Abstract
Most soil bacteria belong to family-level phylogenetic groups with few or no known cultivated representatives. We cultured a collection of 350 isolates from soil by using simple solid media in petri dishes. These isolates were assigned to 60 family-level groupings in nine bacterial phyla on the basis of a comparative analysis of their 16S rRNA genes. Ninety-three (27%) of the isolates belonged to 20 as-yet-unnamed family-level groupings, many from poorly studied bacterial classes and phyla. They included members of subdivisions 1, 2, 3, and 4 of the phylum Acidobacteria, subdivision 3 of the phylum Verrucomicrobia, subdivision 1 of the phylum Gemmatimonadetes, and subclasses Acidimicrobidae and Rubrobacteridae of the phylum Actinobacteria. In addition, members of 10 new family-level groupings of subclass Actinobacteridae of the phylum Actinobacteria and classes Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria of the phylum Proteobacteria were obtained. The high degree of phylogenetic novelty and the number of isolates affiliated with so-called unculturable groups show that simple cultivation methods can still be developed further to obtain laboratory cultures of many phylogenetically novel soil bacteria.
For over 80 years it has been known that there is a large discrepancy between the number of bacterial colonies that form on solid media when soil is used as an inoculum and the total number of bacterial cells actually present in that same soil (9, 23, 40). This discrepancy has limited our understanding of the species diversity of soil bacterial communities. In the past decade, this limitation has been partially overcome through the application of molecular ecological techniques. In particular, comparative analysis of 16S rRNAs or 16S rRNA genes derived from nucleic acids extracted directly from soil has revealed the presence of many new groups of bacteria that were previously undetected in cultivation studies (2, 11-13, 16, 19, 21, 26, 28, 32, 35). Some of these groups appear to be important within soils, at least in terms of relative abundance of 16S rRNAs or 16S rRNA genes. However, these numerically abundant bacteria are rarely, if ever, isolated in cultivation experiments, which instead tend to result in the isolation of bacteria that appear to be minor components of the soil bacterial community (1, 12, 16, 35). As a consequence, traditional cultivation techniques such as plate counting methods have been increasingly considered inadequate, and new, more sophisticated techniques have been developed for the isolation of novel bacteria from complex microbial habitats. These methods include the use of micromanipulators and optical (laser) tweezers (14), the construction of simulated natural environments (24), and cell encapsulation in gel microdroplets (43). Such methods are specialized and unlikely to be adopted by the broader scientific community, and so the challenge of obtaining cultures of the large number of “unculturable” bacterial groups seems a daunting one. The only avenues currently available for the study of uncultured bacteria are cultivation-independent molecular ecological techniques that have proved to be very powerful for the study of bacteria in their natural settings (18). Parallel study of laboratory cultures would, however, strongly complement molecular ecological investigations and enhance research into the roles of soil bacteria and their biotechnological potentials. Assigning functions to bacteria known only by their 16S rRNA genes is a difficult task, and detailed investigations of their physiologies and genomes are even more challenging. The availability of pure cultures greatly simplifies such studies.
We suggest that for many of the “unculturable” groups of bacteria that have been detected in soil by molecular biological techniques, no great innovations are required and that the growth of these microorganisms in pure culture with simple media is straightforward and reproducible. We have previously shown that microorganisms from widely distributed groups with few or no previously isolated representatives can be isolated by simple methods that can be readily implemented (22, 37). Here we report the results of the application of these methods to generate a collection of 350 isolates of soil bacteria.
MATERIALS AND METHODS
Soil sampling.
Soil cores were collected from a rotationally grazed pasture of perennial ryegrass (Lolium perene) and white clover (Trifolium repens) (22, 37) at the Dairy Research Institute, Ellinbank, Victoria, Australia (38°14.55′S, 145°56.11′E). Collection dates were 2 March 2001, 5 April 2001, 31 January 2002, and 27 March 2002. A clean 25-mm-diameter metal corer was used to obtain 100-mm-long soil cores, which were transported intact at ambient temperature in sealed polyethylene bags and processed within 3 h of collection. The upper 30 mm of each core was discarded, and large roots and stones were removed from the remainder, which was then sieved through a sterile brass sieve with a 2-mm aperture size (Endecotts Ltd., London, United Kingdom) and used immediately for dry weight determination and cultivation experiments.
