Abstract
Pseudomonas sp. strain ADP uses the herbicide atrazine as the sole nitrogen source. We have devised a simple atrazine degradation assay to determine the effect of other nitrogen sources on the atrazine degradation pathway. The atrazine degradation rate was greatly decreased in cells grown on nitrogen sources that support rapid growth of Pseudomonas sp. strain ADP compared to cells cultivated on growth-limiting nitrogen sources. The presence of atrazine in addition to the nitrogen sources did not stimulate degradation. High degradation rates obtained in the presence of ammonium plus the glutamine synthetase inhibitor MSX and also with an Nas− mutant derivative grown on nitrate suggest that nitrogen regulation operates by sensing intracellular levels of some key nitrogen-containing metabolite. Nitrate amendment in soil microcosms resulted in decreased atrazine mineralization by the wild-type strain but not by the Nas− mutant. This suggests that, although nitrogen repression of the atrazine catabolic pathway may have a strong impact on atrazine biodegradation in nitrogen-fertilized soils, the use of selected mutant variants may contribute to overcoming this limitation.
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is a herbicide of the s-triazine family used for broad-leaf weed control in both crop and noncrop lands. Its widespread use and high mobility in soil have led to its frequent detection in surface water and groundwater at concentrations exceeding the maximum levels allowed (21, 22, 30, 37). The high incidence of atrazine-contaminated water and the increasing concern about the toxicological and ecotoxicological properties of atrazine (3, 6, 16, 17) have boosted research directed toward bioremediation of atrazine-polluted sites.
A few laboratories have reported the isolation of bacteria with the ability to utilize atrazine, achieving in some cases the complete mineralization of the herbicide (see reference 29 and references therein). The best-characterized atrazine-mineralizing bacterial strain is Pseudomonas sp. strain ADP (23), which uses atrazine as the sole nitrogen source by means of a catabolic pathway comprising six enzymatic steps (25, 40). The complete degradative pathway is encoded in the 108-kbp conjugative catabolic plasmid pADP-1, which was recently sequenced (25). The atzA, atzB, and atzC genes, responsible for the conversion of atrazine to cyanuric acid, are harbored at three distant positions within a large (>40 kbp) unstable region in pADP-1. Loss of one or more of these genes is the cause of the frequent appearance of Atr− (unable to utilize atrazine) mutants in nonselective medium (10). The genes involved in the s-triazine ring cleavage and ammonium release are clustered at a different location in pADP-1, to form the atzDEF operon (25). The atzA, atzB, and atzC genes have been shown to be widespread and plasmid borne in a number of independent isolates from different parts of the world (9, 10, 31, 39, 40).
The influence of nitrogen compounds on the efficiency of atrazine catabolism has been the focus of a number of studies, since most atrazine-degrading bacteria use it as a nitrogen source and agricultural soils are often rich in nitrogen due to routine fertilization. Nitrogen amendments have been shown to have a negative effect on atrazine biodegradation by indigenous populations in soils (1, 2, 4, 12). The effect of nitrogen sources on atrazine degradation has also been tested in pure cultures of degrading organisms, and both nitrogen-repressed and nonresponsive strains have been described (15, 28, 36). Pseudomonas sp. strain ADP has been shown to metabolize atrazine rapidly when previously grown on atrazine or glycine while degradation was significantly slower with cells grown on ammonium, nitrate, or urea (5, 19).
In this paper, we use a simple atrazine degradation assay in resting cell suspensions to characterize nitrogen control of the atrazine catabolic pathway in Pseudomonas sp. strain ADP. In addition, we determine the effect of nitrate amendment in soil on atrazine mineralization by Pseudomonas sp. strain ADP and describe a mutant that, by overriding nitrogen control, mineralizes atrazine efficiently in nitrate-amended soil.
MATERIALS AND METHODS
Bacterial strains and growth media.
Minimal medium for Pseudomonas sp. strain ADP was previously described (24). Sodium succinate (1 g liter−1) was always used as a carbon source. Nitrogen sources were added at a final concentration of 4 mM nitrogen, except for atrazine, which was supplied at the saturating concentration from a reservoir. Briefly, 0.15 ml of a 20-mg liter−1 atrazine solution in methanol was added to the cap of a microcentrifuge tube. The open side of the cap was topped with a piece of cellulose dialysis membrane (Sigma-Aldrich Chemie, Steinheim, Germany), which was sealed in place with the top ring of the same microcentrifuge tube. The sealed reservoir was added to the water in the medium and sterilized by autoclaving. This method provides a continuous supply of atrazine without the problems derived from its low water solubility (31 mg liter−1). Minimal agar plates were prepared as described above but with the addition of 20 g of agar noble (Difco, Detroit, Mich.) liter−1. Minimal atrazine agar plates included 0.4 g of atrazine liter−1, added from a 20-g liter−1 solution in methanol. Precipitation of atrazine crystals gives these plates a turbid appearance. Luria-Bertani (LB) agar was used as rich solid medium and was supplemented with atrazine (0.4 g liter−1) when required. Cells were grown at 30°C, and shaking (∼180 rpm) was used to aerate liquid cultures.
