Abstract
Zoospores play an important role in the infection of plant and animal hosts by oomycetes and other zoosporic fungi. In this study, six fluorescent Pseudomonas isolates with zoosporicidal activities were obtained from the wheat rhizosphere. Zoospores of multiple oomycetes, including Pythium species, Albugo candida, and Phytophthora infestans, were rendered immotile within 30 s of exposure to cell suspensions or cell culture supernatants of the six isolates, and subsequent lysis occurred within 60 s. The representative strain SS101, identified as Pseudomonas fluorescens biovar II, reduced the surface tension of water from 73 to 30 mN m−1. The application of cell suspensions of strain SS101 to soil or hyacinth bulbs provided significant protection against root rot caused by Pythium intermedium. Five Tn5 mutants of strain SS101lacked the abilities to reduce the surface tension of water and to cause lysis of zoospores. Genetic characterization of two surfactant-deficient mutants showed that the transposons had integrated into condensation domains of peptide synthetases. A partially purified extract from strain SS101 reduced the surface tension of water to 30 mN m−1 and reached the critical micelle concentration at 25 μg ml−1. Reverse-phase high-performance liquid chromatography yielded eight different fractions, five of which had surface activity and caused lysis of zoospores. Mass spectrometry and nuclear magnetic resonance analyses allowed the identification of the main constituent as a cyclic lipopeptide (1,139 Da) containing nine amino acids and a 10-carbon hydroxy fatty acid. The other four zoosporicidal fractions were closely related to the main constituent, with molecular massesranging from 1,111 to 1,169 Da.
Microbial compounds that alter the conditions prevailing at a surface or interface are often referred to as adjuvants, bioemulsifiers, or biosurfactants (5, 14, 51). A variety of microorganisms, including bacteria, fungi, and yeasts, have been reported to produce biosurfactants (16, 36). Several of these biosurfactants are well described chemically and categorized into high- and low-molecular-mass compounds. The low-molecular-mass biosurfactants include glycolipids and lipopeptides, such as rhamnolipids and surfactin. The high-molecular-mass compounds include proteins and lipoproteins, or complex mixtures of these polymers. Although biosurfactants are structurally diverse, they all have an amphiphilic nature, i.e., they contain both hydrophobic and hydrophilic groups. The hydrophobic moieties are usually saturated, unsaturated, or hydroxylated fatty acids or apolar amino acids, like leucin and isoleucin. The hydrophilic moieties consist of mono-, di-, or polysaccharides, carboxylic acids, polar amino acids, or peptides (36).
Among the bacterial genera, Pseudomonas spp. have been reported to produce biosurfactants (14, 33, 36). Pseudomonas spp. are common inhabitants of soil and rhizosphere environments and have received considerable attention in the areas of bioremediation of xenobiotics and biological control of plant-pathogenic fungi. In the area of bioremediation, surfactant-producing Pseudomonas spp. have been implicated in facilitating the degradation of ubiquitous pollutants, such as polycyclic aromatic hydrocarbons and n-alkanes (2, 12). In the area of biological control of plant-pathogenic fungi, the potential of biosurfactants produced by Pseudomonas spp. was recently recognized. Rhamnolipids produced by strains of Pseudomonas aeruginosa were shown to be highly effective against plant pathogens, including Pythium aphanidermatum, Plasmopara lactucae-radicis, and Phytophthora capsici (43). Purified rhamnolipids caused cessation of motility and the lysis of entire zoospore populations within <1 min. The introduction of a rhamnolipid-producing strain into a recirculating hydroponic system gave good, although transient, control of P. capsici on pepper (43). Kim et al. (27) confirmed and extended these observations by showing that rhamnolipid B, produced by P. aeruginosa B5, has not only lytic effects on zoospores but also inhibitory activity against the spore germination and hyphal growth of several other pathogens. Mycelial growth of P. capsici and spore germination of Colletotrichum orbiculare were inhibited in vitro, and the diseases caused by these pathogens were suppressed in pepper and cucumber plants, respectively, by application of purified rhamnolipid B to leaves (27).
Several cyclic lipopeptide surfactants with antibiotic properties were recently proposed as biological compounds for the control of plant-pathogenic fungi (29, 31-33, 48). Viscosinamide, produced by soil-inhabiting Pseudomonas sp. strain DR54, was shown to induce encystment of Pythium zoospores and to adversely affect mycelia of Rhizoctonia solani and Pythium ultimum, causing reduced growth and intracellular activity, hyphal swellings, increased branching, and rosette formation (18, 46, 47). The specific cyclic lipopeptide amphisin, produced by Pseudomonas sp. strain DSS73, appeared to play an important role in the surface motility of the producing strain, allowing efficient containment of root-infecting plant-pathogenic fungi (1). Furthermore, in combination with cell wall-degrading enzymes of Trichoderma atroviride, lipodepsipeptides produced by the pathogen Pseudomonas syringae pv. syringae acted synergistically in antagonism to various plant-pathogenic fungi (15). Collectively, these studies clearly indicate the potential of biosurfactant-producing bacteria for crop protection.
In this study, surfactant-producing Pseudomonas isolates were obtained from the rhizosphere of wheat and screened for activity against zoospores of multiple oomycete pathogens, including Pythium species and Phytophthora infestans. The biocontrol ability of the representative strain Pseudomonas fluorescens SS101 was tested in hyacinth flower bulbs against root rot caused by Pythium intermedium. Two genes involved in surfactant production by P. fluorescens strain SS101 were obtained by random Tn5 mutagenesis followed by anchored PCR and subsequent sequencing of the Tn5 flanking regions. Tn5 mutants were characterized phenotypically, and their activities against zoospores of oomycete pathogens were compared to that of their parental strain. The surfactants produced by strain SS101 were isolated by reverse-phase high-pressure liquid chromatography (RP-HPLC), and their activities were assessed in bioassays. The identity of the main surface-active constituent was determined by liquid chromatography-mass spectrometry (LC-MS) and nuclear magnetic resonance (NMR).
MATERIALS AND METHODS
Microorganisms and growth conditions.
The bacterial strains and oomycetes used in this study are listed in Table 1. Naturally occurring fluorescent Pseudomonas spp. were isolated from the roots of wheat on King's medium B agar supplemented with chloramphenicol (13 μg ml−1), ampicillin (40 μg ml−1), and cycloheximide (100 μg ml−1) (KMB+) (41). SW medium (40), containing cetyltrimethylammoniumbromide (CTAB)-methylene blue, was used to screen for surfactant-producing Pseudomonas isolates: isolates that produce anionic surfactants are able to form a halo around the colony after 3 to 5 days of growth on SW medium. All bacterial strains were stored at −80°C in Luria-Bertani or KMB broth supplemented with 40% (vol/vol) glycerol. Spontaneous rifampin-resistant derivatives of Pseudomonas sp. strain SS101 were selected on KMB supplemented with rifampin (100 μg ml−1). Escherichia coli strains were grown in Luria-Bertani broth amended with the appropriate antibiotics (Table 1). Tn5 mutants of strain SS101 with the ability to produce surfactants disrupted are resistant to rifampin (100 μg ml−1) and kanamycin (100 μg ml−1).
TABLE 1.
Microorganisms used in this study
| Microorganism | Reference or sourcea |
|---|---|
| Bacteria | |
| E. coli HB101pRK2013 | 13 |
| E. coli S17λ pir pUT | 9 |
| P. fluorescens SS101 | This study |
| P. fluorescens SV7 | This study |
| P. fluorescens SSB17 | 10 |
| P. putida WCS358 | UU |
| P. aeruginosa PAO1 | 35 |
| P. aeruginosa DSMZ1128 | DSMZ |
| Oomycetes | |
| P. intermedium P52 | PPO |
| P. ultimum var. sporangiiferum 219.65 | CBS |
| A. candida FLS 000 | WU |
| P. infestans 88069 | WU |
UU, P. A. H. M. Bakker, Section Plant Pathology, Utrecht University (Utrecht, The Netherlands); DSMZ, Deutsche Sammlung von Mikroorganismen und Zelculturen (Braunschweig, Germany); PPO, Applied Plant Research, Section Flower Bulbs (Lisse, The Netherlands); CBS, Dutch Collection of Microorganisms (Utrecht, The Netherlands); WU, Laboratory of Phytopathology, Wageningen University (Wageningen, The Netherlands).
All Pythium isolates were routinely grown on potato dextrose agar (Oxoid Ltd., Basingstoke, Hampshire, England). P. infestans was grown on rye agar medium (4). Mycelial plugs of all Pythium isolates and P. infestans were stored in sterile mineral oil at 15°C. The obligate oomycete Albugo candida was allowed to multiply on Brussels sprout plants grown in controlled climatic chambers at 18°C. Leaves infested with A. candida were stored in plastic bags at −20°C.
Production of zoospores.