Cultivation.
Soil samples were dispersed in distilled water, treated by sonication, diluted, and spread onto solid media in petri dishes as described by Janssen et al. (22). The diluent was the VL55 medium (37) without added growth substrates or vitamins for all experiments except those in which petri dishes containing medium based on LN55 medium (see below) were inoculated, for which the diluent was LN55 medium without added growth substrates or vitamins. These cultivation dishes were incubated at 25°C for up to 3 months.
The majority of media were based on VL55 medium (37) but with the xylan replaced by other growth substrates as appropriate (see below) and solidified either with agar (35) or with 0.8% (wt/vol) gellan (Phytagel; Sigma). In some media (labeled LN55), the (NH4)2HPO4 of the VL55 medium base was replaced with KH2PO4 at the same molarity. The growth substrates were added to these media from concentrated stocks that were sterilized by autoclaving (in the case of polymers) or by passage through Minisart sterile filters (Sartorius AG, Göttingen, Germany) with a pore size of 0.22 μm. The growth substrates and their final concentrations in the solidified media were as follows: N-acetylglucosamine, 2 mM; a mixture of d-glucose, d-galactose, and d-xylose, and l-arabinose, 0.5 mM each; a mixture of d-galacturonic acid, d-glucuronic acid, l-ascorbic acid, and sodium d-gluconate, 0.5 mM each (stock adjusted to pH 7 with NaOH); a mixture of sodium acetate, sodium benzoate, sodium l-lactate, and methanol, 0.5 mM each; an amino acid mixture (20) with 0.08 g of l-tryptophan per 100 ml of stock solution added at 10 ml of stock solution per liter of medium; sodium alginate, 0.05% (wt/vol); xanthan, 0.05% (wt/vol); pectin, 0.05% (wt/vol); xylan, 0.05% (wt/vol); and carboxymethylcellulose, 0.05% (wt/vol). Some media were also prepared without an added substrate.
Other media used were dilute nutrient broth solidified with agar or gellan (22), cold-extracted soil extract agar (34) containing only 1/10 of the nutrient solution, Winogradsky's salt solution agar (41), and 10-fold-diluted tryptone soy broth (Oxoid) with bacteriological agar no. 1 (Oxoid) to give a final agar concentration of 15 g per liter.
Cultures were randomly selected and subcultured as described by Sait et al. (37). The cultures reported here originated from plates that had been inoculated with dilutions of soil so that the theoretical amount of dry soil per plate was 1.7 × 10−8 to 2.1 × 10−8 g.
Phylogenetic inference.
16S rRNA gene sequences were determined by the methods described by Sait et al. (37). The partial and nearly complete 16S rRNA gene sequences were imported into an ARB database (http://www.arb-home.de/), aligned, and inserted into a main tree with the parsimony insertion tool. An evolutionary distance tree of the nearly complete sequences and related family and phylum representatives was calculated by using the Olsen substitution model and the neighbor-joining tree-building algorithm (37). Phylum-level representatives used to determine the depths of the phylum groups were removed from the final tree (Fig. 1), but the depth of each phylum is indicated in Fig. 1. The robustness of the inferred topology was confirmed by evolutionary distance and maximum-parsimony bootstrap resampling and by varying the outgroup composition as previously described (10). The data set is available upon request. Family-level assignments were based on monophyly of the isolate sequences with existing families, orders, classes, and phyla. If the sequences were not reproducibly affiliated with recognized families (17) within a higher-level grouping, e.g., the Ellin5078 sequence and related sequences in the class Gammaproteobacteria, novel families were proposed as the most conservative estimates of novel groups with higher rankings than species and genus. Novel families are labeled with the names of representative isolates or cloned 16S rRNA or 16S rRNA gene sequences. The nomenclature for phylogenetic and taxonomic groupings follows that of Garrity et al. (17) except for that for the subdivisions of the phyla Acidobacteria and Verrucomicrobia, which follows the schemes of Hugenholtz et al. (21); that for the subdivisions of the phylum Planctomycetes, which follows the scheme of Fuerst et al. (15); and that for the subdivisions of the phylum Gemmatimonadetes, which follows the scheme of Zhang et al. (44).