Pseudomonas sp. strain ADP (23) was used as a model atrazine-degrading bacterium. This strain is naturally resistant to ampicillin (40 mg/liter). Spontaneous Atr− (deficient in atrazine utilization) mutants were isolated by taking advantage of the intrinsic instability of the atrazine-degradative phenotype (10). Pseudomonas sp. strain ADP was grown to saturation in liquid LB medium containing 1 g of NH4Cl liter−1 and then subcultured in the same medium. After three serial dilutions, cells were plated on LB agar containing 0.4 g of crystallized atrazine liter−1. Colonies not surrounded by clear halos were streaked on minimal agar plates containing cyanuric acid as the sole nitrogen source to test for the cyanuric acid utilization phenotype (Cya). Both Cya+ and Cya− colonies were examined for the presence of atrazine catabolic genes by PCR and restriction analysis of the pADP-1 plasmid. Two derivatives, named MPO100 and MPO101, were chosen for further study. MPO100 is cured from pADP-1, as inferred from its Atr− Cya− phenotype, lack of amplification with specific atzA, atzB, and atzC primers (see reference 9 for primer sequences), absence of plasmid bands in plasmid preparations, and 100-fold-increased conjugation frequency as a recipient of IncP plasmids (incompatible with pADP-1). MPO101 is Atz− Cya+, and PCR and restriction analysis revealed that it harbors a deletion derivative of pADP-1 that lacks virtually the entire unstable region, including the atzA, atzB, and atzC genes and intervening sequences. A spontaneous Nas− (deficient in nitrate assimilation) mutant derivative, named MPO102, was isolated based on chlorate resistance (14) by plating Pseudomonas sp. strain ADP on minimal medium containing KClO3 (10 g liter−1) and then scoring for slow growth on minimal agar plates containing nitrate as the sole nitrogen source. Spontaneous rifampin-resistant (Rifr) derivatives of these strains were isolated when required by mass plating on LB agar containing rifampin (20 mg liter−1).
Plasmid pADP-1 DNA preparation and manipulation.
Small-scale plasmid preparations from Pseudomonas sp. strain ADP were performed by a simple alkaline lysis method, followed by one phenol-chloroform (24:1, vol/vol) extraction and ethanol precipitation (32). Preparations displayed one single slow-migrating band when run on 0.8% agarose gels. Digestion with BamHI, EcoRV, or HindIII yielded band patterns consistent with those expected for pADP-1 according to the published sequence (25).
Resting cell assay of atrazine degradation.
A simple resting cell assay was devised to measure the atrazine degradation of Pseudomonas sp. strain ADP based on a spectrophotometric atrazine chlorohydrolase assay described previously (7). Preinocula were grown to saturation by shaking overnight at 30°C in 3 ml of minimal medium, with ammonium as the sole nitrogen source. Cells were harvested by centrifugation, washed three times with phosphate-buffered saline solution (60 mM sodium-potassium phosphate [pH 7.0], 0.5 g of NaCl liter−1), and resuspended in minimal medium with NH4Cl, NaNO3, l-proline, urea, l-serine, biuret, cyanuric acid, or atrazine as a nitrogen source. In some experiments, atrazine was added as a second nitrogen source. Cultures were subsequently shaken at 30°C for 16 to 20 h to early exponential phase (optical density at 600 nm [OD600], ∼0.3). Cells were harvested by centrifugation, and pellets were washed three times with U buffer (10 mM sodium phosphate [pH 7], 0.1 mM MgSO4) and resuspended in the same buffer to an OD600 of 0.25. Aliquots (5 ml) were placed in a water bath at 30°C and incubated for 2 min, and then 50 μl of 6 mM atrazine was added. Samples (0.5 ml) were withdrawn at 5-min intervals and centrifuged immediately for 3 min in a microcentrifuge at full speed. The atrazine concentration was determined from the absorbance at 225 nm (A225) of the supernatants by using an experimentally derived molar extinction coefficient of 2.71 × 104 liter cm−1 mol−1.
Effect of MSX on atrazine degradation.
The glutamine synthetase inhibitor l-methionine sulfoximine (MSX) was used to inhibit nitrogen assimilation. To determine the appropriate concentration of MSX, Pseudomonas sp. strain ADP was grown in minimal medium with ammonium (1 g of NH4Cl liter−1) as the sole nitrogen source to an OD600 of ∼0.2. Then the culture was split between five flasks and exposed to 0, 0.2, 0.5, 0.8, or 1 mM MSX. The effect of the inhibitor on bacterial growth was determined by monitoring the OD600 of the cultures.
