Abstract
Most studies on the reduction of disease incidence in soil treated with Trichoderma asperellum have focused on microbial interactions rather than on plant responses. This study presents conclusive evidence for the induction of a systemic response against angular leaf spot of cucumber (Pseudomonas syringae pv. lachrymans) following application of T. asperellum to the root system. To ascertain that T. asperellum was the only microorganism present in the root milieu, plants were grown in an aseptic hydroponic growth system. Disease symptoms were reduced by as much as 80%, corresponding to a reduction of 2 orders of magnitude in bacterial cell densities in leaves of plants pretreated with T. asperellum. As revealed by electron microscopy, bacterial cell proliferation in these plants was halted. The protection afforded by the biocontrol agent was associated with the accumulation of mRNA of two defense genes: the phenylpropanoid pathway gene encoding phenylalanine ammonia lyase (PAL) and the lipoxygenase pathway gene encoding hydroxyperoxide lyase (HPL). This was further supported by the accumulation of secondary metabolites of a phenolic nature that showed an increase of up to sixfold in inhibition capacity of bacterial growth in vitro. The bulk of the antimicrobial activity was found in the acid-hydrolyzed extract containing the phenolics in their aglycone form. High-performance liquid chromatography analysis of phenolic compounds showed a marked change in their profile in the challenged, preelicited plants relative to that in challenged controls. The results suggest that similar to beneficial rhizobacteria, T. asperellum may activate separate metabolic pathways in cucumber that are involved in plant signaling and biosynthesis, eventually leading to the systemic accumulation of phytoalexins.
Several recent studies have established the role of selected strains of nonpathogenic plant growth-promoting rhizobacteria (PGPR) and fungi (PGPF) in enhancing plant resistance (30, 31, 35, 36, 41, 42, 47). An example of PGPF are Trichoderma spp., which have recently been shown to induce local and systemic defense responses in cucumber (48, 49) and other agricultural crops, such as cotton, tobacco, lettuce, and bell pepper (1, 9, 20). Induced systemic resistance (ISR) (23, 26, 27), mediated by such nonpathogenic rhizospheric microorganisms, has been demonstrated in several plant species and shown to be effective against bacterial, viral, and fungal disease (30, 35). However, little is known about the molecular basis underlying this type of ISR. Unlike pathogen-induced or chemically induced resistance, some of these defense responses were independent of salicylic acid or pathogenesis-related protein accumulation (35, 43, 45). One of the most studied mechanisms of resistance to infection in plants is the rapid accumulation of antimicrobial, low-molecular-weight secondary metabolites known as phytoalexins (17, 24). Several studies have established the role of phytoalexins in the defense response of cucumber (Cucumis sativus L.) and other cucurbits (6, 11, 12, 18, 19, 33). Early findings suggested that the lignin precursor, coniferyl alcohol, may function as a phytoalexin in cucumber (18). Subsequent analysis of the lignin-like material indicated that it was a p-coumaryl alcohol derivative (40). Recent studies using Milsana (Compo, Munster, Germany) (6) or silicon (11) as elicitors have also demonstrated the involvement of glycosylated phenolics in the defense response of cucumber to powdery mildew. Increased accumulation of the antifungal glycosides of p-coumaric methyl ester and rhamnetin was demonstrated in response to Milsana elicitation. Recently, by use of the same experimental system, an increase in the accumulation of flavonoid compounds was found in correlation with the accumulation of two flavonoid biosynthetic genes, i.e., the chalcone synthase and chalcone isomerase genes (12). Phytoalexin synthesis involves the rapid transcriptional activation of genes encoding a number of key biosynthetic enzymes. Phenylalanine ammonia lyase (PAL) (EC 4.3.1.5) catalyzes the first step in the phenylpropanoid biosynthetic pathway leading to the production of phenolic compounds with a significant range of biological functions, including phytoalexins (17). Hydroperoxide lyase (HPL) is part of the octadecanoic pathway. HPL utilizes some of the lipoxygenase products as its substrates and produces antimicrobial and wound-related molecules known as C6-volatiles. These molecules are related to the jasmonic acid pathway and have been associated with the plant's defense response to pathogens. Some HPL products are signal molecules that have been shown to induce the gene expression and production of phytoalexins (50). Nonetheless, few attempts have been made to study the participation of such secondary metabolites in the ISR afforded by soil microorganisms, including PGPR and PGPF (41, 43). Recently, systemic induction of phytoalexins following treatment with fluorescent pseudomonas was shown in the overall defense response of cucumber (33, 34). The role of such compounds in the defense response afforded by root treatment with the fungal antagonist Trichoderma asperellum (25) remains to be established. In this study, we developed a model system using cucumber as the model plant and Pseudomonas syringae pv. lachrymans as the challenging pathogen. Our goal was to characterize the systemic response induced in cucumber plants following T. asperellum application to the root system and to establish the biochemical nature of that response.
