Abstract
Isolates belonging to six genera not previously known to oxidize CO were obtained from enrichments with aquatic and terrestrial plants. DNA from these and other isolates was used in PCR assays of the gene for the large subunit of carbon monoxide dehydrogenase (coxL). CoxL and putative coxL fragments were amplified from known CO oxidizers (e.g., Oligotropha carboxidovorans and Bradyrhizobium japonicum), from novel CO-oxidizing isolates (e.g., Aminobacter sp. strain COX, Burkholderia sp. strain LUP, Mesorhizobium sp. strain NMB1, Stappia strains M4 and M8, Stenotrophomonas sp. strain LUP, and Xanthobacter sp. strain COX), and from several well-known isolates for which the capacity to oxidize CO is reported here for the first time (e.g., Burkholderia fungorum LB400, Mesorhizobium loti, Stappia stellulata, and Stappia aggregata). PCR products from several taxa, e.g., O. carboxidovorans, B. japonicum, and B. fungorum, yielded sequences with a high degree (>99.6%) of identity to those in GenBank or genome databases. Aligned sequences formed two phylogenetically distinct groups. Group OMP contained sequences from previously known CO oxidizers, including O. carboxidovorans and Pseudomonas thermocarboxydovorans, plus a number of closely related sequences. Group BMS was dominated by putative coxL sequences from genera in the Rhizobiaceae and other α-Proteobacteria. PCR analyses revealed that many CO oxidizers contained two coxL sequences, one from each group. CO oxidation by M. loti, for which whole-genome sequencing has revealed a single BMS-group putative coxL gene, strongly supports the notion that BMS sequences represent functional CO dehydrogenase proteins that are related to but distinct from previously characterized aerobic CO dehydrogenases.
Carbon monoxide occurs in nonurban atmospheres at about 60 to 300 ppb (11, 33, 43). In spite of these low concentrations, CO participates in a number of chemical reactions that determine the oxidative state of the troposphere as well as the residence times of many organic species, including greenhouse gases such as methane (11, 12, 20, 34). Atmospheric CO also serves as a substrate for soil microbes, which remove as much as 15% of the annual CO flux to the atmosphere (7, 21). Although soil microbes play an important role in the global CO budget, the populations involved remain poorly known (7, 8). Limited evidence indicates that fungi, actinomycetes, and bacteria actively consume CO but that “classic” carboxydotrophs play a minimal role in situ (3, 10). However, the role of such carboxydotrophs has only been inferred from laboratory studies that may not accurately reflect capabilities expressed in situ.
Carboxydotrophic bacteria constitute a small but diverse group of aerobes (31), primarily within the α-Proteobacteria and the Firmicutes, that utilize CO as sole carbon and energy sources at high (much higher than 1%) concentrations while expressing relatively low-affinity CO uptake systems (apparent Km, ≥500 ppm). While the ecological roles of these organisms need clarification, several carboxydotrophs have provided important insights about the biology, biochemistry, and molecular biology of CO oxidation (30, 31).
In particular, similarities to authentic CO dehydrogenase have provided a basis for identifying putative CO dehydrogenases in genomes of bacteria not previously reported to oxidize CO, including Burkholderia fungorum LB400, Mesorhizobium loti, Mycobacterium smegmatis, M. tuberculosis, and the marine roseobacterium, Silicibacter pomeroyi, among others. Since genomic data have indicated that the trait for CO oxidation occurs more commonly than previously recognized, a survey of selected taxa was undertaken to establish whether putative CO dehydrogenases were associated with CO oxidation in vivo. In addition, known and putative CO dehydrogenase sequences were aligned and used to design primers for PCR amplification of a 1,260- to 1,290-bp region of the coxL gene. These primers were evaluated using CO oxidizers enriched from terrestrial and marine sources (G. M. King and H. Crosby, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. I-4, p. 247, 2002), including a previously described high-affinity soil isolate (Aminobacter sp. strain COX) (14). The results significantly expand the known diversity of CO-oxidizing bacteria and provide a basis for new ecological studies.
MATERIALS AND METHODS
Culture sources, isolation, and characterization.
Oligotropha carboxidovorans OM5 and M. tuberculosis H37Ra were obtained from the American Type Culture Collection. Bradyrhizobium japonicum USDA 6, M. loti USDA 3471, and Sinorhizobium fredii USDA 205 were obtained from P. van Berkum (U.S. Department of Agriculture, Beltsville, Md.); Bradyrhizobium sp. strain CPP was obtained from T. Wacek (Urbana Laboratories, Urbana, Ill.); B. fungorum LB400 was obtained from J. Haddock (Southern Indiana University, Carbondale, Ind.); Burkholderia sp. strain JS-150 was obtained from J. Spain (Tyndall Air Force Base, Fla.); Burkholderia cepacia and Burkholderia multivorans were obtained from T. Lessie (University of Massachusetts, Amherst); M. avium and M. smegmatis were obtained from M. Glickman (Sloan-Kettering Memorial Cancer Center, New York, N.Y.); Stappia sp. strain CV812-530, Stappia sp. strain CV902-700, S. aggregata, S. stellulata, and Vibrio fischeri were obtained from K. Boettcher (University of Maine, Orono, Maine); and Silicibacter lacuscaerulensis and S. pomeroyi were obtained from M. A. Moran (University of Georgia, Athens, Ga.).
CO-oxidizing bacteria were also isolated from terrestrial and aquatic plant roots and macroalgal tissue. Isolates were obtained by incubating air dried peat, rinsed macroalgae (Ulva lactuca or Ascophyllum nodosum), or soil-free roots of Phaseolus vulgaris (bean), Lupinus perennis (lupine), or Pontederia cordata (pickerelweed) in basal salts medium containing 0.01 to 0.05% yeast extract (14). All of these materials supported CO-oxidizing microbes (24, 36; King and Crosby, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol.). The headspace of enrichment flasks contained 1,000 ppm of CO in air. CO was replenished through several cycles of depletion and addition. Subsamples of medium from flasks that tested positive for CO consumption were transferred to fresh medium and subjected to additional cycles of CO addition and depletion. Positive enrichments were diluted serially and plated onto basal salts agar medium with or without 25 mM pyruvate and incubated in the presence or absence of CO at concentrations up to 20%. Colonies were transferred to liquid medium, grown to stationary phase, and incubated with CO to determine oxidation potentials. Colonies that proved positive for CO oxidation were purified by repeated serial dilution with liquid and solid medium and CO at concentrations of 1,000 ppm. Headspace CO and time courses of CO utilization were assayed by gas chromatography as reported previously (14).