Zoospores of Pythium ultimum var. sporangiiferum and P. intermedium were produced using a modification of a method described by Zhou and Paulitz (52). P. ultimum var. sporangiiferum and P. intermedium were grown for 3 days at 25°C on V8 juice agar (N. V. Campbell Foods, Puurs, Belgium) amended with 10 g of CaCO3 liter−1. A fully grown agar plate was cut into 2-cm-wide strips, and half of the strips were transferred to another petri plate. The agar strips were flooded with 20 ml of sterile water and kept at 25°C. After 1 h, the water was discarded and replaced with the same volume of water. The plates were incubated at 18°C for an additional 4 days, the water was removed and replaced with the same volume of water, and the plates were incubated at 18°C for 2 h for release of zoospores. Zoospores of A. candida were obtained by immersing infected leaves containing zoosporangia in sterile water, followed by incubation for 2 h at 15°C. P. infestans zoospores were obtained by flooding a 14-day-old culture with 15 ml of sterile water, followed by incubation at 4°C for 3 h. Zoospore suspensions of 103 to 105 ml−1 were typically obtained.
Soils.
Three soils were obtained from agricultural fields in The Netherlands. CB, SV, and SSB soils were collected in December 1997 from the upper 50 cm of the soil profile, air dried for a week by passive ventilation, and passed through a 0.5-cm-pore-size mesh screen prior to use. CB, SV, and SSB soils were collected from a polder in the southwest of The Netherlands located 10 km from the city of Bergen op Zoom. These soils were physicochemically similar, containing on average 27% clay, 10% silt, and 51% sand (10). Under field conditions, all of the soils had been cropped continuously to wheat, with the exception of CB soil, which was cropped to wheat and sugar beets in a rotation scheme (1:2). The soil used in the biocontrol assays with hyacinth bulbs was collected from fields near the experimental station of Applied Plant Research (Lisse, The Netherlands). The soil was steamed and left in the open air for 6 months for recolonization. This sandy soil had an organic-matter content of 1% and a pH of 7.0.
Wheat cultivation.
Pots containing 200 g of sieved soil were sown with 15 seeds of wheat (cv. Bussard). The plants were grown in a controlled climatic chamber at 15°C with a 12-h photoperiod. The plants received 50 ml of one-third strength Hoagland's solution (macroelements only) twice a week. After 30 days, the roots were harvested and the remaining soil and excised roots were mixed, returned to the same pot, and sown again with 15 wheat seeds. This process of successively growing wheat and harvesting roots was repeated seven times.
Isolation of surfactant-producing Pseudomonas spp.
For each replicate, roots of five randomly selected wheat plants were harvested and loosely adhering soil was gently removed. One gram of roots plus adhering rhizosphere soil was suspended in 5.0 ml of sterile distilled water, vortexed for 1 min, and sonicated for 1 min in an ultrasonic cleaner. For isolation and enumeration of fluorescent pseudomonads, samples were dilution plated onto KMB+ and the plates were incubated at 25°C for 48 h. Approximately 10% of the colonies of fluorescent pseudomonads were randomly selected and tested for halo formation on SW medium. The experiment had four replicates and was performed twice.
Zoosporicidal activity of surfactant-producing Pseudomonas isolates.
Bacterial isolates able to form a halo around the colony on SW medium were screened for the ability to lyse zoospores. Bacterial suspensions containing 109 CFU ml−1 (optical density at 600 nm [OD600] = 1) were prepared from colonies grown on KMB for 48 h at 25°C. A 10-μl aliquot of the bacterial suspension was mixed on a glass slide with an equal volume of zoospore suspensions of P. ultimum var. sporangiiferum. The behavior of the zoospores was observed under a light microscope (Dialux 20 EB; Ernst Leitz GmbH, Wetzlar, Germany) at ×100 magnification for up to 5 min.
Biocontrol activity against pythium root rot.
Hyacinth bulbs (cv. Pink Pearl) were disinfested with 0.5% formaline, dried, and stored at 17°C in a climate chamber. Four weeks prior to the bioassays, the bulbs were transferred to a 9°C climate chamber. An inoculum of P. intermedium strain P52 was prepared by growing the oomycete for 3 weeks at 24°C in an autoclaved mixture containing 50% sandy soil, 50% river sand, and oatmeal (10 g liter−1). For the assay, sandy soil was mixed with the inoculum at a 1% (vol/vol) rate. P. fluorescens strain SS101 was applied as a bulb treatment or as a soil treatment. For the bulb treatment, hyacinth bulbs were immersed in a suspension of 108 CFU ml−1 for 15 min prior to being planted. For soil treatments, initial bacterial densities were 107 CFU g (fresh weight) of soil−1. Hyacinth bulbs were transferred to plastic pots filled with 2.75 kg of soil treated with Pythium and/or P. fluorescens isolate SS101 or untreated. After the bulbs were planted, the pots were wrapped in plastic bags to maintain high-humidity conditions in the soil for the duration of the experiment. After 8 weeks of plant growth at 9°C in the dark, the bulbs were harvested and the roots were rinsed with tap water. The roots were excised from the bulbs, the fresh weight was determined, and disease caused by P. intermedium P52 was scored visually on a 0-to-5 scale. In this disease index, 0 indicates no disease, 1 indicates 1 to 20% loss of root biomass relative to the control, 2 indicates 21 to 40% loss of root biomass, 3 represents 41 to 60% loss, 4 represents 61 to 80% loss, and 5 represents 81 to 100% loss (50). The disease index data were ranked. After the normal distribution and homogeneity of variances were certified, root weight data and ranked disease index data were analyzed by analysis of variance, followed by Tukey's studentized range test (SAS Institute, Inc., Cary, N.C.).
Biochemical characterization.
Gas chromatography-fatty acid methyl ester (GC-FAME) analysis and API 20NE tests (BioMerieux, S.A., Lyon, France) were performed to classify P. fluorescens strain SS101, one of the isolated Pseudomonas strains that caused lysis of zoospores. API 20NE tests were performed following the recommendations of the supplier. For GC-FAME analysis, isolate SS101 was cultivated on tryptic soy broth agar (Becton Dickinson, Cockeysville, Md.) and incubated for 24 h at 28°C. Cells were collected with a 4-mm-diameter transfer loop (Microbial ID, Inc., Newark, Del.) and processed for extraction of fatty acids following the procedures outlined by the manufacturer. Fatty acid methyl esters were analyzed using a microbial identification system equipped with an HP5890 series II gas chromatograph, HP3365 Chem. Station, and version 3.9 of the aerobe library (Microbial ID, Inc.).
Tn5 mutagenesis and phenotypic characterization of mutants.
Surfactant-deficient mutants of a spontaneous rifampin-resistant derivative of P. fluorescens strain SS101 were obtained by biparental mating with E. coli strain S17 λ pir harboring the mini-Tn5lacZKm element in plasmid pUT (9) according to protocols described by Sambrook and Russel (38). Transformants were selected on KMB supplemented with rifampin (100 μg ml−1) and kanamycin (100 μg ml−1) and subsequently transferred to SW medium. Transformants unable to produce a halo around the colony were selected after 3 to 5 days of growth on SW medium at 25°C. Putative surfactant-deficient mutants were characterized for the ability to cause lysis of zoospores of P. ultimum var. sporangiiferum, P. intermedium, A. candida, and P. infestans as described above. Bacterial cell suspensions used for zoospore lysis tests were vortexed vigorously and checked for foam formation, a phenomenon correlated with surfactant production (51). The drop collapse test was performed as described by Jain et al. (23) and Hildebrand (19). Protease and phospholipase C activities were detected by growing the mutants and the parental strain on skim milk and egg yolk agar, respectively (37). β-Galactosidase activity was tested on KMB plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactosidase) and IPTG (isopropylthio-β-d-galactosidase) according to standard protocols (38). Fluorescence was evaluated by growing the bacterial strains on Pseudomonas agar F plates for 48 h and subsequent observation under near-UV light (360 nm).
DNA isolations.
Total genomic DNA, used for Southern blot analysis and anchored PCR, was extracted from bacterial strains by a modified version of a CTAB-based protocol (3). A 1.5-ml sample of an overnight bacterial culture was centrifuged for 3 min at 14,000 rpm (Eppendorf microcentrifuge), the supernatant was discarded, and the pellet was resuspended in 550 μl of TE buffer (10 mM Tris, 10 mM EDTA, pH 8.0) amended with lysozyme (1.82 mg ml−1) and incubated at 37°C for 30 min. Seventy-five microliters of 10% sodium dodecyl sulfate (SDS) amended with proteinase K (0.86 mg ml−1) was added to the bacterial suspension and thoroughly mixed. After 15 min of incubation at 65°C, 100 μl of 5 M NaCl and 80 μl of CTAB- NaCl (0.3 M CTAB, 0.7 M NaCl) were added. After 10 min of incubation at 65°C, DNA was obtained by extraction with chloroform-isoamyl alcohol (24:1 [vol/vol]), isopropanol precipitation, and subsequent washes with 70% ethanol. The extracted DNA was dissolved in 100 μl of 10 mM Tris (pH 8.0) containing RNase (20 μg ml−1) and stored at −20°C.
Southern hybridization.