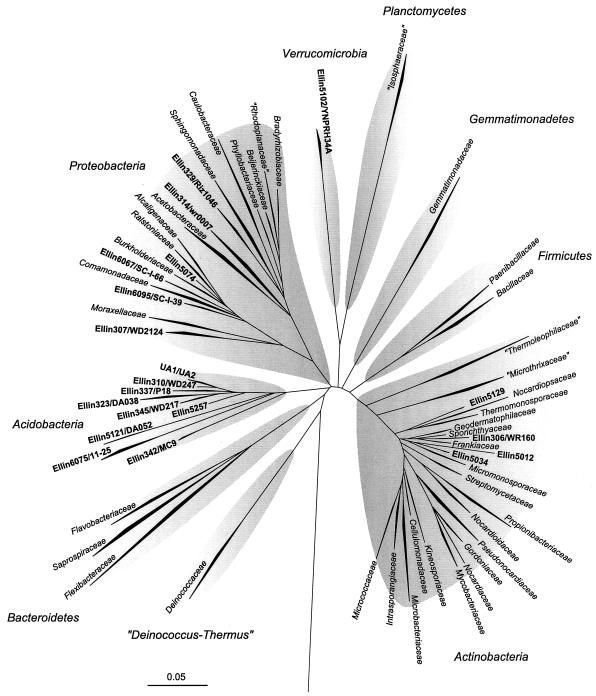
FIG. 1.
Evolutionary distance tree of the bacterial domain showing an overview of culturable bacterial diversity for the Ellinbank soil samples at the family level. All family-level groupings (shown as wedges) were supported by bootstrap resampling (bootstrap proportion values, >75%). The phylogenetic depth of each of the phyla to which the families belong is shaded. Where the new isolates are the first isolated representatives of a proposed family, the family name is shown in boldface type. The 16S rRNA gene sequences of Sulfolobus acidocaldarius (GenBank accession no. D14876) and Methanococcus vannielii (accession no. M36507) and the hot spring 16S rRNA gene sequence pJP27 (accession no. L25852) were used as outgroups (data not shown) in the analysis. Bar, 0.05 change per nucleotide.
Family-level diversity estimates.
Estimates of total numbers of families with culturable members were obtained by using an abundance-base coverage estimator (7) and Chao's richness estimator (6) implemented in EstimateS (version 5; R. K. Colwell; available at http://viceroy.eeb.uconn.edu/estimates).
Nucleotide sequence accession numbers.
All partial 16S rRNA gene sequences obtained in this study have been deposited in the GenBank databases under accession nos. AY234418 to AY234767.
RESULTS AND DISCUSSION
We obtained 350 isolates of bacteria from a series of soil cores collected on different dates from one location. These isolates were subcultured from randomly selected colonies that appeared on plates of a range of media. The plates had been inoculated with the most diluted aliquots of soil that had resulted in colony formation. The soil was substantially root free but still contained fine roots and also contained soil animals and protozoa, and so the exact source of each of the isolates cannot be exactly defined. The isolates were identified by comparative analysis of their 16S rRNA genes and assigned to different families of nine bacterial phyla (Fig. 1) on the basis of reproducible phylogenetic affiliation, i.e., high bootstrap proportions (>75%) supporting family groupings under a range of outgroup configurations (10). 16S rRNA genes were only partially sequenced (ca. 400 to 450 bp) if they had matches >97% identical to full-length 16S rRNA gene sequences with unambiguous family assignments. For example, the 400 bp of sequence information obtained for the 16S rRNA gene of isolate Ellin5005 is 99.8% identical to the corresponding region of the 16S rRNA gene of Bradyrhizobium japonicum, and the isolate is therefore a member of the family Bradyrhizobiaceae. Extended sequence information for the 16S rRNA gene was obtained for isolates that we identified as belonging to phylogenetic groups with few cultivated representatives or for which more sequence information was required to confirm family-level affiliations (see below). A detailed listing of the isolates, including the details of the soil sample from which each was isolated, the medium on which each was isolated, their phylogenetic affiliations, and the GenBank accession numbers of their 16S rRNA gene sequences, is available from the corresponding author.