Identification of isolates to the genus level was accomplished by amplifying and sequencing 16S rRNA genes and by using various biochemical and physiological tests. For 16S rRNA analyses, cultures were harvested from late log to stationary phase, resuspended in phosphate buffer, and then subjected to three cycles of heating at 65°C and freezing at −80°C. DNA was extracted and purified using MoBio spin kits (MoBio Inc.; Solana Beach, Calif.). 16S ribosomal DNA (rDNA) was amplified with primers 27f and 1492r in 50-μl reaction mixtures in an Eppendorf Mastercycler (Brinkmann Inc., Westbury, N.Y.) with MasterTaq DNA polymerase (Brinkmann Inc.) and standard amplification conditions (28, 38). PCR products were visualized with ethidium bromide gel electrophoresis and then purified with MoBio PCR cleanup kits. Purified DNA was sequenced bidirectionally (with an ABI model 377 sequencer) by the University of Maine Sequencing Facility. Nearest phylogenetic neighbors for the isolates were determined using BLAST analysis of the GenBank database or Sequence Aligner software from the Ribosomal Database Project (rdp.cme.msu.edu).
Selected biochemical, physiological, and diagnostic traits for new isolates were determined using API 20 NE test strips (BioMerieux Inc., Marcy L'Etoile, France), GN2 substrate plates (Biolog Inc, Hayward, Calif.), and standard microbiological methods for Gram staining, motility, and oxidase reactions (41). Substrates that supported growth were assessed using basal salts supplemented with 0.05% yeast extract and various carbon and energy sources, including CO (20%)-CO2 (5%) or H2 (20%)-CO2 (5%). Growth was measured spectrophotometrically (600 nm) and compared to that of controls containing yeast extract only. Cultures for these analyses were grown at 30°C with rotary shaking.
Cultures not previously known to oxidize CO (Bradyrhizobium sp. strain CPP, Burkholderia sp. strain JS-150, B. fungorum LB400, B. cepacia, B. multivorans, Cycloclasticus spirillensus, Lutibacterium anuloederans, M. loti, Mycobacterium avium, M. smegmatis, M. tuberculosis H37Ra, S. fredii, S. lacuscaerulensis, S. pomeroyi, Stappia aggregata, S. stellulata, and V. fischeri) were grown to stationary phase at 30°C in stoppered 160-ml serum bottles containing 10 ml of basal salts medium with 0.05% yeast extract and 25 mM pyruvate (14) or Middlebrook 7H9 medium with OADC supplement at 37°C (for M. avium and M. tuberculosis H37Ra). CO was added to bottle headspaces at final concentrations up to 1%. Headspace subsamples were obtained (using a needle and syringe) at intervals; CO concentrations were determined by gas chromatography as previously described (22, 23).
PCR amplification and analysis of coxL gene fragments.
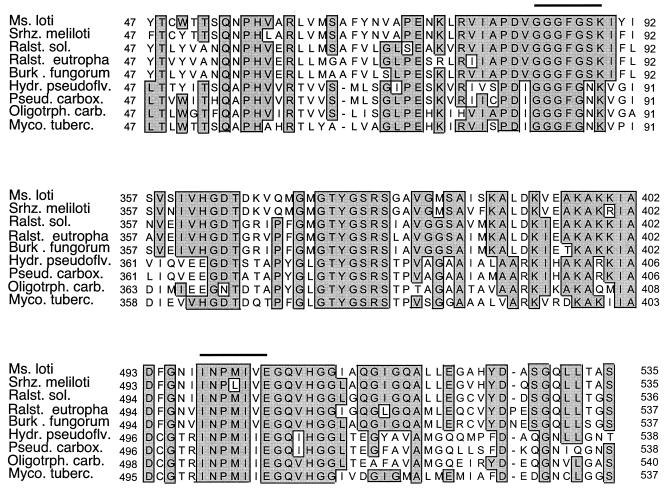
Sequences for the large subunit of authentic CO dehydrogenase genes were obtained from GenBank; putative CO dehydrogenase sequences were obtained from the genome databases for several species (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html): M. loti, M. tuberculosis H37Rv, Pyrobaculum aerophilum, Ralstonia solanacearum, and Sinorhizobium meliloti. Sequences for B. fungorum LB400 and Ralstonia eutropha CH34 were obtained from the Joint Genome Institute (spider.jgi-psf.org/JGI_microbial/html). On the basis of several conserved motifs, inferred amino acid sequences (including that of the active site) (19, 39) were aligned using Clustal X, with manual adjustments as necessary (Fig. 1). Corresponding nucleic acid sequences were used to design two forward primers, OMPf (5′-GGCGGCTT[C/T]GG[C/G]AA[C/G]AAGGT-3′) and BMSf (5′-GGCGGCTT[C/T]GG[C/G]TC[C/G]AAGAT-3′), and a reverse primer, O/Br (5′-[C/T]TCGA[T/C]GATCATCGG[A/G]TTGA-3′). Both forward primers targeted GGFGS(N)K, while the reverse primer targeted INPMIV; these motifs contribute to molybdopterin cytosine dinucleotide (MCD) binding (19, 39). The BMSf-plus-O/Br primer pair was used to amplify putative coxL fragments.
FIG. 1.
