Abstract
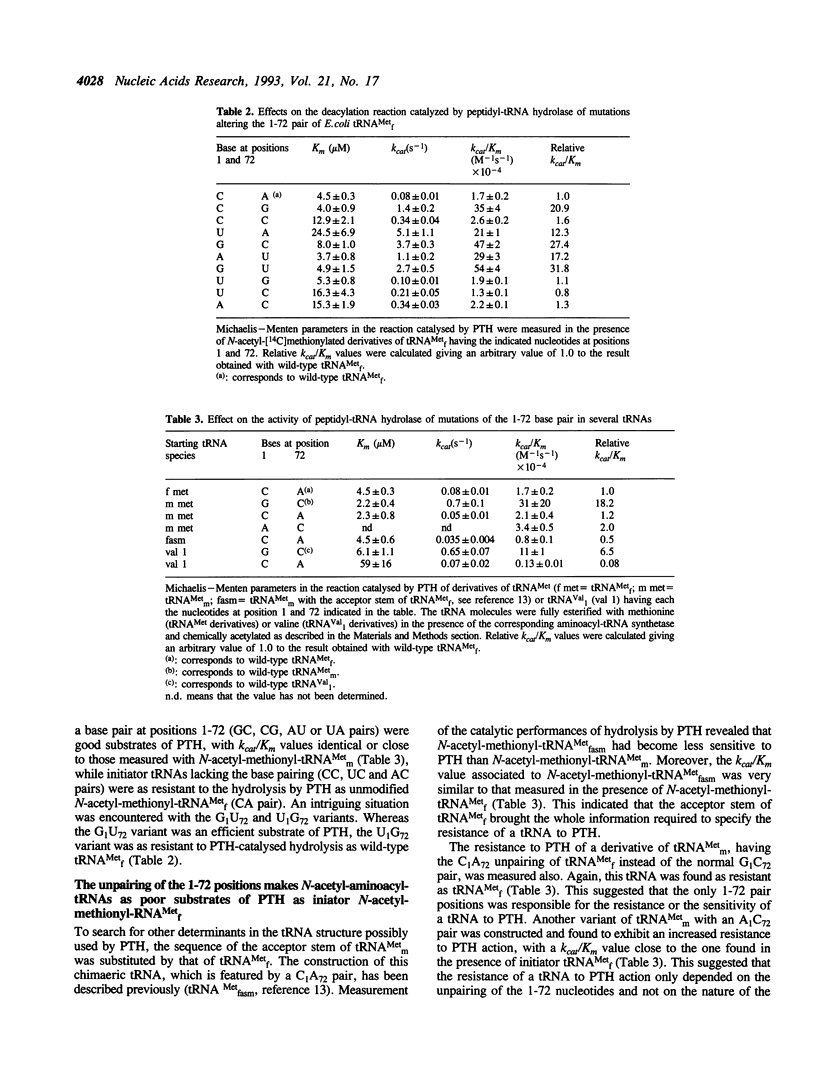
Previous work by Schulman and Pelka (1975) J. Biol. Chem. 250, 542-547, indicated that the absence of a pairing between the bases 1 and 72 in initiator tRNA(fMet) explained the relatively small activity of peptidyl-tRNA hydrolase towards N-acetyl-methionyl-tRNA(fMet). In the present study, the structural requirements for the sensitivity of an N-acetyl-aminoacyl-tRNA to Escherichia coli peptidyl-tRNA hydrolase activity have been further investigated. Ten derivatives of tRNA(fMet) with various combinations of bases at positions 1 and 72 in the acceptor stem have been produced, aminoacylated and chemically acetylated. The release of the aminoacyl moiety from these tRNA derivatives was assayed in the presence of peptidyl-tRNA hydrolase purified from an overproducing strain. tRNA(fMet) derivatives with either C1A72, C1C72, U1G72, U1C72 or A1C72 behaved as poor substrates of the enzyme, as compared to those with C1G72, U1A72, G1C72, A1U72 or G1U72. With the exception of U1G72, it could be therefore concluded that the relative resistance of tRNA(fMet) to peptidyl-tRNA hydrolase did not depend on a particular combination of nucleotides at positions 1 and 72, but rather reflected the absence of a base pairing at these positions. In a second series of experiments, the unpairing of the 1 and 72 bases, created with C-A or A-C bases, instead of G-C in methionyl-tRNA(mMet) or in valyl-tRNA(Val1), was shown to markedly decrease the rate of hydrolysis catalysed by peptidyl-tRNA hydrolase. Altogether, the data indicate that the stability of the 1-72 pair governs the degree of sensitivity of a peptidyl-tRNA to peptidyl-tRNA hydrolase.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atherly A. G., Menninger J. R. Mutant E. coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nat New Biol. 1972 Dec 20;240(103):245–246. doi: 10.1038/newbio240245a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- García-Villegas M. R., De La Vega F. M., Galindo J. M., Segura M., Buckingham R. H., Guarneros G. Peptidyl-tRNA hydrolase is involved in lambda inhibition of host protein synthesis. EMBO J. 1991 Nov;10(11):3549–3555. doi: 10.1002/j.1460-2075.1991.tb04919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Starn T. K., Rundquist C., Crow P., White J., Olin A., Wagner T. Purification and initial characterization of peptidyl-tRNA hydrolase from rabbit reticulocytes. J Biol Chem. 1992 Jan 25;267(3):2073–2079. [PubMed] [Google Scholar]
- Guillon J. M., Mechulam Y., Schmitter J. M., Blanquet S., Fayat G. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992 Jul;174(13):4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon J. M., Meinnel T., Mechulam Y., Lazennec C., Blanquet S., Fayat G. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNA(fMet) formyltransferase. J Mol Biol. 1992 Mar 20;224(2):359–367. doi: 10.1016/0022-2836(92)91000-f. [DOI] [PubMed] [Google Scholar]