MATERIALS AND METHODS
Plant material.
Seeds of cucumber (Cucumis sativus L. cv. Delila) from Gedera Seeds Co. (Gedera, Israel) were used in this experiment. Plant growth medium (PGM) was prepared as described previously and adjusted daily (48).
Aseptic growth system.
Seeds were surface disinfested in 2% NaOCl for 2 min and thoroughly washed with sterile distilled water. They were then put on a sterile gauze sheet and placed in aseptic hydroponic growth containers (48). The growth containers were aerated with air filtered through 0.45-μm-pore-size filters. Plants were grown in a controlled environment: 26°C, 80% relative humidity, light at 300 μE/m2/s, and a circadian cycle of 14 h of light and 10 h of darkness.
Fungal material.
T. asperellum strain T-203 (10, 25) was grown on potato dextrose agar (PDA) (Difco, Detroit, Mich.). Synthetic medium for T. asperellum was prepared according to Okon et al. (32). The inoculum consisted of 1 ml (109 spores, counted by hemocytometer) of 7-day-old T. asperellum cultured on PDA added to a 250-ml flask containing 100 ml of synthetic medium. The flask was shaken at 150 rpm for 24 h at 30°C to allow spore germination. The inoculum was then separated from the growth medium by centrifugation at 10,000 × g at 4°C and washed twice in 100 ml of distilled water. T. asperellum mycelial inoculum was added under aseptic conditions to the PGM of 7-day-old seedlings to a final concentration of about 105 germinated spores/ml of PGM.
Bacterial inoculum.
P. syringae pv. lachrymans was grown in tryptic soy broth overnight at 30°C. Bacterial cells were centrifuged at 10,000 × g, and the pellet was resuspended in sterile phosphate-buffered saline (5 mM, pH 7.2). Challenge was performed 48 h after application of T. asperellum to the PGM. P. syringae pv. lachrymans bacterial suspension (optical density at 600 nm = 0.1) containing 0.01% (wt/vol) surfactant (Tween 20) was applied to the surface of the first and second leaves of cucumber seedlings. Leaves were either rubbed with a sterile cheesecloth pad saturated with bacterial suspension or pipetted onto the leaf with four 10-μl droplets by using a repeating pipettor. Inoculation was performed under aseptic conditions.
Multiplication of P. syringae pv. lachrymans was assessed in challenged leaves at different times after inoculation. Two pools of 1 g of randomly selected samples of 15 leaves taken from 15 plants per treatment were rinsed thoroughly in sterile water and homogenized in a sterile solution of 10 mM phosphate-buffered saline. Dilutions were plated onto Pseudomonas selective King's B agar (22) supplemented with a 1-ml liter−1 solution containing 9 mg of basic fuchsin, 200 mg of cycloheximide, 10 mg of nitrofurantoin, and 23 mg of nalidixic acid. After incubation at 28°C for 24 to 36 h, the number of P. syringae pv. lachrymans CFU per gram of infected leaf tissue was determined.
Microbial bioassay.
P. syringae pv. lachrymans, Agrobacterium tumefaciens, Bacillus megaterium, and Micrococcus luteus (strains were taken from a laboratory collection) were cultured in tryptic soy broth (Difco). Crude phenolic extract was concentrated by Speed-Vac and adjusted to 20 μl with absolute methanol. The concentrated samples were pipetted onto tryptic soy agar plates or PDA plates and dried in a laminar flow hood. Bacterial suspensions (200 μl) were mixed into 3 ml of soft tryptic soy agar and overlaid on the dried plates. Antimicrobial activity of the extract was assayed 48 h after bacterial application and appeared as clear lytic circles on the plates.
RNA isolation.
For RNA analysis, roots, cotyledons, and leaves were harvested at different times after inoculation and stored at −70°C until use. Total RNA was extracted using an EZ-RNA total RNA isolation kit (Biological Industries Co., Beit-Haemek, Israel). RNA was treated with RNase-free DNase I in a solution containing 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2, and 10 mM CaCl2 for 30 min at 37°C (Roche, Penzberg, Germany), followed by phenol-chloroform and chloroform extraction and subsequent ethanol precipitation.
RT.
After treatment with DNase, 1 μg of total RNA was used for a reverse transcription (RT) reaction using Expand reverse transcriptase (Roche) according to the manufacturer's instructions.
Quantitative PCR.