Partial alignments of inferred amino acid sequences of the large subunit of CO dehydrogenase (coxL) and putative coxL. Regions targeted for PCR primer design are indicated by horizontal bars. Regions with similar sequences are indicated with shaded boxes. Abbreviations: Ms. loti, Mesorhizobium loti; Srhz. meliloti, Sinorhizobium meliloti; Ralst. sol., Ralstonia solanacearum; Ralst. eutropha, Ralstonia eutropha; Burk. fungorum, Burkholderia fungorum; Hydr. pseudoflv., Hydrogenophaga pseudoflava; Pseud. carbox., Pseudomonas thermocarboxydovorans; Oligotrph. carb., Oligotropha carboxidovorans; Myco. tuberc., Mycobacterium tuberculosis.
PCRs typically used 50-μl volumes in 200-μl tubes with standard final concentrations for buffer, deoxynucleoside triphosphates, and primers (0.1 μM each); magnesium ion was present at a final concentration of 1.5 mM. DNA extracts from cultures were used at final concentrations of approximately 1 to 10 ng, with separate reactions for each of the two forward primers. Positive controls included DNA extracts from known CO oxidizers (e.g., O. carboxidovorans and B. japonicum) (28); negative controls included extracts from organisms for which no CO oxidation has been observed (Burkholderia sp. strain JS-150, B. cepacia, B. multivorans, M. avium, and two recently described marine species, C. spirillensus and L. anuloederans (5). Templates were denatured at 94°C in an Eppendorf Mastercycler thermocycler (Brinkmann Inc.) for 3 min prior to the addition of 0.5 U of MasterTaq DNA polymerase (Brinkmann Inc.) at 80°C. Each amplification cycle consisted of a 45-s denaturation step at 94°C, a 60-s annealing step with the temperature profile described below, and a 90-s extension at 72°C. Annealing temperatures differed according to a “touchdown” protocol, which was initiated at 62°C for two amplification cycles followed by 1°C stepwise decreases in annealing temperature (two amplification cycles for each step) to a final value of 58°C, which was used for 30 amplification cycles. A final 20-min extension at 72°C completed amplification. PCR products were visualized with ethidium bromide gel electrophoresis and then purified with MoBio PCR cleanup kits. Purified DNA was sequenced bidirectionally (using the PCR primers with an ABI model 377 sequencer) by the University of Maine Sequencing Facility.
Inferred amino acid sequences were derived from partial coxL sequences and aligned (using Clustal X as described above) with corresponding sequences from GenBank and genome databases, with manual adjustments as necessary. Sequences were analyzed using PAUP* 4.0b software (Sinauer Associates, Inc.; Sunderland, Mass.) to determine phylogenetic relationships among taxa. After excluding gapped positions, 348 residues were subjected to a neighbor-joining algorithm (1,000 bootstrap replicates) for tree construction. A putative coxL sequence from P. aerophilum was used as an outgroup. Several additional MCD-binding enzymes with sequences similar to that of CO dehydrogenase (e.g., quinoline oxidase, nicotine dehydrogenase) were included in the analysis.
Nucleotide sequence accession number.
Thesequences described in this work have been deposited in GenBank (see Table 1 for accession numbers).
TABLE 1.
Accession numbers for 16S rDNA and coxL gene sequences obtained from isolates analyzed in this studya
| Isolate | 16S accession no. | BMS coxL accession no. | OMP coxL accession no. |
|---|---|---|---|
| Aminobacter sp. strain COX | NDb | AY307908 | —c |
| Bradyrhizobium japonicum USDA 6 | ND | AY307910 | AY307921 |
| Bradyrhizobium sp. strain CPP | ND | AY307900 | AY307913 |
| Burholderia fungorum LB400 | ND | AY307901 | AY307916 |
| Burkholderia sp. strain LUP | AY307923 | AY307907 | — |
| Mesorhizobium sp. strain NMB1 | AY307926 | AY307906 | — |
| Mycobacterium smegmatis | ND | — | AY307918 |
| Stappia sp. strain M4 | AY307928 | AY307902 | AY307916 |
| Stappia sp. strain M8 | AY307927 | AY307903 | AY307917 |
| Stappia sp. strain CV812-530 | ND | AY307898 | AY307193 |
| Stappia sp. strain CV902-700 | ND | AY307899 | AY307914 |
| Stappia aggregata | ND | AY307904 | AY307918 |
| Stappia stellulata | ND | AY307905 | AY307919 |
| Stenotrophomonas sp. strain LUP | AY307922 | AY307920 | — |
| Xanthobacter sp. strain COX | AY307925 | AY307911 | — |
See text for methods used.
ND, not determined.
Dashes indicate that no coxL product was detected by PCR analysis.
RESULTS
Pure culture CO oxidation.
Newly isolated CO oxidizers from marine and terrestrial sources consumed CO at rates from about 6 to 101 μg of CO mg−1 of protein h−1 for initial concentrations of about 1,000 ppm (Table 2). Similar CO consumption rates were also observed for a number of strains for which a CO oxidation capacity had not been previously reported (rate values represent means ± 1 standard error [in micrograms]): Bradyrhizobium sp. strain CPP, 9 ± 2; B. fungorum LB400, 15 ± 1; M. loti, 17 (n = 2); S. fredii, 21 (n = 2); S. pomeroyi, 17 (n = 2); Stappia sp. strain CV812-530, 13 ± 2; Stappia sp. strain CV902-700, 21 ± 3; and S. stellulata, 32 ± 7. CO was not oxidized by Burkholderia sp. strain JS-150, B. cepacia, B. multivorans, M. avium, S. lacuscaerulensis, or three marine isolates, C. spirillensus, L. anuloederans, and V. fischeri.
TABLE 2.