To test the effect of MSX on atrazine degradation, Pseudomonas sp. strain ADP was grown as described above to an OD600 of 0.2. Then the culture was split between two flasks, and MSX was added to one of them to a final concentration of 0.5 mM. After 6 h of incubation, cells were harvested and resting cell assays of atrazine degradation were performed.
Atrazine mineralization assay in soil microcosms.
Microcosm assays of atrazine mineralization in soil were performed by using 25-ml airtight glass vials containing 5 g (equivalent dry weight) of field-moist sieved (<2.8-mm pore size) agricultural surface soil (38). This soil has a low nitrate content (0.016 mg of nitrogen g of soil−1), as determined by a direct colorimetric method (13). When required, soil was amended with KNO3 (0.5 mg of nitrogen g of soil−1). Spontaneous Rifr derivatives of the Pseudomonas sp. strain ADP wild-type and MPO102 strains were grown to mid-exponential phase in minimal medium with atrazine as the sole nitrogen source. Cells were harvested, washed three times in sterile phosphate-buffered saline solution, and inoculated at a density of 108 CFU g of soil−1. Noninoculated controls received the same volume of autoclaved cell suspension. Water content was adjusted to 60% of the holding capacity of the soil. Uniformly ring-labeled [14C]atrazine was subsequently added to the soil (40 μg of atrazine g of soil−1, 250 Bq g of soil−1). A test tube containing 1 ml of 1 M NaOH was placed inside each vial before sealing. Soil microcosms were incubated at 25°C. In acclimation experiments, atrazine-grown Pseudomonas sp. strain ADP cells were incubated in the soil for 6 days prior to the addition of atrazine. At defined time intervals, the vials were opened, and the amount of 14CO2 evolved was determined by scintillation counting of the NaOH solution. Before resealing and further incubation, fresh NaOH (1 ml) was added to each test tube. Scintillation was performed by using a Beckman LS6000TA liquid scintillation system. To monitor the change in bacterial numbers, a parallel set of microcosms was set up, but radiolabeled atrazine was replaced with cold atrazine. Viable Pseudomonas sp. strain ADP counts were determined by plating dilutions at selected time points on LB plates containing 20 mg of ampicillin liter−1 and 10 mg of rifampin liter−1.
Chemicals.
Uniformly ring-labeled atrazine (7.8 mCi/mmol), cyanuric acid, biuret, urea, and MSX were purchased from Sigma-Aldrich-Riedel de Häen. Technical grade atrazine (>98% purity) was a gift from Novartis (Greensboro, N.C.). All other chemicals used were of reagent grade or better.
RESULTS
A simple assay to measure atrazine degradation by Pseudomonas sp. strain ADP.
We have devised a simple method to measure atrazine degradation by resting cells of Pseudomonas sp. strain ADP. Our assay monitors the evolution of atrazine concentration in cell supernatants by means of A225 measurements (see Materials and Methods for further details). Several control experiments were performed to test the validity of this method. A decrease in A225 was not observed when (i) wild-type atrazine-grown cells were killed by boiling prior to the addition of atrazine; (ii) a Pseudomonas sp. strain ADP derivative (MPO100) cured for the pADP-1 plasmid grown on serine as the sole nitrogen source was used; and (iii) another mutant derivative (MPO101) that lacks the atzA, atzB, and atzC genes in pADP-1 grown on serine or cyanuric acid as the sole nitrogen source was used (data not shown). Living cells of wild-type Pseudomonas sp. strain ADP caused a significant decrease in A225 under all of these conditions (see below). These results indicate that the decrease in A225 correlates with the activity of plasmid-borne genes involved in atrazine catabolism.
Effect of nitrogen sources on atrazine degradation.