Southern blot analysis was performed to determine the number of integrations of the Tn5lacZKm element in the surfactant-deficient mutants of P. fluorescens strain SS101. Genomic DNA of the mutants was digested with 5 U of EcoRI or KpnI (Promega), enzymes without restriction sites in the Tn5lacZKm element (9). The digestions were performed in a total volume of 100 μl containing 2.0 μg of DNA. The digested DNA was precipitated with 4 M LiCl, washed with 70% ethanol, dissolved in 15 μl of sterile distilled water, and separated on 1% agarose gels in 1× Tris-borate-EDTA. DNA transfer from agarose gels to Hybond N+ nylon membranes (Amersham Pharmacia Biotech) was performed according to standard methods (38). The high-stringency conditions consisted of prehybridization for 1.5 h at 65°C, hybridization for 12 h at 65°C, membrane washings (twice each for 5 min with 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]- 0.1% SDS at room temperature and twice each for 30 min with 0.1× SSC-0.1% SDS at 65°C). The 575-bp KM probe, specific for the kanamycin resistance gene contained within the Tn5LacZKm element, was obtained by digoxigenin labeling using primers KM1 (5′-CCCGATGCGCCAGAGTTGTT) and KM2 (5′-TCACCGAGGCAGTTCCATAGG) (Roche Corp., Basel, Switzerland); hybridized probes were immunodetected according to the protocols provided by the supplier.
Anchored PCR and sequencing of Tn5 flanking regions.
DNA flanking the Tn5lacZ-Km insertions in different mutants of P. fluorescens isolate SS101 was obtained by anchored PCR (44). Anchored PCR was performed in three steps. The first consisted of ligation of synthetic anchors to genomic DNA digested with the enzyme ApoI or SmaI. For ApoI (with a 5′ AATT overhang), the anchor was EcoRI-16 (5′-AATTGGCGGTGAGTCC); for SmaI (with a blunt end), the anchor was An-B (5′-TGCGGACT). The second step was a linear PCR with the primer EOF (5′-ACTTGTGTATAAGAGTCAG) or EIR (5′-AGATCTGATCAAGAGACAG) targeting the inverted repeats of the Tn5 transposon. PCR was performed in a 50-μl reaction mixture with the high-fidelity PCR system (Roche Diagnostics, Mannheim, Germany) following the recommendations of the supplier. The third step consisted of a nested PCR with primer RH24 (5′-AGCACTCTCCAGCCTCTCACCGCA) for DNA digested with ApoI or primer VECT24 (5′-AGCACTCTCCAGCCTCTCACCGCC) for DNA digested with SmaI and primer EOF or EIR, respectively. The PCR program consisted of an initial denaturation at 95°C for 5 min, followed by 30 cycles at 94°C for 30 s, initial annealing at 72°C for 40 s with the annealing temperature decreasing by 0.5°C per cycle, and a final extension at 72°C for 2 min. The amplified fragments were purified from agarose gels by using the QIAEX II kit (Qiagen GmbH, Hilden, Germany) and cloned into pCRT7/CT-TOPO (Invitrogen, Breda, The Netherlands). Cloned PCR products were sequenced by BaseClear (Leiden, The Netherlands) with primers T7 (5′-TAATACGACTCACTATAGGG) and V5 (5′-ATCCCTAACCCTCTCCTCGGT). Sequences were trimmed, assembled by using the DNAstar program (DNAstar Inc., Madison, Wis.), and deposited in GenBank. BLASTN and BLASTP searches were conducted on the sequences, and they were aligned by CLUSTAL W version 1.81 (45).
Partial purification of surfactants.
KMB agar plates with cells of P. fluorescens strain SS101 grown for 48 h at 25°C were flooded with sterile demineralized water. The cell suspensions were centrifuged twice (Harrier 18/18 centrifuge) at 6,000 rpm for 20 min at 4°C. The cell culture supernatant was filtered (0.2-μm pore size), acidified to pH 2.0 with 9% HCl, and kept on ice for 1 h (24). The precipitate was collected by centrifugation (6,000 rpm, 30 min, 4°C, Harrier 18/80 centrifuge) and washed twice with sterile acidic (pH 2.0) demineralized water. The precipitate was resuspended in sterile demineralized water, adjusted to pH 8.0 with 0.2 M NaOH, and lyophilized (Labconco Corp., Kansas City, Mo.). The lyophilized extract was stored at −20°C. For determining the critical micelle concentration (CMC), the extract was dissolved in sterile distilled water (pH 8.0) at concentrations of 0, 0.01, 0.1, 1, 10, 25, 50, 100, and 1,000 μg ml−1. Surface tension measurements were carried out with a K6 tensiometer (Krüss GmbH, Hamburg, Germany). Measurements were performed at 25°C, and sterile distilled water was used to calibrate the tensiometer.
HPLC and LC-MS analyses.
All solvents used were HPLC grade. Acetonitrile (MeCN) and methanol were obtained from LAB-SCAN Analytical Sciences (Dublin, Ireland). Ultrapure water was obtained from a combined Seradest LFM 20 Serapur Pro 90 C apparatus (Seral, Ransbach, Germany). All LC solvents were degassed by vacuum filtration over a 0.45-μm-pore-size membrane filter (Type RC; Schleicher & Schuell) prior to use. All HPLC separations were carried out in Alltech end-capped C18 columns (length, 250 mm; particle size, 5 μm; pore size, 100 Å). For analytical and preparative separations, 4.6-mm-diameter columns were used. For LC-MS, 2.1-mm-diameter columns were used. For analytical separations, a Gynkotek (Separations) pump was used in combination with a Kratos Spectroflow UV detector and a Sedere Sedex 55 ELS detector. Freeze-drying was carried out on a Christ Alpha 1-2 freeze-drier.
The LC-MS system consisted of a TSP SpectraSYSTEM, including an SN4000 controller, an LC quaternary pump (P4000), an autosampler (AS3000), a UV2000 detector, and a Finnigan LCQ ion trap mass spectrometer. The mass spectrometer was equipped with a Finnigan electrospray ionization (ESI) interface. Data were processed by the Finnigan Xcalibur software system (ThermoQuest, Breda, The Netherlands). For off-line MS studies, a 0.005% solution of the crude surfactant fraction in MeOH was introduced into the ESI interface by continuous infusion using a syringe pump (Hamilton, Reno, Nev.) at a flow of 3 μl min−1. The averaged spectra were recorded over a period of 3 min: the scan range was m/z 310 to 1,200 at two scans per second. Positive ionization was used. In MS2 experiments, helium was used as the collision gas. Only a single parent ion was kept in resonance (isolation width, m/z 1 to 3), all other ions were ejected from the trap without mass analysis. The ion was then agitated and allowed to fragment by collision-induced dissociation. The collision energy was adjusted experimentally to give a >90% yield of fragmentation by varying the relative collision energy from 10 to 40%. During MSn measurements (n ≥ 3), this procedure was repeated for one of the daughter ions. For the on-line LC-MS studies, 5 μl was injected. The solvent was MeCN-H2O (75:25) containing 0.02% formic acid at 0.2 ml/min. Both positive and negative ionization were used. The mass range was m/z 50 to 1,200.
Ten milligrams of crude surfactant extract was dissolved in 1 ml of MeCN-H2O (6:4) and 2 drops of buffer solution (pH 6.01). After membrane filtration, this solution was fractionated by RP-HPLC. The solvent was MeCN-H2O (7:3) at 1.0 ml/min. Detection took place by UV at 210 nm. Eight fractions were collected on the basis of the UV signal (1, 0 to 3 min; 2, 3 to 15.5 min; 3, 15.5 to 17.2 min; 4, 17.2 to 19.7 min; 5, 19.7 to 21.6 min; 6, 21.6 to 24 min; 7, 24 to 26.4 min; and 8, 26.4 to 31.5 min). Eighteen 50-μl injections were made, and the eight fractions were collected manually in round-bottom flasks. After removal of the MeCN with a rotary evaporator (Büchi) in vacuo, the aqueous solution was lyophilized. White powders were obtained only with fraction 8 containing a measurable amount (≈3 mg). All fractions were dissolved in 1.25 ml of sterile demineralized water (pH 8.0) and tested for various physical and biological properties, including drop collapse, foam formation, and activity against zoospores of P. infestans. The identity of fraction 8 was further studied by NMR.
NMR spectroscopy.
Fraction 8 was dissolved in deuterated methanol (CD3OD; 99.9 atom% D; Isotec) and transferred to an NMR microtube (Shigemi). For comparative NMR studies with the cyclic lipopeptide massetolide A (17), fraction 8 was dissolved in deuterated acetone ((CD3)2CO; 99.8 atom% D; Acros) and transferred to a standard 5-mm-diameter NMR tube, and its 13C NMR spectrum was recorded at 100 MHz. NMR spectra were recorded at a probe temperature of 25°C on a Bruker DPX-400 spectrometer located at the Wageningen NMR Centre (Wageningen, The Netherlands). Chemical shifts are expressed in parts per minute relative to methanol (δ 1H, 3.31; δ 13C, 49.00) or to acetone (δ 1H, 2.05; δ 13C, 29.84). One- and two-dimensional double-quantum-filtered COSY, TOCSY, HMBC, and HMQC spectra were acquired using standard pulse sequences delivered by Bruker (Rheinstetten, Germany). For the 1H-COSY and -TOCSY spectra, 512 experiments of 32 scans each were recorded, resulting in a measuring time of 9 h for each spectrum. The mixing time for the TOCSY was 80 ms. For the [1H,13C]-HMBC and -HMQC experiments 1,024 experiments of 152 and 40 scans each, respectively, were recorded, resulting in measuring times of 69 and 20 h, respectively.