Selected characteristics of root and macroalgal isolates
| Characteristic | Result for isolate:
|
|||||
|---|---|---|---|---|---|---|
| Burkholderia sp. strain LUP | Mesorhizobium sp. strain NMB1 | Stappia sp. strain M4 | Stappia sp. strain M8 | Stenotrophomonas sp. strain LUP | Xanthobacter sp. strain COX | |
| Motility | + | + | + | + | − | − |
| Oxidase reaction | + | + | + | + | − | + |
| Nitrate reduction | − | + | + | + | − | + |
| Denitrification | − | − | + | + | − | − |
| CO uptake ratea | 34 ± 1 | 25 ± 3 | 10 ± 1 | 6 ± 1 | 101 ± 14 | 17 ± 0.3 |
| Hydrogen oxidation | + | − | + | + | − | + |
| Growth substratesb | ||||||
| Sugars | 5/9 | 7/8 | 9/9 | 9/9 | 2/9 | 0/8 |
| Amino acids | 7/8 | 4/8 | 4/8 | 5/8 | 2/8 | 1/4 |
| Organic acids | 15/15 | 6/15 | 13/15 | 11/15 | 6/15 | 10/12 |
| Methanol | − | − | − | − | + | + |
| Sodium required | − | − | + | + | − | − |
CO uptake rates are means ± 1 standard error for triplicate determinations.
Numbers for growth substrates represent positive substrate responses out of total examined.
Isolate characterization.
CO-oxidizing bacteria were obtained from terrestrial sources (peat and plant roots) and marine macroalgal enrichments. Isolates were identified to the genus level primarily on the basis of 16S rDNA sequence results. Nearest phylogenetic neighbors and percent similarities (in parentheses) for the isolates were as follows: B. fungorum LB400 for Burkholderia sp. strain LUP (99.3%); Mesorhizobium sp. strain SH 15003 for Mesorhizobium sp. strain NMB1 (99.9%); Stappia sp. strain CV902-700 for Stappia sp. strain M4 (97.9%); Stappia sp. strain CV812-530 for Stappia sp. strain M8 (99.9%); Stenotrophomonas maltophila sp. strain LMG-957 for Stenotrophomonas sp. strain LUP (99.1%); and Xanthobacter sp. strain INA43/2-2 for Xanthobacter sp. strain COX (98.2).
Genus assignments were supported by the results for Gram reactions (all negative), oxidase reaction, morphology, motility, pigmentation, ability to reduce nitrate, ability to denitrify, sodium requirement, and patterns of heterotrophic substrate utilization (Table 2). In addition, Burkholderia sp. strain LUP oxidized biphenyl, which was consistent with the characteristics of its nearest phylogenetic neighbor, B. fungorum LB400, and Xanthobacter sp. strain COX produced a yellow pigment characteristic of the genus. Stenotrophomonas sp. strain LUP and Xanthobacter sp. strain COX also grew with methanol as a sole carbon and energy source.
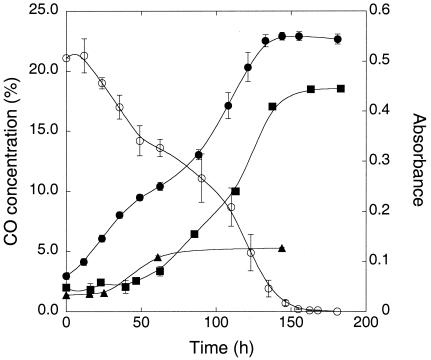
All isolates oxidized CO during stationary phase when incubated with headspace CO concentrations from 1% to <10 ppm (data not shown). However, only one isolate, Xanthobacter sp. strain COX, grew with elevated CO (20%) as a sole carbon and energy source; this isolate also grew with 20% H2[5% CO2 (Fig. 2). The remaining isolates did not grow appreciably with elevated CO under oxic conditions even during extended (several-month) incubations. Further, all isolates except Xanthobacter sp. strain COX appeared to be at least partially inhibited by high CO concentrations, since CO at levels > 1% was consumed slowly if at all.
FIG. 2.
Growthof Xanthobacter sp. strain COX on mineral salts with 0.05% yeast extract-20% CO (•), 20% H2-5% CO2 (▪), or 0.05% yeast extract only (▴) over the time course (in hours) of the experiment. Values represent means of triplicate determinations ± 1 standard error.
coxL PCR and sequence analysis.
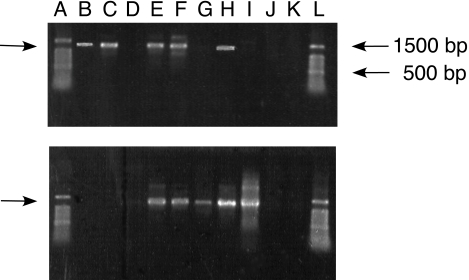
PCR products of the expected size (about 1,260 to 1,290 bp) were obtained from all taxa that oxidized CO (Fig. 3). For certain taxa (e.g., O. carboxidovorans and Xanthobacter sp. strain COX) a single coxL or putative coxL product was obtained from reactions with one or the other of the two forward primers (e.g., OMPf and BMSf), while for other taxa (e.g., B. japonicum and S. stellulata) distinct products were obtained from reactions with each of the two primer sets. PCR results were negative for taxa that did not oxidize CO (e.g., Burkholderia sp. strain JS-150, B. cepacia, B. multivorans, M. avium, C. spirillensus, and L. anuloederans).
FIG. 3.
PCR products from amplification with OMPf (upper panel) and BMSf (lower panel) primers of genomic extracts of CO-oxidizing cultures. Products were visualized with ethidium bromide after gel electrophoresis using 1% agarose. Lanes A and L contain a 100-bp ladder (Promega, Inc.); 500- and 1,500-bp markers are indicated by arrows on the right side of the upper panel. Lane B, Oligotropha carboxidotropha; lane C, M. smegmatis; lane D, M. avium; lane E, B. fungorum LB400; lane F, Stappia stellulata; lane G, Mesorhizobium sp. strain NMB1; lane H, Bradyrhizobium sp. strain CPP; lane I, Aminobacter sp. strain COX; lane J, L. anuloederans; lane K, negative control.
Nucleic acid sequences for PCR products from B. japonicum, B. fungorum LB400, M. tuberculosis H37Ra, M. smegmatis, and O. carboxidovorans were identical or nearly identical (<5 bases differing in about 1,200 bp) to coxL or putative coxL sequences deposited in GenBank or genome databases. For B. japonicum and B. fungorum LB400, coxL sequences from reactions with each of the forward primers matched distinct sequences annotated as putative CO dehydrogenases in the respective genome databases.
