals for this X-ray structure study were obtained by dissolving gallic acid monohydrate in diethyl ether followed by a slow evaporation of the solvent.
Refinement
The structure was solved by direct methods refined by a full-matrix least-squares procedure based on F2. All hydrogen atoms of oxygen atoms were located in a difference Fourier map and restrained to ride on their parent atoms, O—H = 0.82–0.85 Å with Uiso(H) = 1.2Ueq(O). The hydrogen atoms of C-sp2 atom are constrained, C—H = 0.96 Å with Uiso(H) = 1.2Ueq(C), respectively.
Figures
Fig. 1.
Molecular structure of I with thermal ellipsoids plotted at the 30% probability level.
Fig. 2.
The intra- and inter hydrogen bonds of I are shown. Symmetry code: i = x, 1 - y, 1/2 + z; ii = 1/2 - x, 1/2 + y, z - 1/2; iii = 1 - x, -y, -z; iv = x, 1 - y, z - 1/2; v = 1/2 - x, y - 1/2, -z - 1/2; vi = 1/2 - x, 2.5 - y, -z.
Fig. 3.
The packing interactions plotted down the b axis.
Crystal data
| C7H6O5 | F(000) = 704 |
| Mr = 170.12 | Dx = 1.663 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 3476 reflections |
| a = 25.629 (2) Å | θ = 3.3–28.1° |
| b = 4.9211 (4) Å | µ = 0.14 mm−1 |
| c = 11.2217 (9) Å | T = 293 K |
| β = 106.251 (1)° | Hexagon, colourless |
| V = 1358.77 (19) Å3 | 0.30 × 0.19 × 0.11 mm |
| Z = 8 |
Data collection
| Bruker APEX CCD area-detector diffractometer | 1254 independent reflections |
| Radiation source: fine-focus sealed tube | 1172 reflections with I > 2s(I) |
| graphite | Rint = 0.022 |
| Frames, each covering 0.3 ° in ω scans | θmax = 25.5°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2003) | h = −30→30 |
| Tmin = 0.916, Tmax = 1.000 | k = −5→5 |
| 7171 measured reflections | l = −13→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.032 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.091 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.054P)2 + 0.7476P] where P = (Fo2 + 2Fc2)/3 |
| 1254 reflections | (Δ/σ)max < 0.001 |
| 121 parameters | Δρmax = 0.16 e Å−3 |
| 4 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against all reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on all data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.40390 (5) | 0.4849 (2) | −0.00701 (11) | 0.0251 (3) | |
| C2 | 0.40861 (5) | 0.6475 (3) | 0.09733 (11) | 0.0267 (3) | |
| H2A | 0.4380 | 0.6271 | 0.1674 | 0.032* | |
| C3 | 0.36900 (5) | 0.8389 (3) | 0.09502 (11) | 0.0256 (3) | |
| C4 | 0.32438 (5) | 0.8667 (2) | −0.00919 (11) | 0.0249 (3) | |
| C5 | 0.32041 (5) | 0.7058 (2) | −0.11289 (11) | 0.0247 (3) | |
| C6 | 0.36006 (5) | 0.5156 (3) | −0.11223 (12) | 0.0262 (3) | |
| H6A | 0.3575 | 0.4083 | −0.1819 | 0.031* | |
| C7 | 0.44558 (5) | 0.2786 (2) | −0.00657 (11) | 0.0262 (3) | |
| O1 | 0.36873 (4) | 1.0116 (2) | 0.18952 (9) | 0.0367 (3) | |
| H1 | 0.3935 (6) | 0.969 (4) | 0.2539 (15) | 0.044* | |
| O2 | 0.28434 (4) | 1.0485 (2) | −0.01206 (10) | 0.0352 (3) | |
| H2 | 0.2920 (6) | 1.127 (3) | 0.0553 (13) | 0.042* | |
| O3 | 0.27680 (4) | 0.7268 (2) | −0.21619 (9) | 0.0334 (3) | |
| H3 | 0.2610 (7) | 0.872 (3) | −0.2151 (16) | 0.040* | |
| O4 | 0.48474 (4) | 0.2647 (2) | 0.09722 (9) | 0.0392 (3) | |
| H4 | 0.5076 (7) | 0.143 (3) | 0.0928 (17) | 0.047* | |
| O5 | 0.44465 (4) | 0.12950 (19) | −0.09537 (8) | 0.0314 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0237 (6) | 0.0240 (6) | 0.0263 (6) | 0.0027 (5) | 0.0050 (5) | 0.0020 (5) |
| C2 | 0.0244 (6) | 0.0292 (7) | 0.0234 (6) | 0.0041 (5) | 0.0015 (5) | 0.0025 (5) |
| C3 | 0.0284 (6) | 0.0248 (6) | 0.0230 (6) | 0.0007 (5) | 0.0062 (5) | 0.0000 (5) |
| C4 | 0.0224 (6) | 0.0219 (6) | 0.0301 (7) | 0.0036 (5) | 0.0070 (5) | 0.0028 (5) |
| C5 | 0.0214 (6) | 0.0236 (6) | 0.0254 (6) | −0.0008 (5) | 0.0005 (5) | 0.0018 (5) |
