Abstract
Unfolded-protein response (UPR) denotes the upregulation of endoplasmic reticulum (ER)-resident chaperone and foldase genes and numerous other genes involved in secretory functions during the accumulation of unfolded proteins into the ER. Overexpression of individual foldases and chaperones has been used in attempts to improve protein production in different production systems. We describe here a novel strategy to improve foreign-protein production. We show that the constitutive induction of the UPR pathway in Aspergillus niger var. awamori can be achieved by expressing the activated form of the transcription factor hacA. This induction enhances the production of Trametes versicolor laccase by up to sevenfold and of bovine preprochymosin by up to 2.8-fold in this biotechnically important fungus. The regulatory range of UPR was studied by analyzing the mRNA levels of novel A. niger var. awamori genes involved in different secretory functions. This revealed both similarities and differences to corresponding studies in Saccharomyces cerevisiae.
Folding, disulfide bond formation, and subunit assembly, as well as core glycosylation of secreted proteins, takes place in the endoplasmic reticulum (ER). The protein-folding capacity of the ER is able to react to external signals; when unfolded proteins overload the ER, a signal transduction pathway called the unfolded-protein response (UPR) is activated. The UPR controls the expression of genes of several ER-resident chaperones and foldases (reviewed in references 19 and 28) and numerous other genes involved in other secretory functions (38).
The Saccharomyces cerevisiae HAC1 gene encodes the transcription factor of the UPR pathway that binds to a specific region of the promoters of the target genes. An unconventional splicing mechanism involving Ire1p and a tRNA ligase removes a 250-nucleotide (nt) intron from the HAC1 mRNA (37). This splicing removes translational attenuation caused by the intron and causes a replacement of the C-terminal portion of the Hac1p, creating a transcriptionally active Hac1p (9). In higher eukaryotes it has been shown that the XBP1 transcription factor binds to the mammalian ER stress-response element (ERSE) (48), and an unconventional intron is cleaved from the XBP-1 mRNA by Ire1 (6, 22). Another ERSE-binding protein in mammals is ATF-6 (47), an ER transmembrane protein that has been shown to be proteolytically processed upon activation of the UPR (17, 48). Thus, it seems that the UPR activation is more complex in higher eukaryotes than in yeast. We have cloned the functional homologues of the yeast HAC1 from Trichoderma reesei, Aspergillus nidulans (33), and Aspergillus niger var. awamori (unpublished data). Our studies have indicated that the activation of the hac1/hacA genes in filamentous fungi includes two events: splicing of a 20-nt intron analogously to yeast HAC1 and truncation of the mRNA at the 5′ flanking region, removing an upstream open reading frame (33).
Filamentous fungi have been used as protein production hosts because of their high secretion capacities. Aspergillus spp. and the fungus T. reesei are able to secrete several tens of grams per liter of native hydrolytic enzymes, but the production of heterologous proteins, except for those from closely related fungal species, often results in yields of only tens of milligrams per liter (14, 29). There is some indication that problems can occur at different steps of protein synthesis and secretion, translation initiation or elongation, translocation into the ER, folding, transport, processing, or secretion (14).
Many approaches have been used to improve foreign-protein production in various expression systems. These include strain improvement by mutagenesis and screening and genetic modifications such as the deletion of proteases from the production strain (43). Yield improvement has also been obtained by the introduction of multiple copies of expressed genes, the use of strong promoters, gene fusions to well-secreted proteins, the use of native signal sequences, and overexpression of individual ER foldase or chaperone genes (1, 8, 11, 31, 36). Although these strategies have been successful for some heterologous proteins, the production of most foreign proteins still remains problematic. In large-scale protein production, the ER may encounter stress that results from the high level of protein expression. It has been shown that the expression of heterologous proteins can activate the UPR pathway (32).
We describe here a general approach for the improvement of heterologous proteins by overexpressing the UPR pathway regulator. Rather than by overexpressing just one limiting factor in heterologous protein production, we induced some functions throughout the secretory pathway by overexpressing the UPR-induced form of the A. niger var. awamori hacA gene in A. niger var. awamori strains producing a laccase from a basidiomycete fungus Trametes versicolor or bovine chymosin. Basidiomycete laccases are generally proteins whose expression in heterologous hosts can be problematic (18). Calf chymosin has been difficult to express in A. niger but, with the use of efficient strain improvement program, commercial levels of production have been reached (11).
MATERIALS AND METHODS
Construction of strains and plasmids.
The A. niger var. awamori ΔAP3 and ΔAP4 (5) strains are equivalent strains with the pepA gene (encoding the major extracellular aspartic proteinase) deleted and having a pyrG-null mutation. ΔAP3 was transformed with the vector pUCpyrGRG3 to create strain ΔAP3pUCpyrGRG3#11. pUCpyrGRG3 consists of the GRG3 expression cassette (bovine preprochymosin gene between the A. niger glaA promoter and terminator) obtained from pGRG3 (10) and the Neurospora crassa pyr4 gene in pUC19. ΔAP4 was transformed with pGPT-LCC1 to create strain ΔAP4:pGPTlaccase producing Trametes versicolor laccase 1. pGPT-LCC1 is a derivative of pGPTpyrG1 (4) containing the N. crassa pyr4 gene and the A. niger glaA promoter and terminator. To create pGPT-LCC1, the Trametes versicolor lcc1 open reading frame (27) was inserted between the glaA promoter and terminator.