Nucleotide sequence accession numbers.
The sequences obtained in this study were deposited in GenBank under accession numbers AY303770 and AY303771.
RESULTS
Isolation of surfactant-producing Pseudomonas spp.
Wheat plants were grown for seven successive growth cycles in three agricultural soils. Total populations of fluorescent pseudomonads recovered from the rhizosphere on KMB+ ranged from ∼5 × 106 to 2 × 107 CFU g (fresh weight) of roots−1. Putative surfactant-producing Peudomonas spp. were isolated by randomly transferring fluorescent Pseudomonas colonies to SW medium (40). The population densities of surfactant-producing fluorescent Pseudomonas spp. ranged from 1.0 × 105 to 7.0 × 105 CFU g−1 and represented, on average, 1 to 5% of the total population of fluorescent Pseudomonas spp. recovered on KMB+. The total number of fluorescent pseudomonads and the number of surfactant producers remained stable during the seven successive growth cycles of wheat. No significant differences among the population densities of total and surfactant-producing pseudomonads were observed in the three soils (data not shown).
Zoosporicidal activity and phenotypic characterization.
Of a total of 375 randomly selected Pseudomonas isolates that produced a halo on SW medium, 6 isolates were able to lyse zoospores of P. ultimum var. sporangiiferum (Fig. 1). All six isolates were obtained from roots of wheat grown in SSB soil. A cell suspension (109 CFU ml−1) of these isolates caused cessation of zoospore motility within 30 s, and the shape of the zoospores was changed from bean shaped to round (Fig. 1A). The cell contents of the zoospores changed to a granular appearance (data not shown), and within 60 s, lysis of entire zoospore populations occurred (Fig. 1B and C). Identical responses were observed for zoospores from P. infestans, P. intermedium, and A. candida. The lytic effects on zoospores were also obtained with cell culture supernatants of each of the six isolates. Randomly amplified polymorphic DNA (RAPD) analysis with the 10-mer primers M12 and D7 (26) indicated that the six isolates were genotypically identical. The representative strain SS101 was selected for further studies. Biochemical characterization by API 20NE testing and GC-FAME analysis classified strain SS101 as P. fluorescens biovar II. Strain SS101 exhibited multiple characteristics indicative of surfactant production, including halo formation on SW medium, foam formation, drop collapse, and the ability to significantly lower the surface tension of water from ∼73 to 30 mN m−1 (Table 2). The other Pseudomonas strains, including the reference strains Pseudomonas putida WCS358 and P. aeruginosa PAO1 and DSM1128, had similar characteristics; however, none of these isolates caused lysis of zoospores. Cell suspensions of P. putida strain WCS358 lowered the surface tension of water to almost the same level as cell suspensions of strain SS101 but did not adversely affect zoospore behavior. Strain SV7, isolated from the rhizosphere of wheat grown in SV soil (this study), caused cessation of zoospore motility but no subsequent lysis (Table 2).
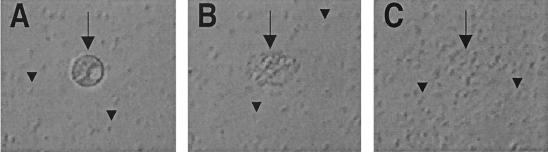
FIG. 1.
Effect of P. fluorescens SS101 on zoospores of P. ultimum var. sporangiiferum. (A) Within 30 s after exposure to strain SS101, zoospores become immotile and round. (B) Within 60 s, lysis of zoospores occurs. (C) Within an additional 10 to 30 s, the remnants of the zoospores completely disappear. The arrows indicate zoospores of P. ultimum var. sporangiiferum, and the arrowheads indicate cells of P. fluorescens strain SS101 (magnification, ×400). The density of the bacterial cell suspension was 109 CFU ml−1, and zoospore suspensions contained 104 zoospores ml−1.
TABLE 2.
Phenotypic characteristics of P. fluorescens strain SS101 and other Pseudomonas strains
| Strain | Haloa | Drop collapseb | Foamc | Surface tension (mN m−1)d | Cessation of zoospore motilitye | Zoospore lysise |
|---|---|---|---|---|---|---|
| Water | −f | − | − | 73.5 | − | − |
| P. fluorescens SS101 | +g | + | + | 29.7 | + | + |
| P. fluorescens SSB17 | − | − | − | 70.5 | − | − |
| P. fluorescens SV7 | + | + | + | 54.0 | + | − |
| P. putida WCS358 | + | + | + | 30.5 | − | − |
| P. aeruginosa PAO1 | + | + | + | 56.5 | − | − |
| P. aeruginosa DSM1128 | + | + | + | 49.5 | − | − |
Halo formation on SW medium after 3 to 5 days of growth at 25°C.
Cell suspensions (OD600 = 1) were tested on an oily surface for the ability to collapse a drop of water.
Foam formation after cell suspensions (OD600 = 1) were vigorously shaken.
Surface tensions of cell suspensions (OD600 = 1).
Zoospore motility and lysis were observed microscopically after bacterial cell suspensions (OD600 = 1) were mixed with zoospore suspensions of P. ultimum var. sporangiiferum (104 zoospores ml−1) in a 1:1 (vol/vol) ratio.
−, negative reaction.
+, positive reaction.
Biocontrol activity of P. fluorescens SS101.
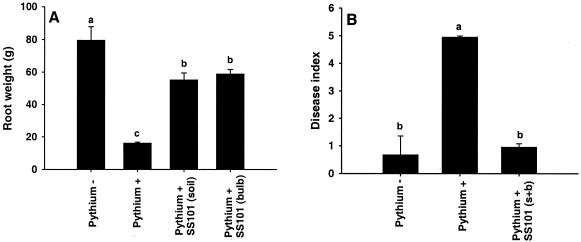
The biocontrol activity of P. fluorescens SS101 against P. intermedium, the causal agent of pythium root rot of hyacinth, was tested in bioassays at 9°C for 8 weeks. P. intermedium caused a significant fourfold reduction in the root biomass of hyacinth (Fig. 2A). Treatment of hyacinth bulbs or soil with cell suspensions of strain SS101 provided substantial protection of hyacinth roots grown in soil infested with the pathogen (Fig. 2A). In a second experiment, simultaneous treatment of bulbs and soil resulted in a reduction of root rot of hyacinth to the same level as observed in the healthy control (Fig. 2B). In both soil and bulb treatments, strain SS101 established population densities in the hyacinth rhizosphere of 3 × 106 and 2 × 105 CFU g (fresh weight) of roots−1 after 4 and 8 weeks of plant growth, respectively.
FIG. 2.
Effect of P. fluorescens SS101 on pythium root rot of hyacinth. An inoculum of P. intermedium P52 was mixed through soil (Pythium +). Soil not treated with P. intermedium (Pythium −) served as a healthy control. P. fluorescens SS101 was applied to soil [SS101(soil)], bulbs [SS101(bulb)], or both [SS101(s + b)]. The density of the cell suspensions of SS101 used for bulb treatment was 108 CFU ml−1, and the initial density of SS101 in soil treatments was 107 CFU g (fresh weight) of soil−1. Plants were grown for 8 weeks at 9°C in the dark. (A) In the first experiment, bulbs were harvested and the root weight was determined. (B) In the second experiment, bulbs were harvested and the disease index was scored on a 0-to-5 scale, where 0 indicates no disease and 5 indicates 81 to 100% loss of root biomass relative to the control. The means of five replicates are shown. Means with the same letter are not statistically different according to Tukey's studentized range test (P = 0.05). The error bars represent the standard errors of the mean. The experiment was repeated four times, and representative results of two experiments are shown.
Tn5 mutagenesis and characterization of surfactant-deficient mutants.
A total of 520 transformants of P. fluorescens SS101 were obtained by mutagenesis with Tn5lacZKm. Five of these transformants were not able to form a halo on SW medium (Table 3). RAPD analysis of these five mutants with the 10-mer primers M12 and D7 (26) confirmed strain integrity. Southern blot analysis of EcoRI- or KpnI-digested genomic DNA with a probe targeting the kanamycin resistance gene showed the presence of a single Tn5 integration for each of the five mutants. The mutants gave negative reactions in the drop collapse test, were unable to form foam after vigorous shaking, did not lower the surface tension of water to the same extent as the wild-type strain SS101, and could no longer affect the motility and cause lysis of zoospores of multiple oomycetes (Table 3). In contrast to the wild type, all five mutants presented β-galactosidase activity, indicating integration of the promoterless Tn5lacZKm element in a coding region. All of the mutants still produced wild-type levels of protease, phospholipase C, and siderophores (based on fluorescence at 360 nm).
TABLE 3.
