Abstract
In the title complex, [Zn(C13H12NO3)2], the ZnII ion is located on a twofold rotation axis and is coordinated by two bidentate Schiff base ligands in a distorted tetrahedral environment. The complex molecules are stacked in columns along the b axis through C—H⋯O hydrogen bonds.
Related literature
For the biological activity and applications of Zn(II) complexes, see: Csaszar et al. (1985 ▶); Greener et al. (1996 ▶); Gultneh et al. (1996 ▶); Aoki & Kimura (2004 ▶). For applications of furfurylamine derivatives, see: Camejo et al. (1992 ▶); Ledovskikh & Camejo (1993 ▶). For a related structure, see; Cai et al. (2010 ▶). 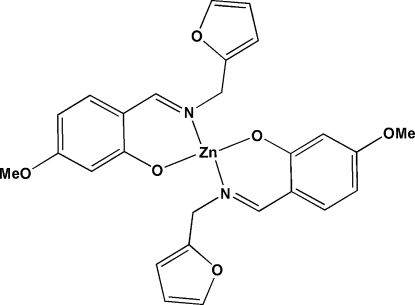
Experimental
Crystal data
[Zn(C13H12NO3)2]
M r = 525.84
Monoclinic,

a = 27.210 (4) Å
b = 5.2244 (7) Å
c = 19.007 (3) Å
β = 119.507 (2)°
V = 2351.5 (5) Å3
Z = 4
Mo Kα radiation
μ = 1.09 mm−1
T = 298 K
0.32 × 0.20 × 0.13 mm
Data collection
Bruker SMART CCD area detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2000 ▶) T min = 0.722, T max = 0.871
6855 measured reflections
2303 independent reflections
2062 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.128
S = 1.01
2303 reflections
160 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.64 e Å−3
Δρmin = −0.31 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811011536/is2670sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011536/is2670Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C9—H9A⋯O1i | 0.97 | 2.32 | 3.292 (6) | 177 |
Symmetry code: (i)  .
.
Acknowledgments
This work was supported by the Education Office of Hubei Province (D20104104).
supplementary crystallographic information
Comment
Zinc(II) complexes with the chelate ligands are extensively investigated as models for the active site of carbonic anhydrase and other hydrolytically active enzymes (Greener et al., 1996; Gultneh et al., 1996). Zinc(II) complexes are also studied on the role of zinc(II) structural properties in protein folding (Aoki & Kimura, 2004). Interestingly, zinc(II) complexes are becoming important in pharmaceutical, dye, plastic industries and for liquid crystal technology (Csaszar et al., 1985). In addition, the furfuryl amine and its derivatives are widely used as bioactive antibacterial agents and as steel corrosion inhibitors in recent research (Ledovskikh & Camejo, 1993; Camejo et al., 1992). By taking the biological importance of furfurylamine into account, we designed the title complex containing nitrogen-oxygen donor atoms coordinated with zinc(II).
The title complex reported here is the mononuclear zinc(II) complex of Schiff-base ligand, derived from the condensation of 4-methoxysalicylaldehyde and furfuryl amine (Fig. 1). The zinc(II) atom has a distorted tetrahedral coordination formed by two N atoms and two O atoms from two Schiff-base ligands. The bond distances of Zn—O and Zn—N are 1.925 (3) and 1.984 (3) Å, respectively. The dihedral angle between the coordination planes (N1/Zn1/O1 and N1A/Zn1A/O1A) is 87.24 (8)° (symmetry code A: - x, y, 1/2 - z), which is slightly larger than that of 84.43 (6)° between the corresponding coordination planes around zinc(II) atom observed in the similar Schiff base zinc(II) complex, bis(3,5-dibromo-N-benzylsalicylaldiminato-N,O)zinc(II) (Cai et al., 2010). The O1—Zn1—O1 angle is 109.27 (18)° and the N1—Zn1—N1 angle is 120.08 (18)°. The other angles subtended at the Zn(II) ion in (ZnN2O2) is in the range of 96.69 (11)–117.49 (12)°.
In the crystal structure, the molecules are linked via intermolecular C—H···O hydrogen bonds forming a column along the b axis (Fig. 2).