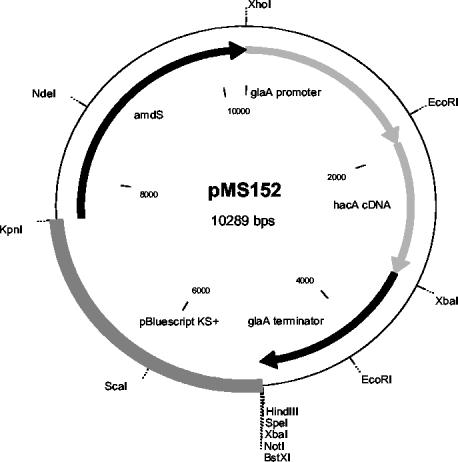
To make a construct for constitutive UPR induction, the induced form of the A. niger var. awamori hacA cDNA was first created by deleting the 20-bp intron and truncating the 5′ flanking region by ca. 150 bp. A truncated hacA PCR fragment was created with the oligonucleotides TCGATTGAATTCGCTGTGTCGACCTACATCACC (forward primer with an EcoRI site) and CGGGGTCGAAATCAACCATA (reverse primer). This fragment was digested with EcoRI and PstI and ligated into EcoRI-NotI-digested pZERO (Invitrogen), together with a 3′-end fragment of hacA digested by PstI and NotI. The hacA gene has two PstI sites precisely at the borders of the 20-bp intron. Thus, the ligation described above created a fragment in which the 5′ end was truncated and the 20-bp intron was removed. This fragment was cloned into the BglII site of the expression vector pGPT-pyrG1 between the glaA promoter and terminator. The hacA expression cassette was cloned as a HindIII-XhoI fragment into the HindIII-XhoI-digested pBluescript II KS(+) (Stratagene) fragment harboring the A. nidulans amdS marker. The hacA overexpression construct (pMS152; Fig. 1) was transformed into the A. niger var. awamori strains ΔAP3pUCpyrGRG3#11 and ΔAP4:pGPTlaccase as described previously (30).
FIG. 1.
Schematic presentation of the pMS152 expression vector containing the A. niger var. awamori hacA gene in constitutively active form. The construction of the vector is explained in the text.
The preprochymosin transformants were cultivated in Clofine special medium (described in international patent application WO 98/31821). The laccase-producing transformants were cultivated in a medium containing (per liter) 8 g of Bacto Soytone (Difco), 12 g of tryptone peptone (Difco), 15 g of (NH4)2SO4, 12.1 g of NaH2PO4 · H2O, and 3.3 g of Na2HPO4 · 7H2O to which were added (per liter) 5 ml of 20% MgSO4 solution, 2 ml of copper citrate solution (110 g of citrate · H2O/liter, 125 g of CuSO4 · 5H2O/liter), 1 ml of Tween 80, 300 ml of 50% maltose solution, and 200 ml of a 100-mg/liter arginine solution after autoclaving. The cultivations were done in 250-ml shake flasks with 70 ml of medium (28°C, 200 rpm).
Nucleic acid methods.
DNA was isolated with the DNAeasy kit (Invitrogen) according to the manufacturer's instructions. Total RNA's were isolated by using the Trizol reagent (Gibco-BRL) as instructed by the manufacturer. Southern and Northern hybridizations were done as described previously (34). The A. niger var. awamori genes encoding secretory functions that were used as probes in Northern hybridizatios, were digested from the plasmid pCMV.SPORT 6 (Life Technologies) with SalI/NotI digestion. The probe fragments were ino1 (GenBank accession no. AY365137), snc1 (AY365139), ktr1 (AY365138), sec61 (AY365136), and nsfA (AF263922). A 950-bp fragment of bipA gene (GenBank accession no. Y08868) was created by PCR with GTCTCCGCCATGGTTCTTGG (forward primer) and GGTGGGCTGGTTATCAGCGG (reverse primer) oligonucleotides with A. niger var. awamori genomic DNA as a template. The lcc1 cDNA (GenBank accession no. U44430) fragment used as a probe was excised from pBK117 plasmid by EcoRI/XhoI digestion. The glaA PCR fragment was created with oligonucleotides CCTGAGCGGCCTCGTCTGCAC (forward primer) and GTCGTATTGCTCGGACATGG (reverse primer) by using pGASKHi vector as a template. The amyB gene fragment encoding α-amylase B from pAMY3 (21) vector with HindIII digestion. Transcript levels were quantified with the PhosphorImager S1 (Molecular Dynamics).
Enzyme activity and protein measurements.