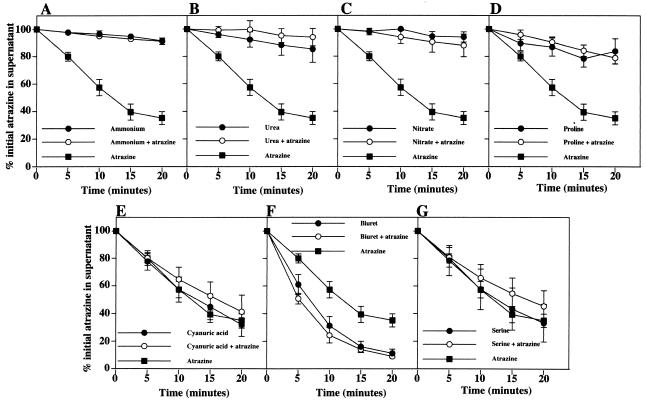
We have used the resting cell assay to address the effect of growth on different nitrogen sources on atrazine degradation by Pseudomonas sp. strain ADP. As expected, atrazine-grown cells exhibited considerable atrazine degradation in our assay (Fig. 1). Cells grown on ammonium as the sole nitrogen source failed to reduce the atrazine concentration in the supernatant significantly (Fig. 1A). This was not a short-term effect of ammonium on atrazine transport or atrazine metabolizing enzymes, since addition of ammonium to the assay buffer did not affect atrazine degradation by atrazine-grown cells (data not shown). The presence of atrazine in addition to ammonium in the growth medium did not stimulate atrazine degradation, suggesting that ammonium-mediated repression operates regardless of the presence of the herbicide in the culture medium. The effect of ammonium was not due to the loss of the catabolic genes, since efficient atrazine degradation was evident when cultures were allowed to exhaust the ammonium by growing to stationary phase (data not shown). Several other nitrogen sources were tested with the same approach. The rate of atrazine degradation by cells grown on urea, proline, or nitrate was low regardless of the presence of atrazine in the growth medium (Fig. 1B to D). However, cyanuric acid, biuret, and serine failed to repress atrazine degradation both in the presence and in the absence of atrazine (Fig. 1E to G). Interestingly, nitrogen sources (ammonium, urea, and proline) that supported fast growth of Pseudomonas sp. strain ADP (doubling time, ∼2 h) repressed atrazine degradation while degradation was efficient with those nitrogen sources on which Pseudomonas sp. strain ADP exhibited slower growth (doubling time, >6 h), such as biuret, atrazine, or serine. Pseudomonas sp. strain ADP had similar doubling times (∼3 h) when grown on nitrate and cyanuric acid, but degradation was observed with cyanuric acid-grown cells and not with nitrate-grown cells. However, this may relate to a specific positive effect of cyanuric acid on the degradative pathway (V. García-González, unpublished data). Taken together, our results suggest that one or more steps in atrazine utilization are repressed under nitrogen-sufficient growth conditions and activated under nitrogen limitation. Activation does not appear to require atrazine or any of its metabolites (Fig. 1G). In addition, atrazine does not stimulate its own catabolism when a repressing nitrogen source is present (Fig. 1A to D).
FIG. 1.
Atrazine degradation in resting cells of Pseudomonas sp. strain ADP grown on different nitrogen sources. The percentage of the initial atrazine concentration remaining in the supernatant at each time point is plotted against time. Each panel represents the results obtained with a different nitrogen source. (A) Ammonium; (B) urea; (C) nitrate; (D) proline; (E) cyanuric acid; (F) biuret; (G) serine. The plot obtained with atrazine-grown cells is displayed in all panels for reference. Closed circles, cells grown on each compound as the sole nitrogen source; open circles, cells grown on each compound plus atrazine as nitrogen sources; closed squares, cells grown on atrazine as the sole nitrogen source. Symbols represent the averages of the results from three to four independent experiments. Error bars represent standard errors of the averages.
Nitrogen incorporation to carbon backbones is required for repression of atrazine degradation.
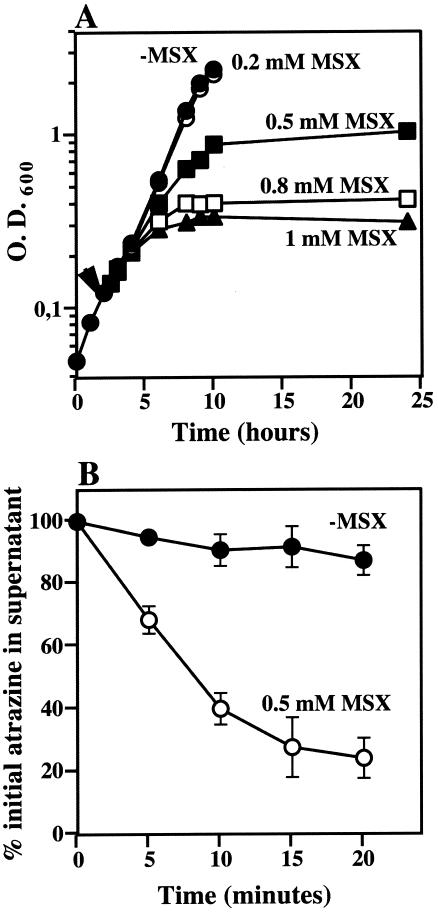
To test whether nitrogen control of atrazine degradation requires assimilation of the nitrogen source, we used MSX, an inhibitor of glutamine synthetase. Treatment of cells growing on ammonium with sublethal concentrations of MSX has been shown to provoke nitrogen starvation in several bacterial species by slowing down ammonium incorporation into carbon backbones and depleting the internal pools of glutamine (8, 34). A concentration of 0.5 mM MSX was chosen, since it had a severe effect on the growth rate but did not cause a complete inhibition of bacterial growth (Fig. 2A). Atrazine degradation by ammonium-grown cells was greatly stimulated when 0.5 mM MSX was also present in the growth medium, as assessed by resting cell assays (Fig. 2B), indicating that slow assimilation is sufficient to override ammonium-mediated repression of the pathway.