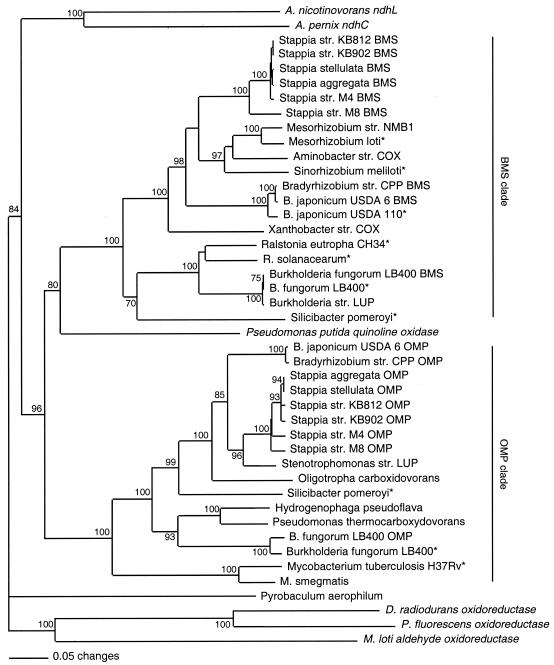
Phylogenetic analysis of inferred amino acid sequences revealed two clades (Fig. 4). One clade, OMP, contained sequences similar to that of O. carboxidovorans coxL; these sequences characteristically contained the active site motif, AYRCSFR (see references 30 and 41). A second clade, BMS, contained putative coxL sequences from several taxa that were not represented in OMP (e.g., M. loti and S. meliloti). Otherwise, the BMS clade was dominated by taxa containing both OMP and BMS coxL sequences. BMS sequences characteristically contained the active site motif, AYRGAGR. Within each clade, taxa tended to form clusters congruent with 16S rRNA phylogeny (Fig. 5). Thus, α-proteobacterial taxa formed a group distinct from those of the β/γ-Proteobacteria and Mycobacteria. Nicotine dehydrogenase and quinoline oxidase sequences were distinct from those of both the OMP and BMS clades. The general topology of the coxL tree was supported strongly by bootstrap analysis and was comparable for both maximum parsimony and maximum likelihood analyses (data not shown). In addition, the coxL tree topology was retained for analyses of full amino acid sequences from select taxa and for analyses of a smaller region (about 150 residues) encompassing the active site and only one of the MCD-binding motifs (data not shown).
FIG. 4.
Neighbor-joining analysis (1,000 bootstrap replicates) of partial inferred amino acid sequences for authentic and putative coxL and inferred amino acid sequences of selected molybdenum hydroxylases (indicated in italic characters in the figure): Aeropyrum pernix nicotine dehydrogenase, chain C, accession number NP148464; Arthrobacter nicotinovorans nicotine dehydrogenase, large subunit, AAK6423; Deinococcus radiodurans oxidoreductase, 285554; M. loti aldehyde oxidoreductase, NP105651; and Pseudomonas putida quinoline 2-oxidoreductase, CAA66830. Sequences derived from genome databases are indicated with asterisks. Bootstrap values < 70% are not shown. A P. aerophilum putative coxL sequence was used as an outgroup.
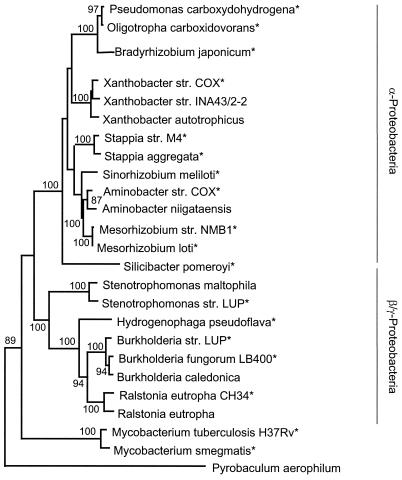
FIG. 5.
Neighbor-joining analysis (1,000 bootstrap replicates, Jukes-Cantor correction) of 1,100 bp of an aligned 16S rDNA sequence for selected CO-oxidizing isolates (indicated by asterisks) and related taxa. Accession numbers for taxa not listed in Table 1: Aminobacter niigataensis, AJ011761; B. japonicum USDA 110, AF363150; B. caledonica, AF215704; B. fungorum LB400, U86373; Hydrogenophaga pseudoflava, AF078770; M. loti, AY278875; M. smegmatis, AJ131761; M. tuberculosis, AJ536031; Oligotropha carboxidotropha, AB099660; R. eutropha CH34, Y10824; R. eutropha, M32021; S. pomeroyi, AF098491; S. aggregata, D88520; Xanthobacter autotrophicus, X94201; and Xanthobacter sp. strain INA43/2-2, AJ306541. Bootstrap values < 50% are not shown. An P. aerophilum sequence was used as an outgroup.
DISCUSSION
Results from enrichment cultures using macroalgal tissues and terrestrial and aquatic plant roots provisionally expand the range of known CO-oxidizing genera to include Burkholderia, Mesorhizobium, Stenotrophomonas, Xanthobacter, and Stappia, a strictly marine genus (Table 1 and Table 2). Genus assignments for the isolates are made on the basis of a high (>97%) degree of similarity between isolate 16S rDNA sequences and sequences of the nearest neighbors in GenBank. The biochemical and physiological characteristics of the various isolates (Table 2) are also consistent with published reports for the respective genera (see, e.g., references 1, 6, 13, 26, 31, 34, 36, 37, 41, and 42), none of which has been recognized before as carboxydotrophic.