Chymosin activity was measured from samples diluted into 1% sodium acetate buffer. A total of 200 μl of the diluted sample was incubated in 5 ml of buffer containing 11% skim milk (Difco) at 30°C. The milk clotting time was correlated to known standards. Laccase activity was measured as described previously (26). α-Amylase activity was measured with the Phadebas amylase test (Pharmacia) according to the manufacturer's instructions. β-Glucosidase activities were measured as described previously (3). The total protein measurements were done from trichloroacetic acid-precipitated samples by using the Bio-Rad protein assay (Bio-Rad).
To study the quantity of the chymosin protein produced, equal amounts of samples from the culture supernatants of the hacA-overexpressing transformants and the controls were analyzed by Western blotting with a chymosin antibody K336 (Novo). Sodium dodecyl sulfate gels were run from culture medium samples containing 10 μg of total protein, and Western detection was carried out with horseradish peroxidase-conjugated secondary antibody (Bio-Rad). The films from the Western filters were scanned by using a GS-710 imaging densitometer (Bio-Rad).
RESULTS
To obtain constitutive UPR induction in A. niger var. awamori, the UPR-induced form of hacA cDNA that lacked the 20-nt intron and had a truncation of ca. 150 bp at the 5′ end was constructed. This gene was cloned into an expression vector under the control of the A. niger glucoamylase glaA promoter. The resulting plasmid (Fig. 1) was transformed into A. niger var. awamori strains producing Trametes versicolor laccase 1 or bovine preprochymosin. The transformants were identified by Southern hybridization (data not shown).
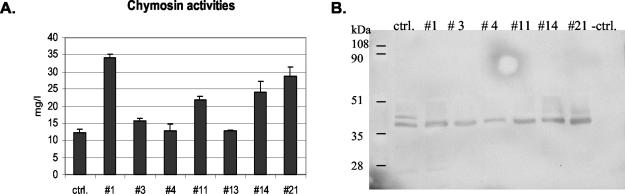
In order to study the effect of constitutive UPR induction on chymosin production, seven transformants shown by Southern hybridization to contain the hacA overexpression construct and the parental strain (ΔAP3pUCpyrGRG3#11) were cultivated in shake flasks, and the chymosin activities were measured from the culture supernatants. The results are shown for the fifth day of cultivation (Fig. 2A). All of the seven transformants studied produced equal or higher chymosin levels than the parental strain. Four transformants produced 1.3- to 2.8-fold more chymosin than the control. Samples from the culture supernatants were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto membrane filters, and probed with chymosin antibody. A band of the size of the mature chymosin was observed in all of the samples. In the parental strain, there were two bands, the smaller one representing the mature chymosin and the bigger one prochymosin. A faint prochymosin signal was also detected in two of the hacA transformants. The results showed that there seems to be more mature chymosin produced by most of the transformants (Fig. 2B).
FIG. 2.
Chymosin activities measured from the culture supernatants of the hacA-overexpressing transformants and the parental strain. (A) Chymosin activities. The results shown are averages (± the standard deviation [±SD]) from two parallel cultures. (B) Western blot analysis of the culture supernatants. The “−ctrl.” lane shows a sample (10 μg of total protein) from the culture supernatant of a laccase-producing control strain that was used as a negative control for the chymosin antibody.
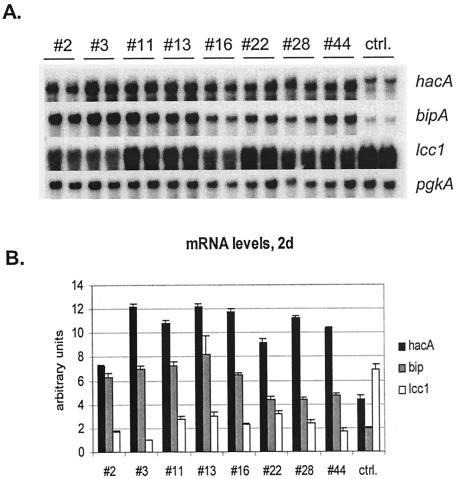
Transformants from the laccase-producing strain were studied in more detail. The strains were cultivated for Northern analysis and measurement of the produced enzyme activities. Northern hybridization from the second culture day of the laccase-producing strains revealed an mRNA of the expected size from the hacA overexpression cassette in all of the transformants studied, in addition to the 1.7-kb band derived from the native hacA gene (Fig. 3). In the 2-day samples derived from the parental strain, a faint hacA band somewhat smaller than the full-length mRNA signal was detected. This suggests that the native UPR pathway might be partially induced in the parental strain, possibly caused by production of laccase. Similarly, the hacA mRNA derived from the overexpression construct was detected in transformants of the preprochymosin strain but not in the parental strain (data not shown).
FIG. 3.
(A) Expression levels of hacA, bipA, and the laccase gene lcc1 in the transformants and parental strain (ctrl.). The results of two parallel cultures are shown for each strain. (B) The signal intensities of the Northern blots were quantified and normalized to the pgkA signal intensities. The panels show the averages (±SD) from the two parallel cultures of each strain.