Properties of wild-type strain P. fluorescens SS101 and its five surfactant-deficient Tn5 mutants
| Property | Valuej
|
|||||
|---|---|---|---|---|---|---|
| SS101 | Tn5 mutant:
|
|||||
| 9.26 | 10.24 | 11.17 | 13.3 | 17.18 | ||
| Halo formationa | + | − | − | − | − | − |
| Drop collapseb | + | − | − | − | − | − |
| Foam formationc | + | − | − | − | − | − |
| Surface tension (mN m−1)d | 29.7 | 64.3 | 56.0 | 60.0 | 56.5 | 62.0 |
| β-Galactosidase activitye | − | + | + | + | + | + |
| Proteasef | + | + | + | + | + | + |
| Phospholipase Cg | + | + | + | + | + | + |
| Fluorescenceh | + | + | + | + | + | + |
| Cessation of zoospore motilityi | + | − | − | − | − | − |
| Zoospore lysisi | + | − | − | − | − | − |
Evaluated on SW medium after 3 to 5 days of growth at 25°C.
Cell suspensions (OD600 = 1) were tested on an oily surface for the ability to cause the collapse of a drop of water.
Foam formation after cell suspensions (OD600 = 1) were vigorously shaken.
Surface tensions of cell suspensions (OD600 = 1).
Evaluated on KMB plates containing X-Gal and IPTG after 48 h of growth at 25°C.
Tested on skim milk plates.
Tested on egg yolk plates.
Tested on Pseudomonas agar Funder near-UV light (360 nm).
Zoospore motility and lysis were observed microscopically after bacterial cell suspensions (OD600 = 1) were mixed with zoospore suspensions of P. ultimum var. sporangiiferum, A. candida, or P. infestans (103 to 104 zoospores ml−1) in a 1:1 (vol/vol) ratio.
+, positive reaction; −, negative reaction.
Identification of genes involved in surfactant biosynthesis.
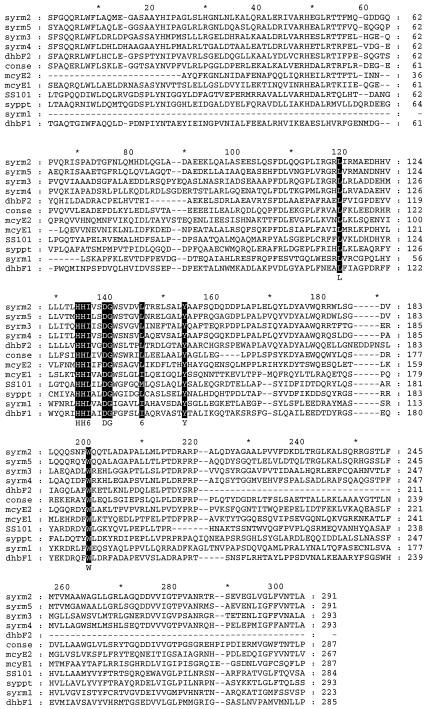
The flanking regions of the Tn5 transposon were cloned and sequenced for the surfactant-deficient mutants 10.24 and 17.18. Sequences of mutant 10.24 yielded an 852-bp fragment with a deduced protein sequence that is similar to those of the condensation domains of peptide synthetase genes from several bacteria and eukaryotes (Fig. 3). The deduced protein sequence showed 54% (161 of 293) similarity with that of a condensation domain in the syringopeptin synthetase gene sypA from P. syringae pv. syringae B301D (39) and various degrees of similarity to those of five condensation domains of the syringomycin synthetase (syrE) gene of strain B301D. The syrE gene encodes a 9,376-amino acid (aa) protein, and the similarities of its different condensation domains to the condensation domain of P. fluorecens SS101 were 48% (109 of 226) for positions 65 to 283, 38% (112 of 288) for positions 4270 to 4557, 36% (107 of 293) for positions 5332 to 5623, 35% (106 of 295) for positions 6388 to 6681, and 39% (117 of 295) for positions 8548 to 8841. The similarities were 44% (121 of 271) for positions 11 to 279 and 32% (71 of 210) for positions 1053 to 1258 of a 1,278-aa protein involved in the synthesis of the siderophore 2,3-dihydroxybenzoate by Bacillus subtilis and 41% (121 of 281) for positions 1990 to 2270 and 45% (83 of 189) for positions 3145 to 3322 of a 3,487-aa protein encoded by the gene mcyE that is responsible for the synthesis of the cyclic heptapeptide microcystin in the bacterium Microcystis aeruginosa K-139. A majority sequence obtained from the alignment of 43 different condensation domains, including proteins involved in the synthesis of gramicidin S by Brevibacillus brevis, surfactin by B. subtilis, and peptide synthetases from several fungi, including Cochliobolus carbonum, Penicillium chrysogenum, Emericella nidulans, and Acremonium chrysogenum, showed that the active-site motif (HHXXXDG) is conserved in all aligned condensation domains, including the partial sequence obtained from strain SS101 (Fig. 3).
FIG. 3.
Alignment of the deduced protein sequences of P. fluorescens SS101 and other condensation domains of peptide synthetase sequences. The sequence of P. fluorescens SS101 was obtained from the Tn5 flanking region of the surfactant-deficient mutant 10.24. syrm1 to syrm5 are condensation domains from the syringomycin synthetase protein of P. syringae pv. syringae B301D (accession number T14593). syppt is a condensation domain of the syringopeptin synthetase protein from P. syringae pv. syringae B301D (AAF99707). conse represents a majority sequence obtained from the alignment of 43 condensation domains of different peptide synthetases (PF00574). dhbF1 and dhbF2 are condensation domains of a peptide synthetase involved in the synthesis of the siderophore 2,3-dihydroxybenzoate by B. subtilis (NP_391076), and mycE1 and mycE2 are condensation domains of a protein responsible for the synthesis of microcystin by M. aeruginosa K139 (BAB12211). The residues in solid boxes have high levels of similarity. Dashes indicate gaps in the sequence.
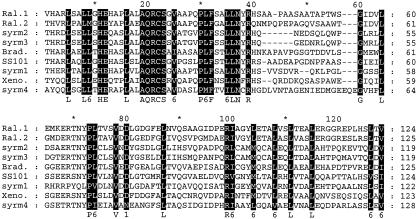
Sequence analysis of the Tn5 flanking region of mutant 17.18 yielded a 453-bp fragment with a deduced protein sequence (Fig. 4) that presents 61% (90 of 145) similarity with a 3,310-aa protein from Bradyrhizobium japonicum 110spc4, 66% (96 of 144) similarity for positions 415 to 558 and 61% (90 of 146) similarity for positions 2550 to 2695 of a 5,953-aa peptide synthase protein of the bacterial pathogen Ralstonia solanacearum GMI1000, 63% (92 of 144) similarity with the 3,316-aa protein XpsB from Xenorhabdus bovienii T228, and similarity with four domains of the 9,376-aa syringomycin synthetase protein from P. syringae pv. syringae B301D (Fig. 4). The similarity levels for these domains were 58% (86 of 146) for positions 1337 to 1478, 57% (84 of 144) for positions 2425 to 2563, 57% (84 of 144) for positions 3512 to 3650, and 56% (85of148) for positions 7794 to 7941 of syringomycin synthetase. A database search and comparison of these sequences strongly suggest that the transposon had integrated into a condensation domain of peptide synthetases; however, the flanking sequence of mutant 17.18 was not long enough to identify conserved active-site motifs of condensation domains.
FIG. 4.
Alignment of deduced protein sequences of P. fluorescens SS101 and other proteins found in the database. For P. fluorescens SS101, the sequence was obtained from the Tn5 flanking region of the surfactant-deficient mutant 17.18. Ral.1 and Ral.2 are different domains of a peptide synthetase protein from R. solanacearum GMI1000 (NP_522203). syrm1 to syrm4 are different domains of the syringomycin synthetase protein from P. syringae pv. syringae B301D (T14593). Brad. is a protein from B. japonicum 110spc4 (AAG61082), and Xeno. is a domain from the XpsB protein from X. bovienii T228 (AAL57600). The residues in solid boxes are highly similar in all aligned sequences. Dashes indicate gaps in the sequence.
Isolation and characterization of surfactants.
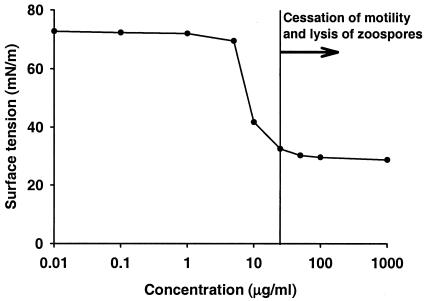
Extraction of the surfactant(s) from the cell culture supernatant of P. fluorescens SS101 yielded a white precipitate, whereas no precipitate was obtained from the five surfactant-deficient mutants. Prior to separation by RP-HPLC, the physical and biological properties of the extract were determined (Fig. 5). The CMC is defined as the solubility of a surfactant in an aqueous phase. At concentrations above the CMC, surfactant molecules form aggregates called micelles, which solubilize oil or water into the other phase, creating a microemulsion. No further drop in the surface tension occurs as the amount of surfactant exceeds the CMC because the emulsion is already saturated (16). The CMC of the partially purified extract from strain SS101 was shown to be ∼25 μg ml−1 (Fig. 5). The effects on zoospores (cessation of motility and lysis) were observed at the CMC and concentrations higher than the CMC (Fig. 5).