The 350 isolates were affiliated with 17 class-level groups and could be assigned to a total of 60 different family-level groups (Table 1). Of these 60 family-level groups, 20 (33%) did not encompass validly named or described species. Ninety-three (27%) of the isolates belonged to these novel family-level groups. A further 15 isolates (4%) fell into four family-level groups that have not been formally described but to which some validly named species clearly belong. We have provisionally designated these families “Microthrixaceae,” “Thermoleophilaceae,” “Isosphaeraceae,” and “Rhodoplanaceae” (Table 1). Altogether, these family-level affiliations represent a high degree of phylogenetic novelty within the collection of 350 isolates (Fig. 1). The number of isolates that are likely to fall into new genera or new species is even higher. However, since genus- and species-level affiliations should be determined on the basis of shared phenotypic characteristics, extensive characterization of the isolates would be required before relationships below the family level could be resolved. Estimates of the total number of families with members that might be culturable from these samples by the approaches employed in this study were 80 (obtained by using an abundance-base coverage estimator) and 84 (obtained by using Chao's richness estimator). This finding suggests that increasing the number of isolates screened would have revealed yet further family-level diversity.
TABLE 1.
Phylogenetic affiliations of 350 randomly selected isolates based on comparative analysis of their 16S rRNA gene sequences
| Phylum | Class, subclass, or subdivision | No. of isolates | No. of new families | No. of isolates in new families | Family grouping | No. of isolates in family grouping |
|---|---|---|---|---|---|---|
| Acidobacteria | Subdivision 1 | 29 | 6 | 29 | “Ellin310/WD247 group” | 3 |
| “Ellin323/DA038 group” | 4 | |||||
| “Ellin337/P18 group” | 3 | |||||
| “Ellin345/WD217 group” | 4 | |||||
| “Ellin5257 group” | 1 | |||||
| “UA1/UA2 group” | 14 | |||||
| Subdivision 2 | 1 | 1 | 1 | “Ellin5121/DA052 group” | 1 | |
| Subdivision 3 | 5 | 1 | 5 | “Ellin342/MC9 group” | 5 | |
| Subdivision 4 | 2 | 1 | 2 | “Ellin6075/11-25 group” | 2 | |
| Actinobacteria | Acidimicrobidae | 3 | 0 | 0 | “Microthrixaceae” | 3 |
| Actinobacteridae | 132 | 4 | 25 | “Ellin306/WR160 group” | 13 | |
| “Ellin5012 group” | 3 | |||||
| “Ellin5034 group” | 5 | |||||
| “Ellin5129 group” | 4 | |||||
| Cellulomonadaceae | 2 | |||||
| Frankiaceae | 1 | |||||
| Geodermatophilaceae | 2 | |||||
| Gordoniaceae | 1 | |||||
| Intrasporangiaceae | 6 | |||||
| Kineosporiaceae | 8 | |||||
| Microbacteriaceae | 5 | |||||
| Micrococcaceae | 4 | |||||
| Micromonosporaceae | 15 | |||||
| Mycobacteriaceae | 22 | |||||
| Nocardiaceae | 6 | |||||
| Nocardioidaceae | 11 | |||||
| Nocardiopsaceae | 8 | |||||
| Propionibacteriaceae | 1 | |||||
| Pseudonocardiaceae | 3 | |||||
| Sporichthyaceae | 1 | |||||
| Streptomycetaceae | 10 | |||||
| Thermomonosporaceae | 1 | |||||
| Rubrobacteridae | 8 | 0 | 0 | “Thermoleophilaceae” | 8 | |
| Bacteroidetes | Flavobacteria | 1 | 0 | 0 | Flavobacteriaceae | 1 |
| Sphingobacteria | 12 | 0 | 0 | Flexibacteraceae | 11 | |
| Saprospiraceae | 1 | |||||
| “Deinococcus-Thermus” | Deinococci | 1 | 0 | 0 | Deinococcaceae | 1 |
| Firmicutes | Bacilli | 6 | 0 | 0 | Bacillaceae | 3 |
| Paenibacillaceae | 3 | |||||
| Gemmatimonadetes | Subdivision 1 | 3 | 0 | 0 | Gemmatimonadaceae | 3 |
| Planctomycetes | “Isosphaerae” | 3 | 0 | 0 | “Isosphaeraceae” | 3 |