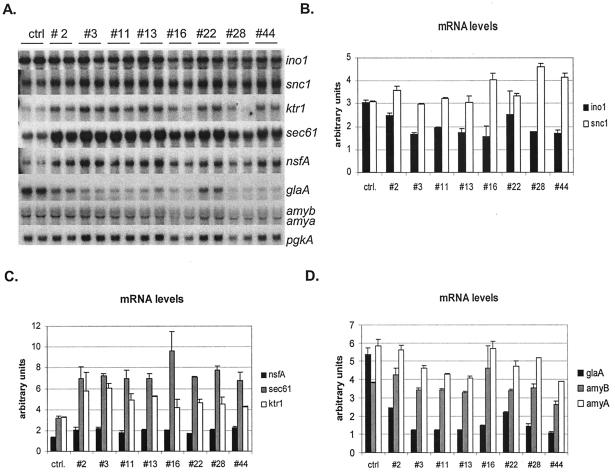
In order to show that the inducing form of hacA had a regulatory effect on the downstream genes of the UPR, Northerns were probed with the bipA gene encoding the major ER chaperone. HacA overexpression led to a significant increase in the bipA transcript levels (two- to fourfold) on the second culture day in all of the hacA-transformants compared to the parental strains (Fig. 3), thus demonstrating that the UPR pathway is induced in the transformants. The results also show that the extent of bipA induction correlates with the expression level of the induced hacA. In samples from later time points (days 5 and 7), the induction of the bipA gene in the hacA transformants was reduced. The induction was 1.25- to 2-fold compared to the parental strain (data not shown). A fragment of the Trametes versicolor lcc1 gene was also used as a probe, and the results show that laccase expression levels were higher in the parental strain than in any of the transformants (Fig. 3). The lower amount of lcc1 mRNA in the transformants may be due to a titration effect of transcription factors since both the laccase gene and hacA was produced under the A. niger glaA promoter (42). The Northerns were also probed with a glaA and α-amylase gene fragments. The results show that the glaA is expressed at ∼2-fold-higher levels in the parental strain than in any of the transformants (Fig. 4D). Probing with the amyB a gene fragment showed two bands. This is because the α-amylase a and b genes are almost identical (21). Quantification and normalization of the signals showed that their expression was not affected by hacA overexpression. This further confirms the titration of the transcription factors for glaA promoter.
FIG. 4.
(A) Expression of ino1, snc1, ktr1, sec61, nsfA, glaA, and amylase genes in the transformants and the parental strain (ctrl). Two parallel cultures are shown for each strain. (B to D) Signal intensities of ino1 and snc1 (B); nsfA, sec61, and ktr1 (C); and glaA, amyA, and amyB (D) genes. The signal intensities of the Northern blots were quantified and normalized to the pgkA signal intensities. The panels show the averages (±SD) from the two parallel cultures of each strain.
It has been shown that the UPR pathway regulates many functions throughout the whole secretory pathway in S. cerevisiae and Arabidopsis spp. (23, 38). Northern blots made from the UPR-induced A. niger var. awamori strains were probed with novel genes encoding different functions in the secretory pathway. The orthologs of these genes in yeast have been shown to be induced by UPR (38). The ino1 encoding inositol-3-phosphate synthase involved in lipid biosynthesis and the snc1 encoding a v-SNARE functioning in exocytosis were not induced in the hacA-overexpressing transformants (Fig. 4A). The ktr1 gene encoding mannosyltransferase involved in protein glycosylation in the Golgi complex and sec61 encoding a major component of the translocon complex were clearly induced in all of the transformants compared to the parental strain (Fig. 4B). The induction of these genes was 1.5- to 3-fold in the transformants. The nsfA gene that is the homologue of yeast SEC18 encoding general fusion factor functioning at different steps of the vesicle transport was also slightly induced in the transformants compared to the parental strain (Fig. 4B).
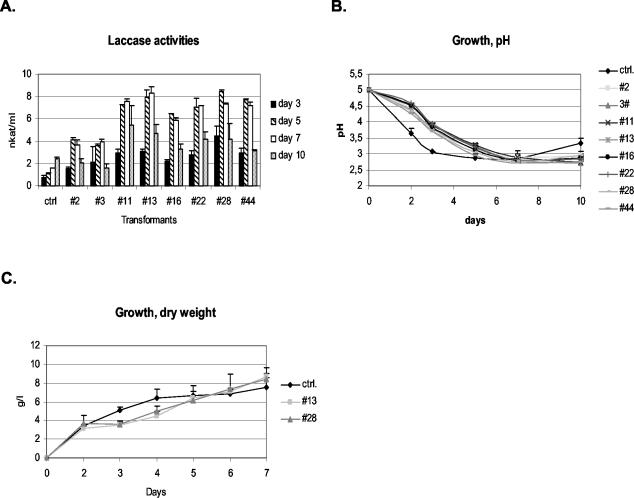
Laccase activity measurements from shake flask cultures in a complex medium showed that all eight hacA transformants studied produced more laccase than the parental strain (Fig. 5A). The laccase levels of the transformants in the fifth day samples were 3- to 7.6-fold higher than those of the parental strain. The transformants produced laccase much more rapidly than the parental strain, reaching the peak already on the fifth day, whereas the parental strain continued to produce more laccase until the end of the culture.