FIG. 5.
Relationships among the concentration of the partially purified extract obtained from P. fluorescens SS101, surface tension, and cessation of zoospore motility and lysis. The partially purified extract was dissolved in sterile demineralized water (pH 8.0) at different concentrations, and surface tension was measured with a tensiometer. At concentrations of 25 μg ml−1 and higher, the extract caused cessation of motility and subsequent lysis of zoospores of P. ultimum var. sporangiiferum. The mean values of two replicates per concentration are shown.
The composition of the extract was investigated by RP-HPLC with both UV and evaporative-light-scattering (ELS) detection. With a gradient of acidified water-acetonitrile, 17 peaks were observed, and the eluates of 18 injections were collected in eight fractions (Fig. 6). Fraction 8 was the main component and accounted for ∼50% of the extract. All fractions, with the exception of fraction 1, were positive in the drop collapse test, indicating a decrease in surface tension (Table 4). Only fractions 5 and 8 were positive in the foam formation test. Fractions 4 to 8 caused cessation of zoospore motility and subsequent lysis. Fraction 8 caused lysis of zoospores in a much shorter time than the other fractions. Zoospores were rendered immotile within 10 s and lysed within 30 s after contact with fraction 8, whereas for the other fractions it took ∼30 s for cessation of motility and ∼60 s for lysis. The lysis caused by fraction 7 was not the characteristic explosion observed with fractions 4, 5, 6, and 8 but resembled leaking at one point in the zoospore membrane (Table 4).
FIG. 6.
RP-HPLC profile of a preparative separation of the crude surface-active extract obtained from cell culture supernatant of P. fluorescens SS101. The solvent used is MeCN-H2O (7:3) containing 0.1% formic acid at 1 ml/min, and detection was performed with UV at 210 nm. Numbers represent fractions.
TABLE 4.
Properties of eight fractions of the surface-active extract obtained by RP-HPLC
| Fraction | RP-HPLC retention time (min) | Haloa | Drop collapseb | Foam formationc | Cessation of zoospore motilityd | Zoospore lysisd |
|---|---|---|---|---|---|---|
| 1 | 0-3 | − | −e | − | − | − |
| 2 | 3-15.5 | − | +f | − | − | − |
| 3 | 15.5-17.2 | − | + | − | − | − |
| 4 | 17.2-19.7 | − | + | − | + | + |
| 5 | 19.7-21.6 | − | + | + | + | + |
| 6 | 21.6-24 | − | + | − | + | + |
| 7 | 24-26.4 | − | + | − | + | ±g |
| 8 | 26.4-31.5 | + | + | + | + | + |
| Water | − | − | − | − | − |
Evaluated on SW medium after 3 days at 25°C. +, present; −, absent.
Dissolved fractions were tested on an oily surface for the ability to cause the collapse of a drop of water.
Foam formation after dissolved fractions were vigorously shaken.
Zoospore motility and lysis were observed microscopically after the dissolved fractions were mixed with zoospore suspensions of P. infestans in a 1:1 (vol/vol) ratio.
−, negative reaction.
+, positive reaction.
Lysis was not complete, and the effect resembled leakage at one point in the zoospore membrane.
Chemical identification of fraction 8.
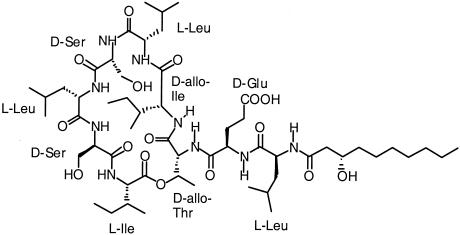
For fractions 2 to 8, there was a good correlation between the UV signal at 210 nm and the ELS signal. None of the peaks showed any absorbance around 254 nm, ruling out the presence of compounds with conjugated double bonds or an aromatic nucleus. Fraction 8 was further investigated by infusion ESI-MS in the positive mode. The major peaks in the mass spectrum were observed at m/z 1140.5 [M + H]+ and 1162.7 [M + Na]+. In view of the isolation procedure, the high surface activity, and data in the literature, this suggested a compound related to viscosin, a cyclic acetylated nonapeptide from Pseudomonas viscosa with mass 1,125.5 (C54H95N9O16) (20). Confirmation of this hypothesis was obtained from MSn studies. Collision-induced dissociation of the [M + H]+ ion signal produced fragments at m/z 1122 (loss of water), 857 (loss of β-hydroxydecanoic acid and leucine), and 728 (loss of β-hydroxydecanoic acid, leucine, and glutamic acid). As further identification by MS was not possible, fraction 8 was studied by various NMR techniques. Careful assignment of all resonances and cross-peaks in the one- and two-dimensional NMR spectra resulted in the assignment of all amino acids and the fatty acid (Table 5. From the HMBC spectrum, a complete sequential assignment of the amino acids and the position of the fatty acid was derived. These data indicated that fraction 8 is a cyclic lipopeptide containing nine amino acids and a 10-carbon hydroxy fatty acid (Fig. 7). The quantities of the other zoosporicidal fractions purified by HPLC were insufficient to determine their structures by NMR. However, on-line LC-ESI-MS in both positive and negative modes showed that all of the compounds were closely related to fraction 8, with molecular masses ranging from 1,111 to 1,169 Da.
TABLE 5.
NMR data for fraction 8, recorded in CD3OD at 400 (1H) and 100 (13C) MHz at 25°C
| Residue | Atom | δ 13Ca | δ 1H, mult., J (in Hz)a,b |
|---|---|---|---|
| LEU-1 | C=O/NH | 176.34 | 8.91, bs |
| α | 54.24 | 3.93, m | |
| β | 40.11 | 1.73, m | |
| 1.73, m | |||
| γ | 25.85 | 1.69, m | |
| δ1 | 23.46 | 0.99, p.n. | |
| δ2 | 22.46 | 0.95, p.n. | |
| GLU-2 | C=O/NH | 177.02 | 9.19, bs |
| α | 57.50 | 4.17, m | |
| β | 27.22 | 2.07, m | |
| γ | ND | 2.47, m | |
| δ | ND | ND | |
| THR-3 | C=O/NH | 175.08 | 8.33, d, 7.5 |
| α | 61.93 | 4.17, m | |
| β | 70.64 | 5.45, dq, 11.0, 6.0 | |
| γ | 18.55 | 1.37, d, 6.0 | |
| ILE-4 | C=O/NH | 174.75 | 7.69, d, 6.0 |
| α | 64.38 | 3.60, d, 11.2c | |
| β | 36.37 | 2.01, m | |
| γ1 | 26.66 | 1.53, m | |
| 1.13, m | |||
| γ2 | 16.96 | 0.98, p.n. | |
| δ | 10.94 | 0.95, p.n. | |
| LEU-5 | C=O/NH | 173.84 | 8.51, d, 6.5 |
| α | 53.92 | 3.80, m | |
| β | 37.66 | 2.03, m | |
| 1.68, m | |||
| γ | 25.78 | 1.64, m | |
| δ1 | 24.07 | 0.94, p.n. | |
| δ2 | 21.4 | 0.89, p.n. | |
| SER 6 | C=O/NH | 173.61 | 7.55, d, 7.4 |
| α | 57.55 | 4.42, dd, 3.5, 2.5c | |
| β | 64.42 | 4.19, dd, 12, 3.5 | |
| 3.95, dd, 12, 2.5 | |||
| LEU-7 | C=O/NH | 175.26 | 7.64, d, 6.5 |
| α | 55.00 | 4.33, dd, 10.4, 3.6c | |
| β | 41.97 | 2.02, m | |
| 1.63, m | |||
| γ | 25.68 | 1.63, m | |
| δ1 | 23.61 | 1.00, p.n. | |
| δ2 | 21.4 | 0.93, p.n. | |
| SER-8 | C=O/NH | 172.99 | 8.29, d, 8.5 |
| α | 57.66 | 4.51, dd, 4.8, 3.5c | |
| β | 63.01 | 3.99, dd, 11.5, 4.8 | |
| 3.75, dd, 11.5, 3.5 | |||
| ILE-9 | C=O/NH | 170.00 | 7.06, d, 9.8 |
| α | 57.66 | 4.61, m | |
| β | 37.46 | 2.02, m | |
| γ1 | 25.54 | 2.0, m | |
| 1.23, m | |||
| γ2 | 16.19 | 0.88, p.n. | |
| δ | 12.34 | 0.89, p.n. | |
| FA | 1 | 175.12 | |
| 2 | 44.70 | 2.43, m | |
| 3 | 69.98 | 4.11, m | |
| 4 | 38.48 | 1.56, m | |
| 5 | 26.74 | 1.31, m | |
| 6 | 30.69 | 1.31, m | |
| 7 | 30.45 | 1.31, m | |
| 8 | 33.00 | 1.29, m | |
| 9 | 23.74 | 1.31, m | |
| 10 | 14.47 | 0.90, p.n. |
In parts per million; ND, not detectable see the text.
p.n., peak splitting not assigned due to overlap; bs, broad singlet; multi, multiplet.