| Proteobacteria | Alphaproteobacteria | 112 | 2 | 16 | “Ellin314/wr0007 group” | 16 |
| “Ellin329/Riz1046 group” | 2 | |||||
| “Rhodoplanaceae” | 1 | |||||
| Acetobacteraceae | 2 | |||||
| Beijerinckiaceae | 1 | |||||
| Bradyrhizobiaceae | 72 | |||||
| Caulobacteraceae | 2 | |||||
| Phyllobacteriaceae | 1 | |||||
| Sphingomonadaceae | 15 | |||||
| Betaproteobacteria | 21 | 3 | 3 | “Ellin5074 group” | 1 | |
| “Ellin6067/SC-I-66 group” | 1 | |||||
| “Ellin6095/SC-I-39 group” | 1 | |||||
| Alcaligenaceae | 1 | |||||
| Burkholderiaceae | 14 | |||||
| Comamonadaceae | 2 | |||||
| Ralstoniaceae | 1 | |||||
| Gammaproteobacteria | 10 | 1 | 9 | “Ellin307/WD2124 group” | 9 | |
| Moraxellaceae | 1 | |||||
| Verrucomicrobia | Subdivision 3 | 1 | 1 | 1 | “Ellin5102/YNPRH34A group” | 1 |
Since the purpose of this study was not to compare the media used, equal numbers of isolates were not obtained for each medium. Also, the high degree of phylogenetic diversity made it difficult to detect any trends correlating with medium type. However, some of the family groupings with large numbers of isolates were made up of representatives obtained with many different media (data not shown). More significant is the fact that many of the isolates belonged to class-level phylogenetic groupings that are poorly studied and are represented by few or no cultured isolates.
Particularly notable is the large number of members of the phylum Acidobacteria (affiliated with subdivisions 1, 2, 3, and 4). On the basis of the occurrence of their 16S rRNA and 16S rRNA genes, members of the phylum Acidobacteria are widely distributed in soils and have been shown to be active members of soil bacterial communities (2, 13, 16, 21, 26-28, 32). To date, only 14 isolates from soil have been reported (22, 25, 29, 36, 37), 11 of which are from the Ellinbank site (22, 37). In this study, we greatly extend the number of reported isolates, with a total of 37 (11%) of our isolates belonging to different family-level groupings in this phylum. The ease with which members of this reportedly unculturable group were isolated shows that they are in fact readily and repeatedly culturable.
Only a single isolate of the phylum Verrucomicrobia was obtained in this study, but it is the first cultured representative of subdivision 3 of this phylum. This subdivision has previously been defined only on the basis of 16S rRNA genes from various habitats (21). The phylum Verrucomicrobia is a recently recognized and little-studied bacterial lineage that is proving to be widespread, often numerically abundant, and active in the environment (2, 4, 11, 21, 27, 33). The lack of isolates of the soil-inhabiting groups has severely hampered investigation of the biology of members of this phylum.
The phylum Gemmatimonadetes is a very recently described bacterial group whose members, on the basis of data generated from surveys of 16S rRNA genes amplified directly from environmental DNA, are widespread in nature, particularly in soil habitats (2, 27, 28, 31, 33, 44, 45). Only one cultured representative of this phylum, Gemmatimonas aurantiaca, an isolate obtained from activated sludge, has previously been reported (44). The three strains obtained in the present study are the first soil isolates and, although they belong to the same subdivision as the single described representative, G. aurantiaca, their 16S rRNA genes are 7 to 8% divergent from those of G. aurantiaca and 5 to 7% divergent from each other. These three isolates therefore extend the phylogenetic coverage of this phylum by cultured representatives.