The primers used for the quantitative PCR experiments were as follows: for Pal1, forward primer 5′-ATGGAGGCAACTTCCAAGGA-3′ and reverse primer 5′-CCATGGCAATCTCAGCACCT-3′, and for HPL, forward primer 5′-TCTCGCCATGACAGGTTCATC-3′ and reverse primer 5′-GAACATTCCAGGTCCATCAGC-3′. PCRs were carried out in 96-well plates (25 μl per well) in a reaction buffer containing 1× SYBR Green PCR Master Mix (PE Applied Biosystems), 120 nM concentrations of primer (for each forward and reverse primer), and 1/20 of the RT reaction mix. Quantitative analysis was performed using the GeneAmp5700 sequence detection system (PE Applied Biosystems) with PCR conditions as follows: 95°C for 15 s and 60°C for 1 min for 40 cycles. The absence of primer-dimer formation was checked in no-template controls. The specificity of primers to cucumber genes was examined by using Trichoderma DNA and reverse-transcribed RNA as templates. The 18S ribosomal cDNA was used as a control reference. Each sample was examined in duplicate. We first normalized the expression of the specific gene versus the control reference by using the formula 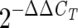 , where the CT (threshold cycle) value is defined as the PCR cycle number that crosses an arbitrary threshold line, ΔCT = CT of the specific gene − CT of the reference gene, and ΔΔCT = ΔCT − the arbitrary constant (the highest ΔCT) (for more elaboration, see Sequence Detector User Bulletin no. 2, PE Applied Biosystems).
, where the CT (threshold cycle) value is defined as the PCR cycle number that crosses an arbitrary threshold line, ΔCT = CT of the specific gene − CT of the reference gene, and ΔΔCT = ΔCT − the arbitrary constant (the highest ΔCT) (for more elaboration, see Sequence Detector User Bulletin no. 2, PE Applied Biosystems).
Phenolics extraction.
Fresh foliar material from 11- and 12-day-old Cucumis sativus L. seedlings was harvested 48 h post-challenge inoculation with P. syringae pv. lachrymans. The experiment included four treatment groups: control plants (not elicited with T. asperellum [T−] and not challenged with P. syringae pv. lachrymans [Psl−]); T. asperellum preelicited, nonchallenged plants (T+Psl−); nonelicited, P. syringae pv. lachrymans-challenged plants (T−Psl+); and T. asperellum-elicited and challenged plants (T+Psl+). Each treatment included a free-phenolic fraction, extracted prior to hydrolysis, and a conjugated fraction consisting of the aglycones released after hydrolysis. All treatments were replicated twice. A modification of the extraction method of Fawe et al. (11) was used to determine free and glycosidic-bound phenolics in the leaf extracts. Harvested foliar material was ground to a fine powder in liquid N2 and extracted in 80% acidified methanol (10 g [fresh weight] per 100 ml). The mixture was kept for 24 h in the dark, and air was replaced with nitrogen to prevent oxidation. The extract was then filtered with glass fiber filters (GF/C; Whatman, Maidstone, England), and the filtrate was concentrated under reduced pressure at 40°C. The aqueous residue was adjusted to pH 2.0 and partitioned against hexane to remove lipophilic compounds, i.e., chlorophyll, carotenoids, lipids, and waxes. The aqueous phase containing the phenolic constituents was further partitioned against ethyl acetate (volume/volume) and then subjected to acid hydrolysis (4 N HCl [vol/vol]) in an autoclave for 20 min. The hydrolysate was cooled and partitioned against ethyl acetate. The two ethyl acetate fractions obtained were dried, and the residues designated as free-phenolic fraction and conjugated-phenolic fraction, respectively, were resuspended in absolute methanol (2.5 g [fresh weight] per ml).
Total phenolic concentration.
Leaf extracts were prepared as described above, and total phenolic concentration was measured in both free and glycosidic-bound phenolic extract by using Folin Ciocalteus reagent (Sigma, St. Louis, Mo.) according to the manufacturer's directions.
High-performance liquid chromatography (HPLC) separation of phytoalexins.
Forty-eight hours post-challenge inoculation with P. syringae pv. lachrymans, leaves from all treatments were collected separately, pooled, and kept at −80°C until use (approximately 40 g [fresh weight]). The phenolic extraction was performed as described. The resultant free- and conjugated-phenolic fractions were subjected to several separation procedure steps to determine their profiles.