Coupling with NH not detectable due to exchange of most of the NH protons.
FIG. 7.
Chemical structure of fraction 8 obtained from cell culture supernatant of P. fluorescens SS101.
DISCUSSION
Oomycetes form a diverse group of eukaryotic fungus-like microorganisms containing saprophytes and a range of economically important pathogens of plants, insects, fish, and animals (25). Many oomycetes reproduce asexually, forming mobile flagellate zoospores under moist conditions. Zoospores locate healthy tissue by responding to chemoattractants released by the plant (52); they accumulate on plant surfaces and encyst in response to, among other things, polysaccharides that interact with receptors on the zoospore surface (8). Thus, zoospore taxis is an essential part of the preinfection process and is a potential target for controlling diseases caused by oomycetes and zoosporic fungi.
In this study, six Pseudomonas isolates were obtained from the rhizosphere of wheat with zoosporicidal activity: zoospores were rendered immotile within 30 s of exposure to cell suspensions or cell culture supernatants of the six isolates, and subsequent lysis occurred within 60 s (Fig. 1). Comparison of strain SS101, identified by GC-FAME analysis as P. fluorescens biovar II, with other Pseudomonas strains and in particular strain WCS358 indicated that reduction of the surface tension alone may not be indicative for causing lysis of zoospores (Table 2). The observed effects on zoospores have also been described for plant saponins (7). For Pseudomonas, similar lytic effects on zoospores have so far been described for rhamnolipids produced by strains of P. aeruginosa (27, 43). Rhamnolipids also were reported to have antifungal activity (27) and to serve as pathogenicity factors during the colonization of human lung tissue (22) and were implicated in affecting attachment of bacterial cells to surfaces and the maintenance of biofilm architecture (6). Southern hybridization and PCR with probes and primers specific for rhlA, rhlB, rhlI, and rhlR, genes involved in the biosynthesis and regulation of rhamnolipid production (34, 35), indicated that these genes are not present in P. fluorescens strain SS101 and the other surfactant-producing Pseudomonas isolates obtained from the wheat rhizosphere (data not shown).
In bioassays, P. fluorescens strain SS101 significantly and consistently controlled pythium root rot of hyacinth (Fig. 2). Pythium root rot is a serious problem in the cultivation of several bulbous crops in The Netherlands and can cause considerable yield losses of Hyacinth, Iris, and Crocus (50). Chemical measures to control this disease do not always provide consistent control, and several of these compounds will be banned in the near future. Given the results of this study, large-scale application of P. fluorescens strain SS101 is currently being explored for controlling pythium root rot. The roles of biosurfactants produced by Pseudomonas spp. in the suppression of plant-pathogenic fungi and oomycetes have so far been inferred from studies with purified compounds or microcosm studies in which in situ production levels were correlated with fungal inhibition (27, 43, 46, 47). As a first step toward determining the roles of the biosurfactants produced by P. fluorescens SS101 in the control of oomycete pathogens, mutants defective in surfactant production were generated. The results of recent biocontrol assays showed that the surfactant-deficient mutant 10.24 was not able to control pythium root rot of hyacinth (M. de Boer and J. M. Raaijmakers, unpublished data). These results may suggest that the biosurfactants produced by strain SS101 play a key role in its biocontrol activity against P. intermedium. To more conclusively show the role of the surfactants in biocontrol by strain SS101, current work focuses on complementation of the mutants, as well as on possible effects that surfactant deficiency may have on rhizosphere colonization and, concomitantly, on biocontrol activity.
Sequencing of the flanking regions of the transposon in two surfactant-deficient mutants, 10.24 and 17.18, led to the localization of the Tn5 elements in condensation domains of peptide synthetases (Fig. 3 and 4). Peptide synthetases are multifunctional enzymes involved in the nonribosomal synthesis of diverse and often complex metabolites, including antibiotics, siderophores, and biosurfactants (11, 21, 28, 30, 42). These enzymes are composed of modules, which contain all enzymatic activities to incorporate one constituent into the final compound. These modules are in a colinear arrangement with the primary structure of the compound and can be divided into domains responsible for single chemical reactions (42). Condensation domains are found as a part of repetitive modules. They are ∼450 aa in length and coincide in frequency with the number of peptide bonds that have to be formed for the linear peptide of the final length (42). Although pleiotropic effects of the mutations in the five mutants described in this study were not observed (Table 3), complementation and expression studies are being initiated to confirm and further characterize the roles of these peptide synthetases in biosurfactant production by strain SS101. Nevertheless, the nature of the disrupted genes is very consistent with the data obtained in chemical identification of the constituents of the surface-active extract of strain SS101.
RP-HPLC, LC-MS, and NMR indicated that the most nonpolar and abundant constituent (fraction 8) in the surface-active extract of strain SS101 was a cyclic lipopeptide containing 9 aa and a 10-carbon fatty acid (Fig. 7). Comparison with values in the literature revealed that fraction 8 is most likely identical to massetolide A, a cyclic lipopeptide isolated from a marine Pseudomonas sp. with activity against Mycobacterium spp. (17). Differences in chemical shifts of 1H and 13C resonances in spectra recorded in methanol (Table 5) from those reported by Gerard et al. (17), recorded in acetone, could be explained by solvent effects. These differences were absent when a 13C NMR spectrum of fraction 8 (data not shown) in acetone-d6 was compared with the spectrum of massetolide A (17). The identical values indicate that not only the basic structure is the same, but also the presence of the different d and l amino acids in the macrocyclic ring. Although there is no doubt about the presence of glutamic acid, based on the mass spectral evidence and the chemical shifts of the α, β, and γ protons and the α and β carbons of this residue, the signals of the γ and δ carbons are missing, and the signal of the β carbon is reduced. This could be explained by the presence of a metal on the δ carboxyl of glutamic acid, influencing the relaxation of the neighboring carbons. On-line LC-ESI-MS in both positive and negative modes showed that the other zoosporicidal fractions, 4 to 7, were closely related to fraction 8, with molecular masses ranging from 1,111 to 1,169 Da. Most likely, several of these compounds are identical to massetolides B to H, described by Gerard et al. (17). Larger-scale isolation and separation will provide proof of this hypothesis.
The CMC of the surface-active extract obtained from P. fluorescens SS101 was ∼25 μg ml−1 (Fig. 6). The CMC, the minimum amount of surfactant required to cause the maximum decrease in surface tension, is an important measure of the surface activity and allows comparison with other surfactants (16). The CMC and the minimum surface tension obtained for the surface-active extract of P. fluorescens SS101 are comparable to those found for rhamnolipids. For example, rhamnolipids produced by P. aeruginosa UG3 lowered the surface tension to 31.4 mN m−1 and achieved a CMC at 30 μg ml−1 (49). A biosurfactant produced by Bacillus licheniformis JF-2, with properties similar to those of surfactin, lowered the surface tension to 27 mN m−1 and reached the CMC at ∼20 μg ml−1 (24). It should be emphasized that the CMC and surface tension reported in this study for strain SS101 are based on a mixture of at least five lipopeptide surfactants. Larger-scale isolation and separation of the individual fractions will allow us to determine CMCs for each of the zoosporicidal constituents.
Acknowledgments
This research was supported by CAPES, Brazil (project no. 1515/96-9), and the Royal Dutch Academy of Arts and Sciences.
We thank G. P. Lelyveld for performing the preparative HPLC separation, C. van de Haar for the freeze-drying of the fractions, and J. A. Boeren (Laboratory of Biochemistry) and F. W. Claassen for their help with the LC-MS analyses. We thank S. Breeuwsma and C. F. Geerds for performing part of the hyacinth bioassays and the RAPD analysis, B. Brandwagt for his help with anchored PCR, and M. Mazzola (U.S. Department of Agriculture-Agricultural Research Service, Wenatchee, Wash.) for GC-FAME analysis of strain SS101.