Experimental
4-Methoxysalicylaldehyde (304 mg, 2 mmol) and furfurylamine (194 mg, 2 mmol) were dissolved in an aqueous methanol solution (25 mL).The mixture was stirred at room temperature for 1 h to give a clear yellow solution, which was added to a solutionof Zn(NO3)2.6H2O (298 mg, 1 mmol) in methanol (10 mL). The mixture was stirred for 30 min at room temperature to give a yellow solution and then filtered. The yellow single crystals suitable for X-ray analysis were obtained by slowly evaporating the above filtrate at room temperature. The crystals were isolated and dried in a vacuum desiccator containing anhydrous CaCl2, in about 71% yield. Anal. Calcd for C26H24ZnN2O6: C 59.38, H 4.60, N 5.33%. Found: C 59.20, H 4.73 N, 5.30%.
Refinement
All the H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms, with C—H distances of 0.93–0.97 Å, and with Uiso(H) = 1.2Ueq(C) or 1.5Ueq(methyl C). A rigid bond restraint was applied for atoms C11 and C12.
Figures
Fig. 1.
The molecular structure of the title compound, with the atom labeling scheme. Displacement ellipsoids are drawn at the 30% probability level. The suffix A corresponds to symmetry code - x, y, 1/2 - z.
Fig. 2.
A packing diagram of the title compound, viewed along the b axis.
Crystal data
| [Zn(C13H12NO3)2] | F(000) = 1088 |
| Mr = 525.84 | Dx = 1.485 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 2246 reflections |
| a = 27.210 (4) Å | θ = 2.5–23.7° |
| b = 5.2244 (7) Å | µ = 1.09 mm−1 |
| c = 19.007 (3) Å | T = 298 K |
| β = 119.507 (2)° | Block, yellow |
| V = 2351.5 (5) Å3 | 0.32 × 0.20 × 0.13 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area detector diffractometer | 2303 independent reflections |
| Radiation source: fine-focus sealed tube | 2062 reflections with I > 2σ(I) |
| graphite | Rint = 0.026 |
| φ and ω scans | θmax = 26.0°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2000) | h = −33→33 |
| Tmin = 0.722, Tmax = 0.871 | k = −6→6 |
| 6855 measured reflections | l = −23→20 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.128 | H-atom parameters constrained |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0784P)2 + 2.4971P] where P = (Fo2 + 2Fc2)/3 |
| 2303 reflections | (Δ/σ)max < 0.001 |
| 160 parameters | Δρmax = 0.64 e Å−3 |
| 1 restraint | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Zn1 | 0.0000 | 0.78566 (12) | 0.2500 | 0.0427 (3) | |
| N1 | 0.02638 (12) | 0.5960 (6) | 0.18435 (18) | 0.0394 (7) | |
| O1 | 0.06578 (11) | 0.9989 (6) | 0.30670 (18) | 0.0549 (7) | |
| O2 | 0.24426 (12) | 1.3537 (7) | 0.3745 (2) | 0.0665 (9) | |
| O3 | −0.03914 (17) | 0.7542 (6) | 0.0242 (2) | 0.0729 (10) | |
| C1 | 0.11535 (15) | 0.8329 (7) | 0.2413 (2) | 0.0403 (8) | |
| C2 | 0.10942 (14) | 1.0010 (7) | 0.2954 (2) | 0.0402 (8) | |
| C3 | 0.15270 (16) | 1.1795 (8) | 0.3399 (3) | 0.0465 (9) | |
| H3 | 0.1490 | 1.2918 | 0.3750 | 0.056* | |
| C4 | 0.20061 (16) | 1.1895 (8) | 0.3319 (3) | 0.0489 (9) | |