FIG. 2.
Effect of MSX on growth and atrazine degradation by resting cells of Pseudomonas sp. strain ADP. (A) Growth curves of Pseudomonas sp. strain ADP growing on ammonium as the sole nitrogen source with the addition of 1 mM MSX (closed triangles), 0.8 mM MSX (open squares), 0.5 mM MSX (closed squares), 0.2 mM MSX (open circles), or no MSX (closed circles, −MSX). The arrowhead indicates the time of addition of MSX. (B) Resting cell assay of atrazine degradation by Pseudomonas sp. strain ADP grown on ammonium (closed circles, −MSX) or ammonium and 0.5 mM MSX (open circles). Symbols represent the averages of the results from four independent experiments. Error bars represent standard errors of the averages.
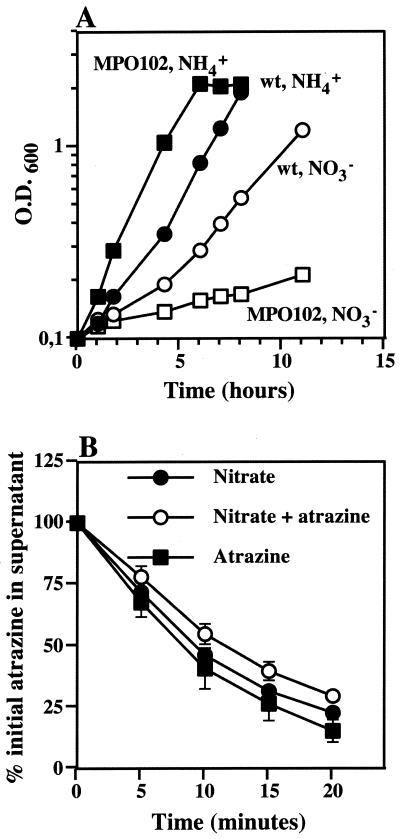
As an alternative means to test nitrogen signaling in Pseudomonas sp. strain ADP, a genetic approach was used. A Nas− mutant (MPO102) was isolated that grows slowly on nitrate but exhibits a growth rate similar to that of the wild type on ammonium (Fig. 3A) and other nitrogen sources (data not shown). Unlike the wild type, MPO102 degraded atrazine efficiently in resting cell assays when grown on nitrate alone or on nitrate and atrazine as nitrogen sources (compare Fig. 3B with 1C). These results strongly suggest that the signaling pathway responsible for nitrogen control of atrazine catabolism requires intracellular incorporation of nitrogen to carbon backbones. In addition, growth-limiting nitrogen assimilation is sufficient to induce atrazine degradation, regardless of the chemical nature of the nitrogen source tested.
FIG. 3.
Growth and atrazine degradation by resting cells of the Nas− MPO102 strain. (A) Growth curve of wild-type (wt) Pseudomonas sp. strain ADP (circles) and MPO102 (squares) growing on ammonium (closed symbols) or nitrate (open symbols) as the sole nitrogen source. (B) Resting cell assay of atrazine degradation by MPO102 grown on nitrate (closed circles), nitrate and atrazine (open circles), and atrazine (closed squares) as nitrogen sources. Symbols represent the averages of the results from three to four independent experiments. Error bars represent standard errors of the averages.
Nitrate addition inhibits atrazine mineralization by Pseudomonas sp. strain ADP in soil.