REFERENCES
- 1.Andersen, J. B., B. Koch, T. M. Nielsen, D. Sørensen, M. Hansen, O. Nybroe, C. Christophersen, J. Sørensen, S. Molin, and M. Givskov. 2003. Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology 149:37-46. [DOI] [PubMed] [Google Scholar]
- 2.Arino, S., R. Marchal, and J. P. Vandecasteele. 1996. Identification and production of a rhamnolipid biosurfactant by a Pseudomonas species. Appl. Microbiol. Biotechnol. 45:162-168. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1992. Short protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Caten, E. C., and J. L. Jinks. 1968. Spontaneous variability of single isolates of Phytophthora infestans. Can. J. Bot. 46:329-348. [Google Scholar]
- 5.Cooper, D. G., and J. E. Zajic. 1980. Surface-active compounds from microorganisms. Adv. Appl. Microbiol. 26:229-253. [Google Scholar]
- 6.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deacon, J. W., and R. T. Mitchell. 1985. Toxicity of oat roots, oat root extracts, and saponins to zoospores of Pythium spp. and other fungi. Trans. Br. Mycol. Soc. 84:479-487. [Google Scholar]
- 8.Deacon, J. W., and S. P. Donaldson. 1993. Molecular recognition in the homing responses of zoosporic fungi, with special reference to Pythium and Phytophthora. Mycol. Res. 97:1153-1171. [Google Scholar]
- 9.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza, J. T., D. M. Weller, and J. M. Raaijmakers. 2003. Frequency, diversity and activity of 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 93:54-63. [DOI] [PubMed] [Google Scholar]
- 11.Devescovi, G., C. Aguilar, M. B. Majolini, J. Marugg, P. Weisbeek, and V. Venturi. 2001. A siderophore peptide synthetase gene from plant-growth-promoting Pseudomonas putida WCS358. Syst. Appl. Microbiol. 24:321-330. [DOI] [PubMed] [Google Scholar]
- 12.Déziel, E., G. Paquette, R. Villemur, F. Lepine, and J. G. Bisaillon. 1996. Biosurfactant production by a soil Pseudomonas strain growing on polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 62:1908-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiechter, A. 1992. Biosurfactants: moving towards industrial application. Trends Biotechnol. 10:208-217. [DOI] [PubMed] [Google Scholar]
- 15.Fogliano, V., A. Ballio, M. Gallo, S. Woo, F. Scala, and M. Lorito. 2002. Pseudomonas lipodepsipeptides and fungal cell wall-degrading enzymes act synergistically in biological control. Mol. Plant-Microbe Interact. 15:323-333. [DOI] [PubMed] [Google Scholar]
- 16.Georgiou, G., S. Lin, and M. M. Sharma. 1992. Surface-active compounds from microorganisms. Bio/Technology 10:60-65. [DOI] [PubMed] [Google Scholar]
- 17.Gerard, J., R. Lloyd, T. Barsby, P. Haden, M. T. Kelly, and R. J. Andersen. 1997. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 60:223-229. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, M., C. Thrane, S. Olsson, and J. Sørensen. 2000. Confocal imaging of living fungal hyphae challenged with the antagonist viscosinamide. Mycologia 92:216-221. [Google Scholar]
- 19.Hildebrand, P. D. 1989. Surfactant-like characteristics and identity of bacteria associated with broccoli head rot in Atlantic Canada. Can. J. Plant Pathol. 11:205-214. [Google Scholar]
- 20.Hiramoto, M., K. Okada, and S. Nagai. 1970. The revised structure of viscosin, a peptide antibiotic. Tetrahed. Lett. 13:1087-1090. [DOI] [PubMed] [Google Scholar]
- 21.Huang, G., L. Zhang, and R. G. Birch. 2001. A multifunctional polyketide-peptide synthetase essential for albicidin biosynthesis in Xanthomonas albilineans. Microbiology 147:631-642. [DOI] [PubMed] [Google Scholar]
- 22.Iglewski, B. 1989. Probing Pseudomonas aeruginosa, an opportunistic pathogen. ASM News 55:303-307. [Google Scholar]
- 23.Jain, D. K., D. L. Collins-Thompson, H. Lee, and J. T. Trevors. 1991. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Methods 13:271-279. [Google Scholar]
- 24.Javaheri, M., G. E. Jenneman, M. J. McInerney, and R. M. Knapp. 1985. Anaerobic production of a biosurfactant by Bacillus licheniformis JF-2. Appl. Environ. Microbiol. 50:698-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamoun, S. 2003. Molecular genetics of pathogenic oomycetes. Eukaryot. Cell 2:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, B. S., J. Y. Lee, and B. K. Hwang. 2000. In vivo control and in vitro antifungal activity of rhamnolipid B, a glycilipid antibiotic, against Phytophthora capsici and Colletotrichum orbiculare. Pest. Manag. Sci. 56:1029-1035. [Google Scholar]
- 28.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 29.Miller, C. M., R. V. Miller, D. Garton-Kenny, B. Redgrave, J. Sears, M. M. Condron, D. B. Teplow, and G. A. Strobel. 1998. Ecomycins, unique antimycotics from Pseudomonas viridiflava. J. Appl. Microbiol. 84:937-944. [DOI] [PubMed] [Google Scholar]
- 30.Mossialos, D., U. Ochsner, C. Baysse, P. Chablain, J.-P. Pirnay, N. Koedam, H. Budzikiewicz, D. U. Fernandez, M. Schäfer, J. Ravel, and P. Cornelis. 2002. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45:1673-1685. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, T. H., C. Christophersen, U. Anthoni, and J. Sørensen. 1999. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 86:80-90. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, T. H., C. Thrane, C. Christophersen, U. Anthoni, and J. Sørensen. 2000. Structure, production characteristics and fungal antagonism of tensin—a new antifungal cyclic lipopeptide from Pseudomonas fluorescens 96.578. J. Appl. Microbiol. 89:992-1001. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, T. H., D. Sørensen, C. Tobiasen, J. B. Andersen, C. Christophersen, M. Givskov, and J. Sørensen. 2002. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl. Envrion. Microbiol. 68:3416-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochsner, U. A., A. Feichter, and J. Reiser. 1994. Isolation, characterization and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding rhamnosyltransferase involved in rhamnolipid synthesis. J. Biol. Chem. 269:19787-19795. [PubMed] [Google Scholar]
- 36.Ron, E. Z., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 37.Sacherer, P., G. Défago, and D. Haas. 1994. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 116:155-160. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Scholz-Schroeder, B., J. D. Soule, S.-E. Lu, L. Grgurina, and D. Gross. 2001. A physical map of the syringomycin and syringopeptin gene clusters localized to approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D. Mol. Plant-Microbe Interact. 12:1426-1435. [DOI] [PubMed] [Google Scholar]
- 40.Siegmund, I., and F. Wagner. 1991. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 5:265-268. [Google Scholar]
- 41.Simon, A., and E. H. Ridge. 1974. The use of ampicillin in a simplified selective medium for the isolation of fluorescent pseudomonads. J. Appl. Bacteriol. 37:459-460. [DOI] [PubMed] [Google Scholar]
- 42.Stachelhaus, T., H. D. Mootz, V. Bergendahl, and M. A. Marahiel. 1998. Peptide bond formation in nonribosomal peptide biosynthesis: catalytic role of the condensation domain. J. Biol. Chem. 273:22773-22781. [DOI] [PubMed] [Google Scholar]
- 43.Stanghellini, M. E., and R. M. Miller. 1997. Biosurfactants: their identity and potential efficacy in the biological control of zoosporic plant pathogens. Plant Dis. 81:4-12. [DOI] [PubMed] [Google Scholar]
- 44.Stuurman, J., M. J. De Vroomen, H. J. J. Nijkamp, and M. J. J. van Haaren. 1996. Single site manipulation of tomato chromosomes in vitro and in vivo using Cre-lox site-specific recombination. Plant Mol. Biol. 32:901-913. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thrane, C., S. Olsson, T. H. Nielsen, and J. Sørensen. 1999. Vital fluorescent stains for detection of stress in Pythium ultimum and Rhizoctonia solani challenged with viscosinamide from Pseudomonas fluorescens DR54. FEMS Microbiol. Ecol. 30:11-23. [Google Scholar]
- 47.Thrane, C., T. H. Nielsen, M. N. Nielsen, J. Sørensen, and S. Olsson. 2000. Viscosinamide-producing Pseudomonas fluorescens DR54 exerts a biocontrol effect on Pythium ultimum in sugar beet rhizosphere. FEMS Microbiol. Ecol. 33:139-146. [DOI] [PubMed] [Google Scholar]
- 48.Thrane, C., M. N. Nielsen, J. Sørensen, and S. Olsson. 2001. Pseudomonas fluorescens DR54 reduces sclerotia formation, biomass development, and disease incidence of Rhizoctonia solani causing damping-off in sugar beet. Microbial Ecol. 42:438-445. [DOI] [PubMed] [Google Scholar]
- 49.Van Dyke, M. I., P. Couture, M. Brauer, H. Lee, and J. T. Trevors. 1993. Pseudomonas aeruginosa UG3 rhamnolipid biosurfactants: structural characterization and their use in removing hydrophobic compounds from soil. Can. J. Microbiol. 39:1071-1077. [DOI] [PubMed] [Google Scholar]
- 50.Van Os, G. J., W. J. M. van Gulik, and W. J. de Boer. 1998. Disease development of Pythium root rot in bulbous Iris and Crocus. Ann. Appl. Biol. 132:227-238. [Google Scholar]
- 51.Zajic, J. E., and W. Seffens. 1984. Biosurfactants. Crit. Rev. Biotechnol. 1:87-107. [Google Scholar]
- 52.Zhou, T., and T. C. Paulitz. 1993. In vitro and in vivo effects of Pseudomonas spp. on Pythium aphanidermatum: zoospore behavior in exudates and on the rhizoplane of bacteria-treated cucumber roots. Phytopathology 83:872-876. [Google Scholar]