Although the genomes of B. fungorum LB400, M. loti, S. meliloti, and S. pomeroyi contain sequences annotated as putative CO dehydrogenases, results presented here are the first to document that these and (or) congeneric taxa actually oxidize CO. These observations and documentation of CO oxidation by M. smegmatis and M. tuberculosis H37Ra (26, 33), for which genome sequences first suggested the possibility of CO oxidation (see, e.g., reference 41), indicate that a number of additional taxa containing putative CO dehydrogenases might oxidize CO. These include P. aerophilum (www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html) and Rhodospirillum rubrum (spider.jgi-psf.org/JGI_microbial/html).
With the exception of Xanthobacter sp. strain COX, neither root nor marine isolates grow with elevated CO (i.e., >1% headspace concentration). Although all isolates oxidize CO at levels < 1%, high concentrations inhibit CO utilization for most of the strains used in this study. This is consistent with previous reports for a high-affinity soil isolate (14) and intact soil (22, 27). Thus, it appears that two groups of CO oxidizers may be distinguished: a CO-tolerant group consisting of isolates that grow with high CO and a CO-sensitive group inhibited by concentrations higher than about 1%. The physiological basis for the distinction between these groups is not yet apparent. However, the latter may be more significant in situ, judging on the basis of responses of soil to elevated CO levels (22, 27).
CO consumption by marine heterotrophs, e.g., Stappia isolates and S. pomeroyi, is significant, because microbial activity represents an important CO sink in the oceans (9, 15, 44) and because CO may represent a significant source of metabolizable carbon. Ammonia oxidizers have been considered dominant CO oxidizers in at least some systems (16-18), but more conventional carboxydotrophs such as those documented here may contribute substantially, as occurs in soils where the role of ammonia oxidizers appears minimal (see, e.g., references 22 and 27).
Thus far, all known Stappia isolates, including two strains from oysters (4), oxidize CO and possess genes for ribulose-1,5-bisphosphate (G. M. King, unpublished results), which plays a central role in lithotrophic carbon fixation. This suggests that the genus Stappia may be carboxydotrophic and function lithotrophically, mixotrophically, or heterotrophically. CO utilization appears more variable in the genus Silicibacter, with a marine isolate, S. pomeroyi, oxidizing CO while a nonmarine isolate, S. lacuscaerulensis, shows no uptake. Genome sequence and physiological results further indicate that S. pomeroyi does not contain rubisco or fix CO2 into biomass (M. A. Moran, personal communication). This suggests the possibility that CO can serve as an energy source for S. pomeroyi and perhaps other strains. However, since three other marine isolates assayed in this study, C. spirillensus, L. anuloederans, and V. fischeri, do not oxidize CO, the distribution and functional roles of CO oxidation obviously differ markedly.
Although a prior survey indicated that Xanthobacter does not oxidize CO (35), results presented here suggest otherwise. However, it is unclear whether CO oxidation occurs rarely within the genus or whether it is associated with hydrogen lithotrophy or methylotrophy, traits expressed by Xanthobacter sp. strain COX (Table 2). CO oxidation by Burkholderia was also previously unknown, but results for Burkholderia sp. strain LUP and B. fungorum LB400 clearly expand the physiological diversity of the genus. Isolation from Hawaiian volcanic deposits of a CO-oxidizing strain similar to Burkholderia caledonica (King, unpublished) suggests that the trait may be relatively common within the Burkholderia graminis clade defined by Coeyne et al. (6). Nonetheless, the negative results for B. cepacia, B. multivorans, and Burkholderia sp. strain JS-150 obtained in this study indicate that the capacity for CO oxidation differs considerably within the genus.
Physiological and molecular data indicate that members of the genus Stenotrophomonas oxidize CO. This represents only the second documentation of CO oxidation by a γ-proteobacterium. Thus far, α-Proteobacteria dominate the known carboxydotrophs. This may reflect the limited nature of surveys to date or more fundamental aspects of the evolution and ecology of CO oxidation (30, 31). A comparable situation exists for symbiotic nitrogen fixation, which is limited almost exclusively to α-Proteobacteria, with only one example of a β-proteobacterial legume symbiont (32).
Primers developed from known and putative coxL sequences may prove useful in developing new insights about CO dehydrogenase distribution and evolution. PCR products obtained from CO-oxidizing strains in this study yield sequences that form two closely related but phylogenetically distinct clades. The OMP clade contains several well-known carboxydotrophs (e.g., O. carboxidovorans and H. pseudoflava) plus newly identified CO oxidizers.
In contrast, newly documented CO oxidizers dominate the BMS clade, which is characterized by putative coxL for which inferred amino acid sequences are about 70% similar to OMP sequences. The active site of BMS sequences, AYRGAGR, differs distinctly from the OMP active site, AYRCSFR, but is similar to that of several other molybdenum hydroxylases, a family of diverse enzymes that includes CO dehydrogenases, quinoline oxidase, aldehyde oxidoreductase, and nicotine dehydrogenases among others (19, 40). Nonetheless, both OMP coxL and BMS putative coxL are phylogenetically distinct from other molybdenum hydroxylases (Fig. 5).
Although the M. loti genome sequence indicates that BMS putative cox genes differ from OMP cox genes in their structural arrangements (subunit orders of MLS[medium, large, small] with no flanking accessory genes versus MSL with flanking accessory genes, respectively) (19, 39, 40), three lines of evidence suggest that the BMS genes code for functional CO dehydrogenases. First, the M. loti genome appears to contain only putative BMS cox genes; no OMP gene sequences have been documented. Thus, CO oxidation by M. loti can be reasonably ascribed to BMS genes. Second, some of the isolates described here, e.g., Mesorhizobium sp. strain NMB1 and Xanthobacter sp. strain COX, oxidize CO (Table 2; Fig. 2) but yielded PCR products only from the BMS primer (Fig. 3). This observation is also consistent with BMS genes coding for functional CO dehydrogenases. Third, BMS putative coxL in the genomes of B. fungorum LB400, M. loti, S. meliloti, and S. pomeroyi is associated with putative coxM and coxS genes, which provide for the synthesis of functional CO dehydrogenase.