- Janiak F., Dell V. A., Abrahamson J. K., Watson B. S., Miller D. L., Johnson A. E. Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu.GTP: evidence for a new functional role for elongation factor Tu in protein biosynthesis. Biochemistry. 1990 May 8;29(18):4268–4277. doi: 10.1021/bi00470a002. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kössel H. Purification and properties of peptidyl-tRNA hydrolase from Escherichia coli. Biochim Biophys Acta. 1970 Mar 19;204(1):191–202. doi: 10.1016/0005-2787(70)90502-2. [DOI] [PubMed] [Google Scholar]
- Kössel H., RajBhandary U. L. Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J Mol Biol. 1968 Aug 14;35(3):539–560. doi: 10.1016/s0022-2836(68)80013-0. [DOI] [PubMed] [Google Scholar]
- Lee C. P., Dyson M. R., Mandal N., Varshney U., Bahramian B., RajBhandary U. L. Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., Seong B. L., RajBhandary U. L. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J Biol Chem. 1991 Sep 25;266(27):18012–18017. [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Fayat G., Blanquet S. Involvement of the size and sequence of the anticodon loop in tRNA recognition by mammalian and E. coli methionyl-tRNA synthetases. Nucleic Acids Res. 1992 Sep 25;20(18):4741–4746. doi: 10.1093/nar/20.18.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Fayat G. Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res. 1988 Aug 25;16(16):8095–8096. doi: 10.1093/nar/16.16.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Lazennec C., Blanquet S., Fayat G. Critical role of the acceptor stem of tRNAs(Met) in their aminoacylation by Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 1993 Jan 5;229(1):26–36. doi: 10.1006/jmbi.1993.1005. [DOI] [PubMed] [Google Scholar]
- Mellot P., Mechulam Y., Le Corre D., Blanquet S., Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989 Aug 5;208(3):429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- Menninger J. R. Accumulation of peptidyl tRNA is lethal to Escherichia coli. J Bacteriol. 1979 Jan;137(1):694–696. doi: 10.1128/jb.137.1.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen H. U., Røll T., Grunberg-Manago M., Clark B. F. Specific interaction of initiation factor IF2 of E. coli with formylmethionyl-tRNA f Met. Biochem Biophys Res Commun. 1979 Dec 14;91(3):1068–1074. doi: 10.1016/0006-291x(79)91989-2. [DOI] [PubMed] [Google Scholar]
- Schofield P., Zamecnik P. C. Cupric ion catalysis in hydrolysis of aminoacyl-tRNA. Biochim Biophys Acta. 1968 Feb 26;155(2):410–416. doi: 10.1016/0005-2787(68)90185-8. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. The structural basis for the resistance of Escherichia coli formylmethionyl transfer ribonucleic acid to cleavage by Escherichia coli peptidyl transfer ribonucleic acid hydrolase. J Biol Chem. 1975 Jan 25;250(2):542–547. [PubMed] [Google Scholar]