The HPLC system (Thermo Separation Products, Riviera Beach, Fla.) consisted of an auto sampler (AS3000), injector (100 μl), column oven (30), pump (P3000), and diode array detector (UV6000). A reverse-phase C18 column (250 by 4.6 mm; Luna 2; Phenomenex, Torrance, Calif.), with a precolumn of similar resin, was employed. Elution was performed using phosphate buffer (50 mM, pH 2.5) and methanol at a flow rate of 1 ml min−1. A linear-gradient program was developed as follows: C18 column (time [in minutes]/methanol [percent]) = 0/0, 6/0, 31/95, 32/0. In all instances, the software was programmed to show peaks at their maximum absorbance. For each treatment, HPLC analyses were repeated at least twice, showing similar results. The data presented are from a representative experiment.
Tissue processing for electron microscopy.
Leaf samples (2 to 5 mm2) were collected from the necrotic zone 48 h post-challenge inoculation with P. syringae pv. lachrymans. Samples were then fixed by immersion in 3% (vol/vol) glutaraldehyde in 0.1 M Na-cacodylate buffer (pH 7.2) for 2 h at room temperature. The samples were postfixed with 1% (wt/vol) osmium tetroxide in the above buffer for 1 h at 4°C and then dehydrated in a graded ethanol series and embedded in Epon 812. Ultrathin sections (0.1 μm) were cut with a diamond knife and collected on Formvar-coated nickel grids. The sections were contrasted with uranyl acetate and lead citrate for immediate examination in a JEOL 1200 EX transmission electron microscope, operating at 80 kV.
Statistical analysis.
The effects of T. asperellum treatment on the diameter and total area of necroses caused by P. syringae pv. lachrymans were analyzed by one-way analysis of variance using the Excel program (Microsoft Corp., Bothell, Wash.). The effect of T. asperellum on antimicrobial activity of the crude phenolic extract and on total phenolics concentration was determined using the Student's t test function of Excel.
RESULTS
Effect of T. asperellum on ISR.
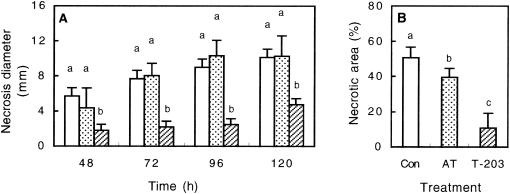
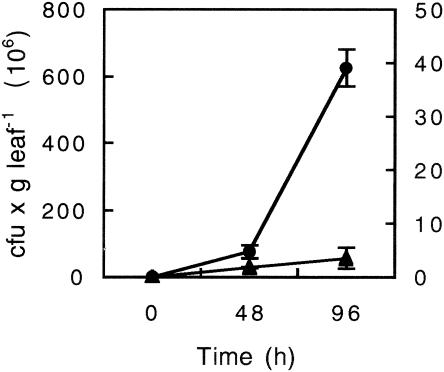
To test T. asperellum-mediated ISR, angular leaf spot of cucumber, P. syringae pv. lachrymans, was used to challenge leaves of 12-day-old cucumber seedlings at 48 h postapplication of T. asperellum to the root system. An aseptic hydroponic growth system was used in which spatial separation of the root-inducing agent from the leaf pathogen was verified. Four days post-challenge inoculation with P. syringae pv. lachrymans, the noninduced (T−Psl+) plants displayed necrotic lesions surrounded by extensively spreading chlorosis, whereas plants pretreated with T. asperellum (T+Psl+) showed significantly lower symptoms. Protection against P. syringae pv. lachrymans was quantified by assessing necrosis diameter and the proportion of total necrotic area per leaf. A 50% reduction in the average necrosis diameter was observed 5 days postchallenge, while total necrotic area was reduced by 80% in the T+Psl+ plants relative to that in T−Psl+ plants (Fig. 1). Application of autoclaved T. asperellum mycelial inoculum to the roots appeared to have only a minor effect on disease reduction (Fig. 1B). Proliferation of P. syringae pv. lachrymans in the challenged leaves was assessed and found to be significantly lower in the T+Psl+ plants than in the T−Psl+ plants. Ninety-six hours postchallenge, bacterial cell densities in the T−Psl+ treatment exceeded those of the T+Psl+ treatment by about 200-fold. Thus, the reduction in disease symptoms appeared to be associated with inhibition of bacterial multiplication (Fig. 2).
FIG. 1.
ISR to P. syringae pv. lachrymans in 14-day-old cucumber leaves at 48 to 120 h post-challenge inoculation, 96 h from application of T. asperellum to the root system. Disease severity is expressed as necrosis diameter (A) or proportion of necrotic area relative to total leaf area (B). Shown are results for noninoculated plants (white columns), plants treated with autoclaved inoculum of T. asperellum (AT) (dotted columns), and plants treated with mycelial inoculum of T. asperellum (T-203) (striped columns). Columns headed by the same letter are not significantly different (α ≤ 0.05, one-way analysis of variance). Error bars represent standard deviations (SD) from results for 20 replicate plants that received the same treatment.