Even within well-characterized phyla for which hundreds of isolates have been described, significant phylogenetic novelty was also observed in our collection of 350 isolates. The majority of soil-inhabiting members of the phylum Actinobacteria belong to three subclasses, the Rubrobacteridae, the Acidimicrobidae, and the Actinobacteridae. Members of these major soil-inhabiting groups were isolated. The subclass Rubrobacteridae contains only a few known, thermophilic isolates in the genera Rubrobacter (5, 39) and Thermoleophilum (42), in addition to two very recently described genera of soil bacteria (see below). The subclass Acidimicrobidae consists of a handful of isolates belonging to the genus Acidimicrobium, obtained from acid mine drainage sites (8), and the candidate genus “Microthrix,” obtained from wastewater (3). To date, only six soil-derived isolates of the subclass Rubrobacteridae and no soil-derived isolates of the subclass Acidimicrobidae have been reported, despite members of both groups being abundant in soils worldwide (2, 16, 19, 28, 35). Four of the six previous soil isolates of the subclass Rubrobacteridae came from the Ellinbank sample site (22, 37), while the other two isolates have been assigned to the newly described species Conexibacter woesei (30) and Solirubrobacter pauli (38). In this study, we cultured eight further isolates of the subclass Rubrobacteridae and the first three soil isolates of the subclass Acidimicrobidae, thus greatly extending the range of cultured representatives of these groups.
In addition to members of the poorly characterized subclasses Rubrobacteridae and Acidimicrobidae of the phylum Actinobacteria, 25 isolates were found to belong to four new families of the well-studied subclass Actinobacteridae of the same phylum. Members of the subclass Actinobacteridae, represented by at least 129 recognized and named genera in 40 named families (17), are important sources of antibiotics and other bioactive compounds and have been the subject of intensive and extensive searches for novel strains. Obtaining isolates from phylogenetically novel groups within the subclass Actinobacteridae is a significant objective of such searches, since such isolates are likely to yield chemically different secondary metabolites that may be active antibiotics to which widespread resistance does not yet exist. In our study, the ease with which multiple isolates of four novel families (encompassing 7% of the isolates) were obtained in culture suggests that there is still considerable scope for culturing phylogenetically distinct members of this group. Four novel families extend the family-level diversity of this subclass by 10%.
The phylum Proteobacteria is represented by a large number of described species, including at least 429 named genera in 72 named families (17). Even so, analysis of soil bacterial communities by directly surveying 16S rRNA or 16S rRNA genes has revealed the presence of many clades at the genus, family, and order levels that are not represented by named species (2, 13, 16, 26, 28, 32). Thirty (9%) of our isolates belong to six novel families of this well-studied phylum, thus expanding the known family-level diversity even within this well-represented phylum by 8%.
Our earlier studies, based on only two medium types (22, 37), found that members of previously unstudied groups of soil bacteria can be isolated by traditional plate cultivation methods. This extended study shows that such isolation is reproducible, with many of the isolates from the earlier studies falling into the same family-level groupings as the isolates obtained in this study (data not shown). In addition, increasing the number of isolates studied has yielded further novel bacteria. Many were the first cultivated representatives of new family-level groupings. Analysis of our collection also reveals that had we screened more isolates, yet more family-level diversity would have been obtained in culture. It is to be expected that some of these isolates would also be representatives of further so-called unculturable or as-yet-uncultivated bacteria.
In conclusion, we feel that it is not correct to assert that the majority of soil bacteria cannot be studied by cultivation-dependent methods. Instead, it should be recognized that many of these bacteria will in fact be readily culturable. Whether the effort to cultivate them is warranted depends greatly on the aims of individual investigations. The approach we have taken allows the random isolation of novel bacteria, some of which may be of interest for specific reasons. This isolation will allow investigation of the properties of these novel bacteria. Culture-based approaches are therefore still a powerful qualitative tool that can be used in conjunction with molecular ecological methods to investigate the functions of many as-yet-uncultivated bacteria.
Acknowledgments
This work was supported by a grant from the Australian Research Council.