FIG. 5.
(A) Laccase activities measured from the culture supernatants of the hacA-overexpressing transformants and the parental strain. The results shown are averages (±SD) from two parallel cultures. (B and C) pH values (±SD) from two parallel cultures (B) and dry weights (±SD) from three parallel cultures (C) of laccase-producing strains. For the pH measurements, the strains were grown in 50-ml shake flask cultures. For the dry-weight measurements, two hacA-overexpressing transformants and the parental strain were grown in 350-ml shake flask cultures.
The growth and native protein production of the laccase-producing strains were analyzed in detail. The pH was measured from the shake flask cultures described above and showed that the parental strain acidified the culture medium more rapidly than any of the transformants (Fig. 5B), indicating that it was growing faster. To verify this, two transformants and the parental strain were grown in larger shake flask cultures in which the growth could be studied by dry-weight measurements. The hacA transformants grew more slowly than the parental strain but reached a slightly higher final biomass (Fig. 5C). This is in accordance with the previously reported result that the constitutive UPR induction retards growth in yeast (9, 20).
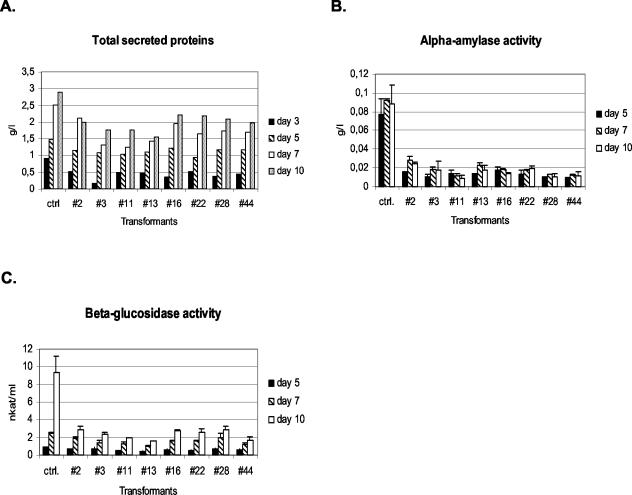
The level of total secreted protein was measured from the shake flask cultures, and the parental strain produced more secreted protein than any of the transformants (Fig. 6A). The transformants that produced the most laccase secreted the least total protein. At the end of the cultures the difference between the parental strain and the transformants was 1.5- to 1.9-fold. The production of specific enzyme activities, i.e., α-amylase and β-glucosidase, was also measured. All hacA transformants produced three- to sevenfold less α-amylase than the control in all time points tested (Fig. 6B) and also less β-glucosidase, although this was only obvious at a later stage of culture (Fig. 6C).
FIG. 6.
(A) Amounts of total secreted protein measured from the culture supernatants of the hacA-overexpressing transformants of the laccase-producing strain and the parental strain grown in 50-ml shake flask cultures. (B and C) α-Amylase (B) and β-glucosidase (C) activities measured form the supernatants of the same cultures. The results (±SD) are from two parallel cultures.
DISCUSSION
The synthesis and secretion processes of proteins in filamentous fungi have been studied widely to understand the problems connected to heterologous protein production. Although the information about the fungal secretion pathway has increased remarkably in recent years (reviewed in reference 8), there still is only limited knowledge of the factors affecting protein production.
One rate-limiting step in the secretion of foreign proteins is in the ER. Limiting amounts of chaperones and foldases involved in proper folding and assembly of proteins may lead to accumulation of misfolded proteins into the ER. It has been shown that this may cause routing of the proteins into the degradation pathway (2, 13). For this reason, the effects of foldase and chaperone expression have been studied in relation to heterologous protein production. Overexpression protein disulfide isomerase (PDI) or KAR2/Bip can increase the production of some heterologous proteins in S. cerevisiae (15, 16, 31, 35, 36). Work along the same lines has been carried out also in filamentous fungi. Overproduction of Bip in strains expressing cutinase variants from Fusarium solani pisi did not affect the protein production capacity in A. awamori (40, 41). On the other hand, overproduction of another ER lumenal chaperone, calnexin, has been shown to increase the production of Phanerochaete chrysosporium manganese peroxidase in A. niger up to fivefold (7). Thaumatin production in A. awamori was improved by PDIA overexpression up to fivefold. The highest improvement was observed in a strain where the relative PDIA level was between two and four compared to a strain with one copy of pdiA gene (24). On the other hand, overexpression of pdiA (25) or the PDI-related gene prpA (44) in A. niger had no effect on heterologous protein production. Also, overexpression of another foldase, cyclophilin, in A. niger did not increase tissue plasminogen activatorproduction (45). This would suggest that the particular secreted protein under investigation greatly affects the outcome.