| C6 | 0.0260 (6) | 0.0252 (6) | 0.0256 (6) | 0.0021 (5) | 0.0042 (5) | −0.0027 (5) |
| C7 | 0.0238 (6) | 0.0264 (7) | 0.0263 (6) | 0.0028 (5) | 0.0037 (5) | 0.0014 (5) |
| O1 | 0.0420 (6) | 0.0387 (6) | 0.0251 (5) | 0.0130 (4) | 0.0020 (4) | −0.0060 (4) |
| O2 | 0.0309 (5) | 0.0345 (6) | 0.0369 (5) | 0.0131 (4) | 0.0038 (4) | −0.0051 (4) |
| O3 | 0.0265 (5) | 0.0293 (5) | 0.0346 (5) | 0.0063 (4) | −0.0074 (4) | −0.0050 (4) |
| O4 | 0.0332 (5) | 0.0433 (6) | 0.0324 (5) | 0.0189 (4) | −0.0054 (4) | −0.0078 (4) |
| O5 | 0.0292 (5) | 0.0322 (5) | 0.0289 (5) | 0.0101 (4) | 0.0019 (4) | −0.0037 (4) |
Geometric parameters (Å, °)
| C1—C6 | 1.3918 (17) | C5—O3 | 1.3706 (14) |
| C1—C2 | 1.3951 (18) | C5—C6 | 1.3800 (18) |
| C1—C7 | 1.4726 (17) | C6—H6A | 0.9300 |
| C2—C3 | 1.3796 (18) | C7—O5 | 1.2325 (15) |
| C2—H2A | 0.9300 | C7—O4 | 1.3093 (15) |
| C3—O1 | 1.3606 (15) | O1—H1 | 0.844 (14) |
| C3—C4 | 1.3949 (17) | O2—H2 | 0.821 (14) |
| C4—O2 | 1.3551 (15) | O3—H3 | 0.824 (14) |
| C4—C5 | 1.3874 (18) | O4—H4 | 0.850 (15) |
| C6—C1—C2 | 120.64 (11) | O3—C5—C4 | 121.15 (11) |
| C6—C1—C7 | 119.36 (11) | C6—C5—C4 | 120.10 (11) |
| C2—C1—C7 | 120.00 (11) | C5—C6—C1 | 119.77 (12) |
| C3—C2—C1 | 119.02 (11) | C5—C6—H6A | 120.1 |
| C3—C2—H2A | 120.5 | C1—C6—H6A | 120.1 |
| C1—C2—H2A | 120.5 | O5—C7—O4 | 121.64 (11) |
| O1—C3—C2 | 125.17 (11) | O5—C7—C1 | 123.91 (11) |
| O1—C3—C4 | 114.22 (11) | O4—C7—C1 | 114.45 (11) |
| C2—C3—C4 | 120.60 (11) | C3—O1—H1 | 110.2 (12) |
| O2—C4—C5 | 118.66 (11) | C4—O2—H2 | 107.7 (12) |
| O2—C4—C3 | 121.50 (11) | C5—O3—H3 | 109.9 (12) |
| C5—C4—C3 | 119.85 (11) | C7—O4—H4 | 110.8 (12) |
| O3—C5—C6 | 118.73 (11) | ||
| C6—C1—C2—C3 | 0.27 (19) | O2—C4—C5—C6 | −178.95 (11) |
| C7—C1—C2—C3 | −179.80 (11) | C3—C4—C5—C6 | 1.06 (19) |
| C1—C2—C3—O1 | −179.47 (12) | O3—C5—C6—C1 | −178.09 (11) |
| C1—C2—C3—C4 | 1.08 (19) | C4—C5—C6—C1 | 0.28 (19) |
| O1—C3—C4—O2 | −1.25 (18) | C2—C1—C6—C5 | −0.96 (19) |
| C2—C3—C4—O2 | 178.26 (11) | C7—C1—C6—C5 | 179.12 (11) |
| O1—C3—C4—C5 | 178.74 (11) | C6—C1—C7—O5 | 0.77 (19) |
| C2—C3—C4—C5 | −1.75 (19) | C2—C1—C7—O5 | −179.16 (12) |
| O2—C4—C5—O3 | −0.62 (18) | C6—C1—C7—O4 | −179.37 (11) |
| C3—C4—C5—O3 | 179.39 (11) | C2—C1—C7—O4 | 0.70 (17) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1 | 0.82 (1) | 2.19 (2) | 2.6625 (14) | 117 (1) |
| O3—H3···O2 | 0.82 (1) | 2.35 (2) | 2.7464 (14) | 110 (1) |
| O1—H1···O5i | 0.84 (1) | 1.89 (2) | 2.7324 (13) | 176 (2) |
| O3—H3···O3ii | 0.82 (1) | 2.04 (2) | 2.8167 (9) | 157 (2) |
| O4—H4···O5iii | 0.85 (2) | 1.81 (2) | 2.6570 (13) | 175 (2) |
Symmetry codes: (i) x, −y+1, z+1/2; (ii) −x+1/2, y+1/2, −z−1/2; (iii) −x+1, −y, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2397).
References
- Billes, F., Mohammed-Ziegler, I. & Bombicz, P. (2007). Vib. Spectrosc. 43, 193–202.
- Bruker (1998). SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2003). SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Gomes, C. A., Girao da Cruz, T., Andrade, J. L., Milhazes, N., Borges, F. & Marques, M. P. M. (2003). J. Med. Chem. 46, 5395–5401. [DOI] [PubMed]
- Jiang, R.-W., Ming, D.-S., But, P. P. H. & Mak, T. C. W. (2000). Acta Cryst. C56, 594–595. [DOI] [PubMed]
- Lu, Y., Jiang, F., Jiang, H., Wu, K., Zheng, X., Cai, Y., Katakowski, M., Chopp, M. & To, S. S. T. (2010). Eur. J. Pharmacol. 641, 102–107. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Okabe, N., Kyoyama, H. & Suzuki, M. (2001). Acta Cryst. E57, o764–o766.
- Priscilla, D. H. & Prince, P. S. M. (2009). Chem. Biol. Interact. 179, 118–124. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811007471/fj2397sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811007471/fj2397Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report