We thank Cameron Gourley and Sharon Aarons (Dairy Research Institute, Ellinbank, Victoria, Australia) for help with access to the sampling site. Michelle Sait contributed many valuable comments during manuscript preparation.
REFERENCES
- 1.Axelrood, P. E., M. L. Chow, C. S. Arnold, K. Lu, J. M. McDermott, and J. Davies. 2002. Cultivation-dependent characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:643-654. [DOI] [PubMed] [Google Scholar]
- 2.Axelrood, P. E., M. L. Chow, C. C. Radomski, J. M. McDermott, and J. Davies. 2002. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:655-674. [DOI] [PubMed] [Google Scholar]
- 3.Blackall, L. L., E. M. Seviour, M. A. Cunningham, R. J. Seviour, and P. Hugenholtz. 1994. “Microthrix parvicella” is a novel, deep branching member of the actinomycetes subphylum. Syst. Appl. Microbiol. 17:513-518. [Google Scholar]
- 4.Buckley, D. H., and T. M. Schmidt. 2001. Environmental factors influencing the distribution of rRNA from verrucomicrobia in soil. FEMS Microbiol. Ecol. 35:105-112. [DOI] [PubMed] [Google Scholar]
- 5.Carreto, L., E. Moore, M. F. Nobre, R. Wait, P. W. Riley, R. J. Sharp, and M. S. da Costa. 1996. Rubrobacter xylanophilus sp. nov., a new thermophilic species isolated from a thermally polluted effluent. Int. J. Syst. Bacteriol. 46:460-465. [Google Scholar]
- 6.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 7.Chazdon, R. L., R. K. Colwell, J. S. Denslow, and M. R. Guariguata. 1998. Statistical methods for estimating species richness of woody regeneration in primary and secondary rain forests of N. E. Costa Rica, p. 285-309. In F. Dallmeier and J. A. Comiskey (ed.), Forest biodiversity research, monitoring and modeling: conceptual background and Old World case studies. Parthenon Publishing, Paris, France.
- 8.Clark, D. A., and P. R. Norris. 1996. Acidimicrobium ferrooxidans gen. nov., sp. nov.: mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology 142:785-790. [DOI] [PubMed] [Google Scholar]
- 9.Cutler, D. W., and L. M. Crump. 1935. Problems in soil microbiology. Longmans, Green and Co., London, United Kingdom.
- 10.Dalevi, D., P. Hugenholtz, and L. L. Blackall. 2001. A multiple-outgroup approach to resolving division-level phylogenetic relationships using 16S rDNA data. Int. J. Syst. E vol. Microbiol. 51:385-391. [DOI] [PubMed] [Google Scholar]
- 11.Felske, A., and A. D. L. Akkermans. 1998. Prominent occurrence of ribosomes from an uncultured bacterium of the Verrucomicrobiales cluster in grassland soils. Lett. Appl. Microbiol. 26:219-223. [DOI] [PubMed] [Google Scholar]
- 12.Felske, A., A. Wolterink, R. van Lis, W. M. de Vos, and A. D. L. Akkermans. 1999. Searching for the predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 13.Felske, A., W. M. de Vos, and A. D. L. Akkermans. 2000. Spatial distribution of 16S rRNA levels from uncultured acidobacteria in soil. Lett. Appl. Microbiol. 31:118-122. [DOI] [PubMed] [Google Scholar]
- 14.Fröhlich, J., and H. König. 2000. New techniques for isolation of single prokaryotic cells. FEMS Microbiol. Rev. 24:567-572. [DOI] [PubMed] [Google Scholar]
- 15.Fuerst, J. A., H. G. Gwilliam, M. Lindsay, A. Lichanska, C. Belcher, J. E. Vickers, and P. Hugenholtz. 1997. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity, G. M., M. Winters, and D. B. Searles. 2001. Taxonomic outline of the procaryotic genera, release 1.0. Springer, New York, N.Y.