While some isolates contain only BMS or OMP genes, several contain both, including all Bradyrhizobium strains, B. fungorum LB400, S. pomeroyi, and all Stappia strains (Fig. 3 and 4). The co-occurrence of BMS and OMP coxL sequences raises questions about the expression and physiological and ecological roles of CO dehydrogenases. Whether both are expressed in the same organism and under what conditions are not known. However, BMS and OMP coxL may affect whole-cell affinities for CO, since Aminobacter sp. strain COX, a high-affinity CO oxidizer (14), may contain only BMS coxL while O. carboxidovorans, a low-affinity CO oxidizer (10), contains only the OMP sequence. In strains that possess both sequence types, expression may be related to available CO concentrations, with OMP expressed when concentrations are relatively high. Thus, differences among strains in affinities for CO may reflect which coxL sequence types are present. The availability of primers for each offers options for exploring this question and for addressing responses of CO oxidizers to both ambient and elevated CO levels.
Finally, the topologies of OMP coxL and BMS putative coxL sequences parallel the topology of 16S rRNA, with two exceptions (Fig. 5). First, S. pomeroyi BMS coxL groups with β-proteobacterial coxL sequences and its 16S rRNA groups with sequences of α-Proteobacteria. Second, OMP coxL from Stenotrophomonas sp. strain LUP, a γ-proteobacterium, falls within a group of α-proteobacterial sequences (Fig. 4). These exceptions notwithstanding, it does not appear that lateral gene transfer plays a prominent role in the distribution of the trait for CO oxidation among major bacterial lineages. Obviously, isolation and analysis of additional cultures will help clarify the evolution of the cox system.
In summary, the phylogenetic and taxonomic diversity of aerobic CO-oxidizing bacteria has been significantly expanded through enrichment and isolation of CO oxidizers and through culture-based assays of taxa containing putative coxL but not previously known to oxidize CO. Previous reports have identified 12 genera with approximately as many species that oxidize CO (2, 29, 30). Direct assays of CO uptake indicate that seven additional genera contain CO oxidizers. Results from molecular studies presented here and from genome sequencing indicate that other taxa, including certain archaea (e.g., Pyrobaculum and Sulfolobus), likely oxidize CO aerobically as well. Collectively, these results suggest that the ability to utilize CO occurs widely among aerobic bacteria, perhaps because CO can serve as an energy supplement when organic substrates are limiting. This notion is consistent with recent observations that CO utilization is associated with microbial community development on volcanic deposits (25) and that populations of CO oxidizers in these systems appear to be very diverse (K. E. Dunfield and G. M. King, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., N-250, p. 443,2003).
Acknowledgments
I thank J. Rich for his efforts to enrich and initially characterize Xanthobacter sp. strain COX while a student at the University of Maine; I thank Heidi Crosby for her efforts to enrich and characterize several plant isolates while an REU student, and I acknowledge the technical support of K. Hardy and K. Roache with these isolates. I thank K. Dunfield for assistance with rhizobial cultures and PCR analyses. I gratefully acknowledge generous gifts of cultures from K. Boettcher, J. Haddock, M. Glickman, T. Lessie, M. A. Moran, J. Spain, P. van Berkum, and T. Wacek.
This work was supported in part by funds from the National Science Foundation (grants 0085495 and 0089900).
Contribution 384 from the Darling Marine Center.
REFERENCES
- 1.Assih, E. A., A. S. Ouattara, S. Thierry, J.-L. Cayhol, M. Labat, and H. Macarie. 2002. Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int. J. Syst. E vol. Microbiol. 52:559-568. [DOI] [PubMed] [Google Scholar]
- 2.Auling, G., J. Busse, M. Han, H. Hennecke, R. Kroppenstedt, A. Probst, and E. Stackenbrandt. 1988. Phylogenetic heterogeneity and chemotaxonomic properties of certain gram-negative aerobic carboxydobacteria. Syst. Appl. Microbiol. 10:264-272. [Google Scholar]
- 3.Bartholomew, G. W., and M. Alexander. 1982. Microorganisms responsible for the oxidation of carbon monoxide in soil. Environ. Sci. Technol. 16:300-301. [DOI] [PubMed] [Google Scholar]
- 4.Boettcher, K. J., B. J. Barber, and J. T. Singer. 2001. Additional evidence that juvenile oyster disease is caused by a member of the Roseobacter group, and colonization of non-infected animals by Stappia stellulata-like strains. Appl. Environ. Microbiol. 66:3924-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, W.-K., and G. M. King. 2001. Isolation, characterization and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl. Environ. Microbiol. 67:5585-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coenye, T., S. Laevens, A. Willems, M. Ohlen, W. Hannant, J. R. W. Govan, M. Gillis, E. Falsen, and P. Vandamme. 2001. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. E vol. Microbiol. 51:1099-1107. [DOI] [PubMed] [Google Scholar]
- 7.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad, R., and W. Seiler. 1980. Role of microorganisms in the consumption and production of atmospheric carbon monoxide by soil. Appl. Environ. Microbiol. 40:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad, R., and W. Seiler. 1982. Utilization of traces of carbon monoxide by aerobic oligotrophic microorganisms in ocean, lake and soil. Arch. Microbiol. 132:41-46. [Google Scholar]
- 10.Conrad, R., O. Meyer, and W. Seiler. 1981. Role of carboxydobacteria in consumption of atmospheric carbon monoxide by soil. Appl. Environ. Microbiol. 42:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crutzen, P. J., and L. T. Gidel. 1983. A two-dimensional photochemical model of the atmosphere. 2: The tropospheric budgets of the anthropogenic chlorocarbons, CO, CH4, CH3Cl and the effect of various NOx sources on tropospheric ozone. J. Geophys. Res. 88:6641-6661. [Google Scholar]