| C5 | 0.20638 (16) | 1.0283 (9) | 0.2781 (3) | 0.0550 (10) | |
| H5 | 0.2382 | 1.0385 | 0.2720 | 0.066* | |
| C6 | 0.16505 (16) | 0.8560 (9) | 0.2348 (3) | 0.0505 (10) | |
| H6 | 0.1694 | 0.7483 | 0.1993 | 0.061* | |
| C7 | 0.2397 (2) | 1.5304 (9) | 0.4279 (3) | 0.0675 (13) | |
| H7A | 0.2080 | 1.6411 | 0.3976 | 0.101* | |
| H7B | 0.2736 | 1.6309 | 0.4545 | 0.101* | |
| H7C | 0.2345 | 1.4386 | 0.4675 | 0.101* | |
| C8 | 0.07515 (15) | 0.6467 (7) | 0.1911 (2) | 0.0417 (8) | |
| H8 | 0.0851 | 0.5491 | 0.1591 | 0.050* | |
| C9 | −0.00946 (17) | 0.4112 (8) | 0.1224 (2) | 0.0508 (9) | |
| H9A | −0.0260 | 0.2943 | 0.1446 | 0.061* | |
| H9B | 0.0134 | 0.3118 | 0.1061 | 0.061* | |
| C10 | −0.05497 (17) | 0.5418 (8) | 0.0510 (2) | 0.0517 (10) | |
| C11 | −0.1095 (2) | 0.4911 (15) | 0.0025 (3) | 0.0929 (17) | |
| H11 | −0.1303 | 0.3563 | 0.0065 | 0.111* | |
| C12 | −0.1294 (3) | 0.6947 (16) | −0.0583 (4) | 0.102 (2) | |
| H12 | −0.1661 | 0.7157 | −0.1007 | 0.123* | |
| C13 | −0.0861 (3) | 0.8419 (15) | −0.0423 (3) | 0.103 (2) | |
| H13 | −0.0874 | 0.9855 | −0.0721 | 0.123* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Zn1 | 0.0333 (4) | 0.0510 (4) | 0.0525 (4) | 0.000 | 0.0278 (3) | 0.000 |
| N1 | 0.0346 (15) | 0.0452 (16) | 0.0426 (16) | 0.0013 (13) | 0.0222 (13) | 0.0020 (13) |
| O1 | 0.0396 (15) | 0.0674 (18) | 0.0709 (18) | −0.0102 (13) | 0.0372 (14) | −0.0200 (15) |
| O2 | 0.0414 (16) | 0.070 (2) | 0.083 (2) | −0.0157 (14) | 0.0268 (16) | −0.0085 (18) |
| O3 | 0.079 (2) | 0.070 (2) | 0.064 (2) | 0.0066 (17) | 0.0316 (19) | 0.0094 (16) |
| C1 | 0.0326 (18) | 0.047 (2) | 0.045 (2) | 0.0031 (15) | 0.0226 (16) | 0.0047 (16) |
| C2 | 0.0301 (17) | 0.0457 (19) | 0.047 (2) | 0.0029 (14) | 0.0205 (15) | 0.0023 (16) |
| C3 | 0.038 (2) | 0.047 (2) | 0.054 (2) | −0.0002 (16) | 0.0232 (18) | −0.0007 (17) |
| C4 | 0.0342 (19) | 0.052 (2) | 0.056 (2) | −0.0027 (16) | 0.0188 (18) | 0.0068 (18) |
| C5 | 0.0336 (19) | 0.074 (3) | 0.066 (3) | 0.0003 (18) | 0.0304 (19) | 0.005 (2) |
| C6 | 0.037 (2) | 0.065 (3) | 0.057 (2) | 0.0034 (18) | 0.0289 (18) | −0.002 (2) |
| C7 | 0.059 (3) | 0.060 (3) | 0.068 (3) | −0.018 (2) | 0.019 (2) | −0.003 (2) |
| C8 | 0.0398 (19) | 0.047 (2) | 0.044 (2) | 0.0051 (15) | 0.0252 (17) | 0.0000 (16) |
| C9 | 0.052 (2) | 0.045 (2) | 0.060 (2) | −0.0084 (18) | 0.031 (2) | −0.0046 (19) |
| C10 | 0.043 (2) | 0.065 (3) | 0.050 (2) | −0.0058 (18) | 0.0245 (18) | −0.014 (2) |
| C11 | 0.050 (3) | 0.155 (5) | 0.079 (3) | −0.021 (3) | 0.036 (3) | −0.049 (3) |
| C12 | 0.064 (4) | 0.167 (7) | 0.056 (3) | 0.042 (4) | 0.015 (3) | −0.015 (3) |
| C13 | 0.119 (6) | 0.113 (5) | 0.055 (3) | 0.052 (5) | 0.027 (4) | 0.014 (3) |
Geometric parameters (Å, °)
| Zn1—O1i | 1.925 (3) | C4—C5 | 1.391 (6) |
| Zn1—O1 | 1.925 (3) | C5—C6 | 1.357 (6) |
| Zn1—N1 | 1.984 (3) | C5—H5 | 0.9300 |
| Zn1—N1i | 1.984 (3) | C6—H6 | 0.9300 |
| N1—C8 | 1.296 (4) | C7—H7A | 0.9600 |
| N1—C9 | 1.463 (5) | C7—H7B | 0.9600 |