- 18.Gray, N. D., and I. M. Head. 2001. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3:481-492. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, A. J., J. Bowyer, M. P. Holley, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2002. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Hudson, J. A., K. M. Schofield, H. W. Morgan, and R. M. Daniel. 1989. Thermonema lapsum gen. nov., sp. nov., a thermophilic gliding bacterium. Int. J. Syst. Bacteriol. 39:485-487. [Google Scholar]
- 21.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, V. 1968. The plate count technique, p. 158-170. In T. R. G. Gray and D. Parkinson (ed.), The ecology of soil bacteria. Liverpool University Press, Liverpool, United Kingdom.
- 24.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 25.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaig, A. E., S. J. Grayston, J. I. Prosser, and L. A. Glover. 2001. Impact of cultivation on characterisation of species composition of soil bacterial communities. FEMS Microbiol. Ecol. 35:37-48. [DOI] [PubMed] [Google Scholar]
- 30.Monciardini, P., L. Cavaletti, P. Schumann, M. Rohde, and S. Donadio. 2003. Conexibacter woesii gen. nov., sp. nov., a novel representative of a deep evolutionary line of descent within the class Actinobacteria. Int. J. Syst. E vol. Microbiol. 53:569-576. [DOI] [PubMed] [Google Scholar]
- 31.Mummey, D. L., and P. D. Stahl. 2003. Candidate division BD: phylogeny, distribution and abundance in soil ecosystems. Syst. Appl. Microbiol. 26:228-235. [DOI] [PubMed] [Google Scholar]
- 32.Nogales, B., E. R. B. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 33.Ochsenreiter, T., D. Selezi, A. Quaiser, L. Bonch-Osmolovskaya, and C. Schleper. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787-797. [DOI] [PubMed] [Google Scholar]
- 34.Olsen, R. A., and L. R. Bakken. 1987. Viability of soil bacteria: optimization of plate-counting technique and comparison between total counts and plate counts within different size groups. Microb. Ecol. 13:59-74. [DOI] [PubMed] [Google Scholar]
- 35.Rheims, H., A. Felske, S. Seufert, and E. Stackebrandt. 1999. Molecular monitoring of an uncultured group of the class Actinobacteria in two terrestrial environments. J. Microbiol. Methods 36:65-75. [DOI] [PubMed] [Google Scholar]
- 36.Rösch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 38.Singleton, D. R., M. A. Furlong, A. D. Peacock, D. C. White, D. C. Coleman, and W. B. Whitman. 2003. Solirubrobacter pauli gen. nov., sp. nov., a mesophilic bacterium within the Rubrobacteridae related to common soil clones. Int. J. Syst. E vol. Microbiol. 53:485-490. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, K., M. D. Collins, E. Iijima, and K. Komagata. 1988. Chemotaxonomic characterization of a radiotolerant bacterium, Arthrobacter radiotolerans: description of Rubrobacter radiotolerans gen. nov., comb. nov. FEMS Microbiol. Lett. 52:33-40. [Google Scholar]
- 40.Wellington, E. M. H., P. Marsh, J. E. M. Watts, and J. Burdon. 1997. Indirect approaches for studying soil microorganisms based on cell extraction and culturing, p. 311-329. In J. D. van Elsas, J. T. Trevors, and E. M. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 41.Winding, A., S. J. Binnerup, and J. Sørensen. 1994. Viability of indigenous soil bacteria assayed by respiratory activity and growth. Appl. Environ. Microbiol. 60:2869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yakimov, M. M., H. Lünsdorf, and P. N. Golyshin. 2003. Thermoleophilum album and Thermoleophilum minutum are culturable representatives of group 2 of the Rubrobacteridae (Actinobacteria). Int. J. Syst. E vol. Microbiol. 53:377-380. [DOI] [PubMed] [Google Scholar]
- 43.Zengler, K., G. Toledo, M. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15684-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., Y. Sekiguchi, S. Hanada, P. Hugenholtz, H. Kim, Y. Kamagata, and K. Nakamura. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. E vol. Microbiol. 53:1155-1163. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, J., B. Xia, H. Huang, D. S. Treves, L. J. Hauser, R. J. Mural, A. V. Palumbo, and J. M. Tiedje. 2003. Bacterial phylogenetic diversity and a novel candidate division of two humid region, sandy surface soils. Soil Biol. Biochem. 35:915-924. [Google Scholar]