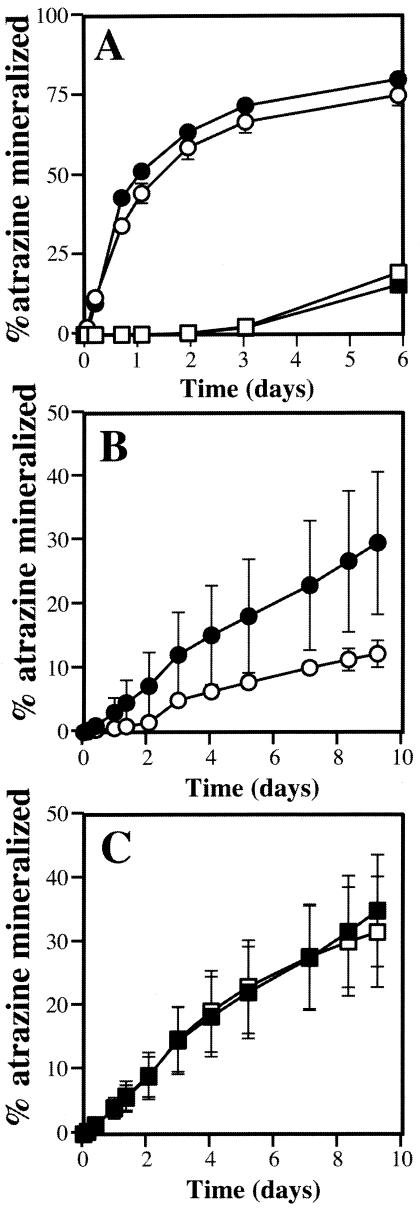
Nitrogen control of atrazine utilization may pose a limitation to degradation of the herbicide by Pseudomonas sp. strain ADP in nitrogen-rich agricultural soils. To address this issue, soil microcosm experiments were designed in which the rate of radiolabeled atrazine mineralization by Pseudomonas sp. strain ADP was determined in nonsterile low-nitrogen soil samples. In initial control experiments, the addition of wild-type atrazine-grown Pseudomonas sp. strain ADP to unamended soil greatly stimulated atrazine mineralization compared to a control to which heat-killed cells were added (Fig. 4A). Interestingly, a low level of atrazine mineralization was evident in the control vials, indicating that atrazine-mineralizing organisms were part of the natural flora in the soil. Atrazine mineralization by Pseudomonas sp. strain ADP was rapid, with more than 50% of the label released within the first 24 h after atrazine addition. Similar results were obtained with the MPO102 mutant (Fig. 4A). To determine the effect of nitrate on atrazine mineralization in soil, Pseudomonas sp. strain ADP cells were inoculated into nitrate-amended and unamended soil microcosms and allowed to acclimate to the soil environment for 6 days prior to the addition of atrazine (Fig. 4, B). Atrazine mineralization by Pseudomonas sp. strain ADP in the amended soil was decreased threefold compared to the unamended control, indicating that atrazine mineralization in soil is repressed by nitrate. Mineralization rates were lower than in the previous experiment, likely due to the decrease in the Pseudomonas sp. strain ADP population in the soil during the acclimation period (half-life, ∼3.1 days).
FIG. 4.
Atrazine mineralization by Pseudomonas sp. strain ADP in soil microcosms. (A) Atrazine mineralization by wild-type Pseudomonas sp. strain ADP (closed symbols) and MPO102 (open symbols). Cells were grown on atrazine as the sole nitrogen source and were inoculated directly to unamended soil (circles) or heat killed prior to addition (squares). (B) Atrazine mineralization by wild-type Pseudomonas sp. strain ADP after 6 days of adaptation to unamended (closed circles) or nitrate-amended (0.5 mg of NO3N g−1) soil (open circles).(C) Atrazine mineralization by MPO102 after 6 days of adaptation to unamended (closed squares) or nitrate-amended (open squares) soil. Symbols represent the averages of the results from three independent experiments. Error bars represent standard errors of the averages.
Atrazine mineralization by the MPO102 mutant is not repressed by added nitrate.
To test whether atrazine degradation by the MPO102 mutant is affected by the presence of nitrate in a soil environment, atrazine-grown MPO102 was also acclimated for 6 days in unamended and nitrate-amended soil microcosms. Radiolabeled atrazine was then added, and mineralization was monitored (Fig. 4C). The atrazine mineralization rate in the unamended soil was similar to that observed with wild-type Pseudomonas sp. strain ADP (compare unamended controls in Fig. 4B and C). Interestingly, there was no significant difference between mineralization rates in unamended and nitrate-amended soils when MPO102 was used. This difference between the wild-type and mutant strains is not due to increased survival of MPO102, since its half-life in nitrate-amended soil was slightly lower than that of the wild-type strain (2.3 days). These results indicate that atrazine degradation by MPO102 is not repressed by nitrate in the soil.
DISCUSSION
In this study, we have found that the presence of preferential nitrogen sources in the environment is detrimental to atrazine degradation by Pseudomonas sp. strain ADP. These results confirm and extend the observations of Bichat et al. (5). These authors noted that atrazine degradation by cells previously grown on ammonium, nitrate, or urea as the sole nitrogen source was slow compared to atrazine- or glycine-grown cells. Our data clearly show that certain nitrogen sources have an inhibitory effect on the catabolic pathway that is not prevented by the simultaneous presence of atrazine in the medium. This and the fact that efficient atrazine degradation can be achieved with compounds unrelated to the atrazine degradative pathway, i.e., serine (Fig. 1G) or glycine (5) as sole nitrogen sources indicate that neither atrazine nor any of its metabolites are required for induction of the pathway.