We have studied the effect of constitutive activation of the UPR pathway on the production of heterologous protein in A. niger var. awamori. It was first demonstrated that the expression of a UPR target gene, bipA, was elevated in the transformants expressing the UPR-activated form of hacA (Fig. 3). The extent of bipA induction would appear to correlate with the expression level of the induced hacA. In the 2-day samples derived from the parental strain, a faint hacA band somewhat smaller than the full-length mRNA signal can be detected. This indicates that the UPR pathway may be induced in the parental strain, possibly by expression of the laccase gene. However, a remarkable increase in the bipA expression levels was achieved by the hacA overexpression.
By expressing the induced form of hacA in a strain producing Trametes versicolor laccase 1 (27), we were able to increase not only the levels of mRNAs encoding an ER-resident chaperone but also the production of both laccase and chymosin compared to the parental strain. Interestingly, even though the laccase mRNA levels were lower in all of the transformants studied compared to the parental strain (Fig. 3), the transformants still secreted more active laccase into the culture medium (Fig. 5A). Thus, the posttranscriptional events of laccase production in the hacA-overexpressing strains were actually enhanced more than what can be concluded just from the secreted activity data.
It would appear that there is an optimal level of hacA expression with respect to laccase production, since the highest levels of laccase were secreted by transformants with intermediate levels of UPR induction. In the present study, we saw more pronounced improvement of laccase production compared to chymosin. This can be due to the intrinsic nature of the expressed proteins that cause problems in their secretion at different stages of secretion. The Western analysis from the culture supernatants of the chymosin-producing strains showed that there seems to be more of mature chymosin produced by the hacA transformants (Fig. 2B). Thus, it is possible that the hacA overexpression enhances the production of the correctly folded chymosin, which is more prone to autocatalytic processing to give mature chymosin. It should be noted that the hacA expression cassette was not directed to a specific locus in the genome. Therefore, it may be that the variations seen in the effects on the production of the two model proteins originate from the site at which the expression cassette was integrated. In order to further optimize this method, screening of the transformants for best producers, perhaps by an automated system, would help in finding the optimal level of the UPR induction versus foreign-protein production.
It has been observed that at least some secreted proteins in filamentous fungi are attached to the cell wall and not secreted efficiently to the culture medium (46). Also a glucoamylase-green fluorescent protein fusion protein has been shown to be partially retained within the cell wall (12). Therefore, we made a simple plate assay with the chromogenic substrate ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] for laccase production and were able to detect similar differences in the laccase activities between the transformants and the parental strain that were observed in the culture supernatants of shake flask cultures. This indicates that the differences seen in the laccase production between the transformants and the parental strain are due to the enhanced secretion of laccase and not, for example, due to the altered structure of the cell walls.
According to pH (Fig. 5B) and dry-weight measurements (Fig. 5C), the growth rate of the transformants with constitutive UPR induction was slower than that of the parental strain. Even so, the rate of laccase production of the transformants was by far faster than the production rate of the parental strain (Fig. 5A). Therefore, the constitutive UPR induction could bring about a remarkable improvement in the specific production rate of laccase (per biomass), a very important measure describing a biotechnical process.
At the same time with enhanced laccase production, the production of native proteins was lessened in the transformants with constitutive UPR induction (Fig. 6). This may be in part due to the slower growth observed in the HACA transformant strains compared to the parental strain. Another factor that should be taken into account is that the α-amylase activity test also measures glucoamylase activity. Thus, since both hacA and Trametes versicolor laccase were expressed from the glaA promoter and since glaA gene is expressed at a lower level in the transformants, the results can be explained by the titration of the regulatory factors as mentioned earlier. On the other hand, it has been shown that induction of UPR in the filamentous fungus T. reesei results in downregulation of the genes encoding secreted proteins (27a). The results presented in the present study do not show such downregulation, since the genes encoding α-amylase were not repressed in the hacA transformants.
It has been shown in S. cerevisiae that the UPR pathway regulates the transcription of approximately 380 genes. Of the functionally characterized genes, 103 are involved in secretion or biogenesis of secretory organelles. As could be expected, these included genes for ER-resident chaperones and foldases. More surprising was that a set of genes encoding for protein functions throughout the whole secretory pathway were induced (38). In our strategy to improve the heterologous protein production in filamentous fungi, the aim was to upregulate the ER folding machinery, as well as other genes from the secretory pathway, by constitutive UPR induction. The results from Northern analysis support the idea that, by constitutive UPR induction, functions throughout the secretory pathway can be induced in A. niger var. awamori. This is seen in the induction of nsfA, sec61, and ktr1 genes that encode functions at different levels of the secretory pathway. In contrast, the expression of ino1 and snc1 genes in A. niger var. awamori was not induced in the hacA transformants. Yeast INO1 is among the genes most strongly induced by UPR; the induction at its highest was ∼10-fold. The gene SNC1 is also induced by UPR in yeast, although the level of induction was lower (ca. two- to threefold higher than in the control). These results indicate that there are both similarities and differences in the range of UPR induction between yeast and A. niger var. awamori.