- 12.Daniel, J. S., and S. Solomon. 1998. On the climate forcing of carbon monoxide. J. Geophys. Res. 103:13249-13260. [Google Scholar]
- 13.de Lajudie, P., A. Willems, G. Nick, F. Moreira, F. Molouba, B. Hoste, U. Torck, M. Neyra, M. D. Collins, K. Lindström, B. Dreyfus, and M. Gillis. 1998. Characterization of tropical tree rhizobia and description of Mesorhizobium plurifarium sp. nov. Int. J. Syst. E vol. Microbiol. 48:369-382. [DOI] [PubMed] [Google Scholar]
- 14.Hardy, K., and G. M. King. 2001. Enrichment of a high-affinity CO oxidizer in Maine forest soil. Appl. Environ. Microbiol. 67:3671-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, J. E., and T. S. Bates. 1996. Sources and sinks of carbon monoxide in the mixed layer of the tropical South Pacific Ocean. Global Biogeochem. Cycles 10:347-359. [Google Scholar]
- 16.Jones, R. D. 1991. Carbon monoxide and methane distribution and consumption in the photic zone of the Sargasso Sea. Deep-Sea Res. 38:625-635. [Google Scholar]
- 17.Jones, R. D., and R. Y. Morita. 1983. Carbon monoxide oxidation by chemolithotrophic ammonium oxidizers. Can. J. Microbiol. 29:1545-1551. [Google Scholar]
- 18.Jones, R. D., R. Y. Morita, and R. P. Griffiths. 1984. Method for estimating chemolithotrophic ammonium oxidation using carbon monoxide. Mar. Ecol. Prog. Ser. 17:259-269. [Google Scholar]
- 19.Kang, B. S., and Y. M. Kim. 1999. Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J. Bacteriol. 181:5581-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil, M. A. K. 1999. Atmospheric carbon monoxide. Chemosphere: Global Change Sci. 1:ix. [Google Scholar]
- 21.King, G. M. 1999. Characteristics and significance of atmospheric carbon monoxide consumption by soils. Chemosphere: Global Change Sci. 1:53-63. [Google Scholar]
- 22.King, G. M. 1999. Attributes of atmospheric carbon monoxide oxidation by Maine forest soils. Appl. Environ. Microbiol. 65:5257-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, G. M. 2000. Impacts of land use on atmospheric carbon monoxide consumption by soils. Global Biogeochem. Cycles 14:1161-1172. [Google Scholar]
- 24.King, G. M. 2001. Aspects of carbon monoxide production and consumption by marine macroalgae. Mar. Ecol. Prog. Ser. 224:69-75. [Google Scholar]
- 25.King, G. M. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69:4067-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, G. M.Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 27.King, G. M., and M. Hungria. 2002. Soil-atmosphere CO exchanges and microbial biogeochemistry of CO transformations in a Brazilian agricultural ecosystem. Appl. Environ. Microbiol. 68:4480-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 29.Lorite, M. J., J. Tachil, J. Sanjuan, O. Meyer, and E. J. Bedmar. 2000. Carbon monoxide dehydrogenase activity in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 66:1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer, O., K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck, and M. Kraut. 1990. Utilization of carbon monoxide by aerobes: recent advances. FEMS Microbiol. Rev. 87:253-260. [Google Scholar]
- 31.Morsdorf, G., K. Frunzke, D. Gadkari, and O. Meyer. 1992. Microbial growth on carbon monoxide. Biodegradation 3:61-82. [Google Scholar]
- 32.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 33.Park, S. W., E. H. Hwang, H. Park, J. A. Kim, J. Heo, K. H. Lee, T. Song, E. Kim, Y. T. Ro, S. W. Kim, and Y. M. Kim. 2003. Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 185:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prather, M. J., R. Derwent, D. Ehhalt, P. Fraser, E. Sanhueza, and X. Zhou. 1995. Other trace gases and atmospheric chemistry, p. 73-126. In J. T. Houghton, L. G. Meira Filho, J. Bruce, H. Lee, B. A. Callender, E. Haites, N. Harris, and K. Maskell (ed.), Climate change 1994. Cambridge University Press, Cambridge, United Kingdom.
- 35.Rainey, F. A., and J. Wiegel. 1996. 16S ribosomal DNA sequence analysis confirms the close relationship between the general Xanthobacter, Azorhizobium, and Aquabacter and reveals a lack of phylogenetic coherence among Xanthobacter species. Int. J. Syst. Bacteriol. 46:607-610. [Google Scholar]
- 36.Rich, J. J., and G. M. King. 1998. Carbon monoxide oxidation by bacteria associated with the roots of freshwater macrophytes. Appl. Environ. Microbiol. 64:4939-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rüger, H.-J., and M. G. Höfle. 1992. Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int. J. Syst. Bacteriol. 42:133-143. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Santiago, B., U. Schübel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115-124. [DOI] [PubMed] [Google Scholar]
- 40.Schübel, U., M. Kraut, G. Mörsdorf, and O. Meyer. 1995. Molecular characterization of the gene cluster coxMSL encoding the molybdenum-containing carbon monoxide dehydrogenase of Oligotropha carboxidovorans. J. Bacteriol. 177:2197-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 42.Uchino, Y., A. Hirata, A. Yokota, and J. Sugiyama. 1998. Reclassification of marine Agrobacterium species: proposals of Stappia stellulata gen. nov., comb. nov., Stappia aggregata sp. nov., nom. rev., Ruegeria atlantica, gen. nov., comb. nov., Ruegeria gelatinovora comb. nov., Ruegeria algicola comb. nov., and Ahrensia kieliense gen. nov., sp. nov., nom. rev. J. Gen. Appl. Microbiol. 44:201-210. [DOI] [PubMed] [Google Scholar]
- 43.Watson, R. T., H. Rodhe, H. Oeschger, and U. Siegenthaler. 1990. Greenhouse gases and aerosols, p. 1-40. In J. T. Houghton, G. J. Jenkins, and J. J. Ephraums (ed.), Climate change: the IPCC scientific assessment. Cambridge University Press, Cambridge, United Kingdom.
- 44.Zafiriou, O. C., S. S. Andrews, and W. Wang. 2003. Concordant estimates of oceanic carbon monoxide source and sink processes in the Pacific yield a balanced global “blue-water” CO budget. Glob. Biogeochem. Cycles 17:1015. [Google Scholar]