General nitrogen control is a well-characterized regulatory phenomenon in a number of gram-positive and gram-negative organisms (26). Our results suggest that atrazine degradation may be subjected to general nitrogen control resembling that described in enterobacteria: (i) regulation is likely to occur primarily at the gene expression level, since addition of ammonium to the assay buffer had no inhibitory effect on atrazine removal by atrazine-grown cells; (ii) atrazine catabolism is repressed in the presence of nitrogen sources that support fast growth of the strain, suggesting preferential nitrogen assimilation from these nitrogen sources; and (iii) nitrogen status is sensed from changes in intracellular pools of metabolites, rather than from the nitrogen species present in the medium (11, 18, 33). Nitrogen control in the enterobacteria requires an alternative form of RNA polymerase holoenzyme that contains σ54 (the product of rpoN) (26). Interestingly, an rpoN mutant of Pseudomonas putida KT2440 can grow normally on the atrazine degradation-repressing nitrogen sources ammonium, urea, and proline but fails to grow on the nonrepressing nitrogen sources serine and glycine (20). The only exception to this correlation is nitrate, which cannot be utilized as a nitrogen source by the P. putida rpoN mutant but represses atrazine degradation in Pseudomonas sp. strain ADP. However, comparison of the growth rates of the two organisms suggests that nitrate may be a growth-limiting nitrogen source for P. putida KT2440 but not for Pseudomonas sp. strain ADP. It is tempting to speculate that a conserved σ54-dependent mechanism is responsible for general nitrogen control in the enterobacteria and the pseudomonads. In that regard, it is worth noting that genes homologous to the signal transduction and regulatory elements for general nitrogen control in the enterobacteria have been found in the sequenced genomes of Pseudomonas aeruginosa PAO1 and P. putida KT2440 (27, 35).
Many xenobiotic-degrading bacterial strains that perform well in laboratory media turn out to be poor degraders in a natural setting (42). One possible explanation for this phenomenon is the presence of environmental traits that have a negative impact on the expression of the degradative pathway. Our results indicate that nitrogen control of atrazine metabolism is functional under soil conditions and may therefore limit the potential of Pseudomonas sp. strain ADP for atrazine bioremediation in nitrogen-fertilized agricultural soils. On the other hand, the absence of nitrate repression in the MPO102 mutant and the fact that nitrate is the major nitrogen species in well-aerated soils (41) suggest that MPO102 may be a more suitable strain for bioremediation of atrazine-contaminated agricultural soils with a high nitrogen content.
Acknowledgments
We thank Inés Canosa for critical review of the manuscript.
This work was supported by the European Union project QLK3-CT-1999-00041 and by a fellowship of the F.P.U. program of the Spanish Ministerio de Educación y Cultura awarded to V.G.-G.
REFERENCES
- 1.Abdelhafid, R., S. Houot, and E. Barriuso. 2000. Dependence of atrazine degradation on C and N availability in adapted and non-adapted soils. Soil Biol. Biochem. 32:389-401. [Google Scholar]
- 2.Abdelhafid, R., S. Houot, and E. Barriuso. 2000. How increasing availabilities of carbon and nitrogen affect atrazine behaviour in soils. Biol. Fertil. Soils 30:333-340. [Google Scholar]
- 3.Allran, J. W., and W. H. Karasov. 2000. Effects of atrazine and nitrate on the northern leopard frog (Rana pipiens) larvae exposed in the laboratory from posthatch through metamorphosis. Environ. Toxicol. Chem. 19:2850-2855. [Google Scholar]
- 4.Alvey, S., and D. E. Crowley. 1995. Influence of organic amendments on biodegradation of atrazine as a nitrogen source. J. Environ. Qual. 24:1156-1162. [Google Scholar]
- 5.Bichat, F., G. K. Sims, and R. L. Mulvaney. 1999. Microbial utilization of heterocyclic nitrogen from atrazine. Soil Sci. Soc. Am. J. 63:100-110. [Google Scholar]
- 6.Biradar, D. P., and A. L. Rayburn. 1995. Chromosomal damage induced by herbicide contamination at concentrations observed in public water supplies. J. Environ. Qual. 24:1222-1225. [Google Scholar]
- 7.Bouquard, C., J. Ouazzani, J.-C. Prom, Y. Michel-Briand, and P. Plésiat. 1997. Dechlorination of atrazine by a Rhizobium sp. isolate. Appl. Environ. Microbiol. 63:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenchley, J. E. 1973. Effect of methionine sulfoximine and methionine sulfone on glutamate synthesis in Klebsiella aerogenes. J. Bacteriol. 114:666-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Souza, M. L., J. Seffernick, B. Martinez, M. J. Sadowsky, and L. P. Wackett. 1998. The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 180:1951-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza, M. L., L. P. Wackett, and M. J. Sadowsky. 1998. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 64:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engleman, E. G., and S. H. Francis. 1978. Cascade control of E. coli glutamine synthetase. II. Metabolite regulation of the enzymes in the cascade. Arch. Biochem. Biophys. 191:602-612. [DOI] [PubMed] [Google Scholar]
- 12.Entry, J. A., K. G. Mattson, and W. H. Emmingham. 1993. The influence of nitrogen on atrazine and 2, 4-dichlorophenoxyacetic acid mineralization in grassland soils. Biol. Fertil. Soils 16:179-182. [Google Scholar]
- 13.Foster, J. C. 1995. Soil nitrogen, p. 79-87. In K. Alef and P. Nannipieri (ed.), Methods in applied soil microbiology and biochemistry. Academic Press, New York, N.Y.