| O1—C2 | 1.306 (4) | C7—H7C | 0.9600 |
| O2—C4 | 1.362 (5) | C8—H8 | 0.9300 |
| O2—C7 | 1.422 (6) | C9—C10 | 1.479 (6) |
| O3—C13 | 1.360 (7) | C9—H9A | 0.9700 |
| O3—C10 | 1.375 (5) | C9—H9B | 0.9700 |
| C1—C2 | 1.422 (5) | C10—C11 | 1.332 (6) |
| C1—C6 | 1.422 (5) | C11—C12 | 1.465 (10) |
| C1—C8 | 1.423 (5) | C11—H11 | 0.9300 |
| C2—C3 | 1.411 (5) | C12—C13 | 1.311 (11) |
| C3—C4 | 1.386 (6) | C12—H12 | 0.9300 |
| C3—H3 | 0.9300 | C13—H13 | 0.9300 |
| O1i—Zn1—O1 | 109.27 (18) | C1—C6—H6 | 118.6 |
| O1i—Zn1—N1 | 117.49 (12) | O2—C7—H7A | 109.5 |
| O1—Zn1—N1 | 96.69 (11) | O2—C7—H7B | 109.5 |
| O1i—Zn1—N1i | 96.69 (11) | H7A—C7—H7B | 109.5 |
| O1—Zn1—N1i | 117.49 (12) | O2—C7—H7C | 109.5 |
| N1—Zn1—N1i | 120.08 (18) | H7A—C7—H7C | 109.5 |
| C8—N1—C9 | 117.3 (3) | H7B—C7—H7C | 109.5 |
| C8—N1—Zn1 | 120.4 (3) | N1—C8—C1 | 128.1 (3) |
| C9—N1—Zn1 | 122.1 (2) | N1—C8—H8 | 115.9 |
| C2—O1—Zn1 | 125.6 (2) | C1—C8—H8 | 115.9 |
| C4—O2—C7 | 118.4 (3) | N1—C9—C10 | 111.1 (3) |
| C13—O3—C10 | 107.1 (5) | N1—C9—H9A | 109.4 |
| C2—C1—C6 | 117.5 (3) | C10—C9—H9A | 109.4 |
| C2—C1—C8 | 125.7 (3) | N1—C9—H9B | 109.4 |
| C6—C1—C8 | 116.7 (3) | C10—C9—H9B | 109.4 |
| O1—C2—C3 | 117.7 (3) | H9A—C9—H9B | 108.0 |
| O1—C2—C1 | 123.4 (3) | C11—C10—O3 | 110.5 (5) |
| C3—C2—C1 | 118.9 (3) | C11—C10—C9 | 133.4 (5) |
| C4—C3—C2 | 120.8 (4) | O3—C10—C9 | 116.0 (4) |
| C4—C3—H3 | 119.6 | C10—C11—C12 | 104.7 (6) |
| C2—C3—H3 | 119.6 | C10—C11—H11 | 127.6 |
| O2—C4—C3 | 123.4 (4) | C12—C11—H11 | 127.6 |
| O2—C4—C5 | 115.9 (4) | C13—C12—C11 | 107.5 (5) |
| C3—C4—C5 | 120.7 (4) | C13—C12—H12 | 126.2 |
| C6—C5—C4 | 119.2 (3) | C11—C12—H12 | 126.2 |
| C6—C5—H5 | 120.4 | C12—C13—O3 | 110.1 (7) |
| C4—C5—H5 | 120.4 | C12—C13—H13 | 124.9 |
| C5—C6—C1 | 122.9 (4) | O3—C13—H13 | 124.9 |
| C5—C6—H6 | 118.6 |
Symmetry codes: (i) −x, y, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C9—H9A···O1ii | 0.97 | 2.32 | 3.292 (6) | 177 |
Symmetry codes: (ii) −x, y−1, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2670).
References
- Aoki, S. & Kimura, E. (2004). Chem. Rev. 104, 769–788. [DOI] [PubMed]
- Bruker (2000). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cai, Y., Wang, W., Qin, X., Li, Y. & Chen, W. (2010). Z. Kristallogr. New Cryst. Struct. 225, 365–366.
- Camejo, J. J., Marrero, R., Gonzalez, C., Dominguez, J. A. & Castro, I. S. D. (1992). Cana. Azucar. 26, 47–52.
- Csaszar, J., Morvay, J. & Herczeg, O. (1985). Acta Phys. Chem. 31, 717–722.
- Greener, B., Moore, M. H. & Walton, P. H. (1996). J. Chem. Soc. Chem. Commun. pp. 27–28.
- Gultneh, Y., Ahvazi, B., Blaise, D., Butcher, R. J. & Jasinski, J. (1996). Inorg. Chim. Acta, 241, 31–38.
- Ledovskikh, V. M. & Camejo, J. J. (1993). Zashch. Met. 29, 597–603.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811011536/is2670sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011536/is2670Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