In another study we obtained improved foreign-protein production by constitutive UPR induction in S. cerevisiae. The overexpression of the UPR-induced form of yeast HAC1 or T. reesei hac1 caused a clear increase in the production of Bacillus amyloliquefaciens α-amylase (39). The experiments reported here indicate that similar induction mechanism of the UPR pathway exists in A. niger var. awamori and in yeast and that, by manipulating the UPR pathway, the protein production can be improved in both organisms. Thus, it would seem that our strategy can be used more generally for the improvement of protein production. The difference between the two organisms is that in yeast the production of a native protein, invertase, was also increased, whereas we did not detect a beneficial effect in A. niger var. awamori on the production of native proteins. This may be due to the low secretion capacity of yeast, in which fine-tuning of the secretory machinery can cause a clear effect on the production of native proteins, as well as differences in the regulation of the secretory machinery as described above.
Acknowledgments
Riitta Nurmi is acknowledged for excellent technical assistance.
The study was partially supported by the Finnish Technology Agency (Tekes).
REFERENCES
- 1.Archer, D. B., D. J. Jeenes, and D. A. Mackenzie. 1994. Strategies for improving heterologous protein production from filamentous fungi. Antonie Leeuwenhoek 65:245-250. [DOI] [PubMed] [Google Scholar]
- 2.Archer, D. B., D. A. MacKenzie, D. J. Jeenes, and I. N. Roberts. 1992. Proteolytic degradation of heterologous proteins expressed in Aspergillus niger. Biotechnol. Lett. 14:357-362. [Google Scholar]
- 3.Bailey, M. J., and K. M. H. Nevalainen. 1981. Induction, isolation, and testing of stable Trichoderma reesei mutants with improved production of solubilizing cellulase. Enzyme Microb. Technol. 3:153-157. [Google Scholar]
- 4.Berka, R. M., and C. C. Barnett. 1989. The development of gene expression systems for filamentous fungi. Biotechnol. Adv. 7:127-154. [DOI] [PubMed] [Google Scholar]
- 5.Berka, R. M., M. Ward, L. J. Wilson, K. J. Hayenga, K. H. Kodama, L. P. Carlomagno, and S. A. Thompson. 1990. Molecular cloning and deletion of the gene encoding aspergillopepsin A from Aspergillus awamori. Gene 86:153-162. [DOI] [PubMed] [Google Scholar]
- 6.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 7.Conesa, A., D. Jeenes, D. B. Archer, C. A. van den Hondel, and P. J. Punt. 2002. Calnexin overexpression increases manganese peroxidase production in Aspergillus niger. Appl. Environ. Microbiol. 68:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conesa, A., P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 9.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. [DOI] [PubMed] [Google Scholar]
- 10.Cullen, D., G. L. Gray, L. J. Wilson, K. J. Hayenga, M. H. Lamsa, M. W. Rey, S. Norton, and R. B. Berka. 1987. Controlled expression and secretion of bovine chymosin in Asperillus nidulans. Bio/Technology 5:369-376. [Google Scholar]
- 11.Dunn-Coleman, N. S., P. Bloebaum, R. M. Berka, E. Bodie, N. Robinson, G. Armstrong, M. Ward, M. Przetak, G. L. Carter, and R. LaCost. 1991. Commercial levels of chymosin production by Aspergillus. Bio/Technology 9:976-981. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, C. L., D. B. Archer, D. J. Jeenes, J. H. Doonan, B. Wells, A. P. Trinci, and G. D. Robson. 2000. A glucoamylase::GFP gene fusion to study protein secretion by individual hyphae of Aspergillus niger. J. Microbiol. Methods 42:39-48. [DOI] [PubMed] [Google Scholar]
- 13.Gouka, R. J., P. J. Punt, J. G. Hessing, and C. A. van den Hondel. 1996. Analysis of heterologous protein production in defined recombinant Aspergillus awamori strains. Appl. Environ. Microbiol. 62:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouka, R. J., P. J. Punt, and C. A. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations, and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Harmsen, M. M., M. I. Bruyne, H. A. Raue, and J. Maat. 1996. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 16.Hayano, T., M. Hirose, and M. Kikuchi. 1995. Protein disulfide isomerase mutant lacking its isomerase activity accelerates protein folding in the cell. FEBS Lett. 377:505-511. [DOI] [PubMed] [Google Scholar]
- 17.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jönsson, L. J., M. Saloheimo, and M. Penttilä. 1997. Laccase from the white-rot fungus Trametes versicolor: cDNA cloning of lcc1 and expression in Pichia pastoris. Curr. Genet. 32:425-430. [DOI] [PubMed] [Google Scholar]
- 19.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 20.Kawahara, T., H. Yanagi, T. Yura, and K. Mori. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8:1845-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korman, D. R., F. T. Bayliss, C. C. Barnett, C. L. Carmona, K. H. Kodama, T. J. Royer, S. A. Thompson, M. Ward, L. J. Wilson, and R. M. Berka. 1990. Cloning, characterization, and expression of two alpha-amylase genes from Aspergillus niger var. awamori. Curr. Genet. 17:203-212. [DOI] [PubMed] [Google Scholar]
- 22.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez, I. M., and M. J. Chrispeels. 2003. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15:561-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moralejo, F. J., A. J. Watson, D. J. Jeenes, D. B. Archer, and J. F. Martin. 2001. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genomics 266:246-253. [DOI] [PubMed] [Google Scholar]
- 25.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. Van Den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niku-Paavola, M. L., E. Karhunen, P. Salola, and V. Raunio. 1988. Ligninolytic enzymes of the white-rot fungus Phlebia radiata. Biochem. J. 254:877-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong, E., W. B. Pollock, and M. Smith. 1997. Cloning and sequence analysis of two laccase complementary DNAs from the ligninolytic basidiomycete Trametes versicolor. Gene 196:113-119. [DOI] [PubMed] [Google Scholar]
- 27a.Pakula, T., M. Laxell, A. Huuskonen, J. Uusitalo, M. Saloheimo, and M. Pentillä. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei: evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem., in press. [DOI] [PubMed]
- 28.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 29.Penttilä, M. 1998. Heterologous protein production in Trichoderma, p. 365-382. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Glioclandium, vol. 2. Taylor & Francis, Ltd., London, United Kingdom.