- 14.Garzón, A., J. Li, A. Flores, J. Casadesús, and V. Stewart. 1992. Molybdenum cofactor (chlorate-resistant) mutants of Klebsiella pneumoniae M5al can use hypoxanthine as the sole nitrogen source. J. Bacteriol. 174:6298-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebendinger, N., and M. Radosevich. 1999. Inhibition of atrazine degradation by cyanazine and exogenous nitrogen in bacterial isolate M91-3. Appl. Microbiol. Biotechnol. 51:375-381. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, T., K. Haston, M. Tsui, A. Hoang, C. Haeffele, and A. Vonk. 2003. Atrazine-induced hermaphroditism at 0.1 ppb in American leopard frogs (Rana pipiens): laboratory and field evidence. Environ. Health Perspect. 111:568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes, T. B., A. Collins, M. Lee, M. Mendoza, N. Noriega, A. A. Stuart, and A. Vonk. 2002. Hermaphroditic, demasculinized frogs after exposure to the herbicide atrazine at low ecologically relevant doses. Proc. Natl. Acad. Sci. USA 99:5476-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda, T. P., A. E. Shauger, and S. Kustu. 1996. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259:589-607. [DOI] [PubMed] [Google Scholar]
- 19.Katz, I., C. G. Dosoretz, R. T. Mandelbaum, and M. Green. 2001. Atrazine degradation under denitrifying conditions in continuous culture of Pseudomonas ADP. Water Res. 35:3272-3275. [DOI] [PubMed] [Google Scholar]
- 20.Köhler, T., S. Harayama, J. L. Ramos, and K. N. Timmis. 1989. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J. Bacteriol. 171:4326-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolpin, D. W., and S. J. Kalkhoff. 1993. Atrazine degradation in a small stream in Iowa. Environ. Sci. Technol. 27:134-139. [Google Scholar]
- 22.Kolpin, D. W., E. M. Thurman, and D. A. Goolsby. 1996. Occurrence of selected herbicides and their metabolites in near-surface aquifers of the midwestern United States. Environ. Sci. Technol. 30:385-390.
- 23.Mandelbaum, R. T., L. P. Wackett, and D. L. Allan. 1995. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandelbaum, R. T., L. P. Wackett, and D. L. Allan. 1993. Mineralization of the s-triazine ring of atrazine by stable bacterial mixed cultures. Appl. Environ. Microbiol. 59:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 28.Radosevich, M., S. J. Traina, Y. L. Hao, and O. H. Tuovinen. 1995. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl. Environ. Microbiol. 61:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralebitso, T. K., E. Senior, and H. W. van Verseveld. 2002. Microbial aspects of atrazine degradation in natural environments. Biodegradation 13:11-19. [DOI] [PubMed] [Google Scholar]
- 30.Richards, R. P., and D. B. Baker. 1993. Pesticide concentration patterns in agricultural drainage networks in the Lake Erie basin. Environ. Toxicol. Chem. 12:13-36. [Google Scholar]
- 31.Rousseaux, S., A. Hartmann, and G. Soulas. 2001. Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 36:211-222. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Senior, P. J. 1975. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J. Bacteriol. 123:407-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, C. J., R. B. Hespell, and M. P. Bryant. 1981. Regulation of urease and ammonia assimilatory enzymes in Selenomonas ruminantium. Appl. Environ. Microbiol. 42:89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 36.Struthers, J. K., K. Jayachandran, and T. B. Moorman. 1998. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl. Environ. Microbiol. 64:3368-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tappe, W., J. Groeneweg, and B. Jantsch. 2002. Diffuse atrazine pollution in German aquifers. Biodegradation 13:3-10. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, J. P., B. Wilson, M. S. Mills, and R. G. Burns. 2002. Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol. Biochem. 34:387-401. [Google Scholar]
- 39.Topp, E., H. Zhu, S. M. Nour, S. Houot, M. Lewis, and D. Cuppels. 2000. Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol. 66:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 41.Wild, A. 1988. Plant nutrients in soil: nitrogen, p. 652-695. In A. Wild (ed.), Russell's soil conditions and plant growth. Longman, Harlow, Essex, United Kingdom.
- 42.Young, C. S., and R. G. Burns. 1993. Detection, survival and activity of bacteria added to soil, p 1-63. In J.-M. Bollag and G. Stotzky (ed.), Soil biochemistry, vol. 8. Marcel Dekker, New York, N.Y.