- 30.Penttilä, M., H. Nevalainen, M. Rattö, E. Salminen, and J. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, A. S., V. Hines, and K. D. Wittrup. 1994. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology 12:381-384. [DOI] [PubMed] [Google Scholar]
- 32.Saloheimo, M., M. Lund, and M. E. Penttilä. 1999. The protein disulphide isomerase gene of the fungus Trichoderma reesei is induced by endoplasmic reticulum stress and regulated by the carbon source. Mol. Gen. Genet. 262:35-45. [DOI] [PubMed] [Google Scholar]
- 33.Saloheimo, M., M. Valkonen, and M. Penttilä. 2003. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47:1149-1161. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schultz, L. D., H. Z. Markus, K. J. Hofmann, D. L. Montgomery, C. T. Dunwiddie, P. J. Kniskern, R. B. Freedman, R. W. Ellis, and M. F. Tuite. 1994. Using molecular genetics to improve the production of recombinant proteins by the yeast Saccharomyces cerevisiae. Ann. N. Y. Acad. Sci. 721:148-157. [DOI] [PubMed] [Google Scholar]
- 36.Shusta, E. V., R. T. Raines, A. Pluckthun, and K. D. Wittrup. 1998. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16:773-777. [DOI] [PubMed] [Google Scholar]
- 37.Sidrauski, C., P. Walter. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90:1031-1039. [DOI] [PubMed] [Google Scholar]
- 38.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 39.Valkonen, M., M. Penttilä, and M. Saloheimo. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Gemeren, I. A., A. Beijersbergen, C. A. van den Hondel, and C. T. Verrips. 1998. Expression and secretion of defined cutinase variants by Aspergillus awamori. Appl. Environ. Microbiol. 64:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Gemeren, I. A., P. J. Punt, A. Drint-Kuyvenhoven, M. P. Broekhuijsen, A. van't Hoog, A. Beijersbergen, C. T. Verrips, and C. A. van den Hondel. 1997. The ER chaperone encoding bipA gene of black aspergilli is induced by heat shock and unfolded proteins. Gene 198:43-52. [DOI] [PubMed] [Google Scholar]
- 42.Verdoes, J. C., A. D. van Diepeningen, P. J. Punt, A. J. Debets, A. H. Stouthamer, and C. A. van den Hondel. 1994. Evaluation of molecular and genetic approaches to generate glucoamylase overproducing strains of Aspergillus niger. J. Biotechnol. 36:165-175. [DOI] [PubMed] [Google Scholar]
- 43.Ward, M. 1989. Heterologous gene expression in Aspergillus. EMBO-ALKO Workshop on Molecular Biology of Filamentous Fungi. ALKO, Espoo, Finland.
- 44.Ward, M., L. J. Wilson, K. H. Kodama, M. W. Rey, and R. M. Berka. 1990. Improved production of chymosin in Aspergillus by expression as a glucoamylase-chymosin fusion. Bio/Technology 8:435-440. [DOI] [PubMed] [Google Scholar]
- 45.Wiebe, M. G., A. Karandikar, G. D. Robson, A. P. Trinci, J. L. Candia, S. Trappe, G. Wallis, U. Rinas, P. M. Derkx, S. M. Madrid, H. Sisniega, I. Faus, R. Montijn, C. A. van den Hondel, and P. J. Punt. 2001. Production of tissue plasminogen activator (t-PA) in Aspergillus niger. Biotechnol. Bioeng. 76:164-174. [DOI] [PubMed] [Google Scholar]
- 46.Witteveen, C. F. B., M. Veenhuis, and J. Visser. 1992. Localization of glucose oxidase and catalase activities in Aspergillus niger. Appl. Environ. Microbiol. 58:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida, H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741-33749. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]