FIG. 2.
Multiplication of P. syringae pv. lachrymans in challenged leaves assessed at the indicated time points postinoculation. Control plants (T−Psl+) were treated with distilled water 48 h prior to challenge with P. syringae pv. lachrymans (•); plants treated with T. asperellum (T+Psl+) were elicited 48 h prior to challenge with P. syringae pv. lachrymans (▴). Data points are means (CFU/gram) ± standard errors (SE) for two sets of 20 randomly selected leaves, and values are the averages from two independent experiments.
To verify whether T. asperellum remained spatially separated for the duration of the experiment, its presence in the shoots was assessed by plating serial dilutions of leaf homogenates into Trichoderma selective growth medium. Trichoderma was not detected in leaf homogenates at any time during the study. In addition, using dual-culture assays in petri dishes, we found that T. asperellum is unable to antagonize P. syringae pv. lachrymans directly.
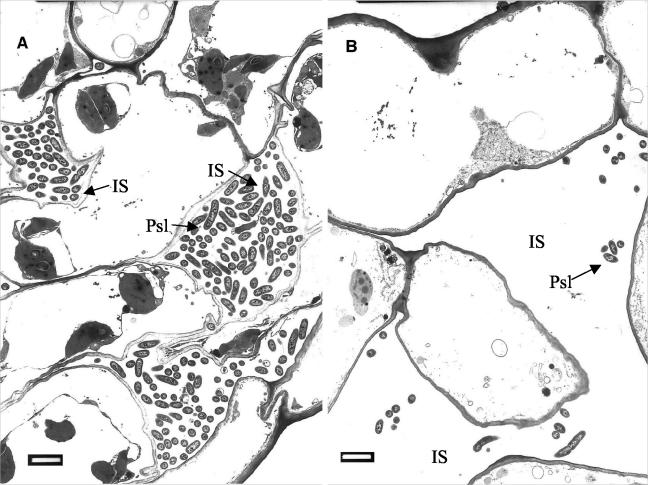
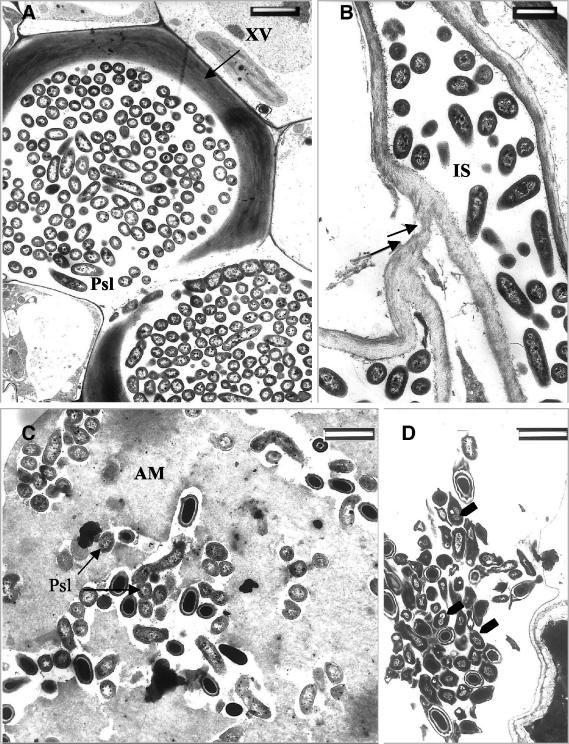
Transmission electron microscopy (TEM) of ultrathin sections from the T−Psl+ treatment group revealed proliferation of P. syringae pv. lachrymans 96 h postchallenge. P. syringae pv. lachrymans invaded the intercellular spaces of the leaves (Fig. 3A). At the same time point, significantly fewer bacterial cells were observed in the T+Psl+ plants (Fig. 3B). In the nonelicited plants, P. syringae pv. lachrymans invaded the xylem vessels, which appeared to be densely colonized with the bacteria (Fig. 4A). In some cases, host cell walls were disrupted and digested by the pathogen (Fig. 4B). Closer examination of the invaded areas revealed the occurrence of host reactions at sites of bacterial penetration. An amorphous matrix showing various degrees of compactness appeared in the intercellular spaces of the T. asperellum-preelicited plants (Fig. 4C). Bacterial cells trapped in this material appeared embedded and exhibited morphological alterations. Apparently, their development was halted (Fig. 4C and D). Some cells contained densely stained osmiophilic material and were severely affected, as may be deduced from the aggregation of their contents (Fig. 4D). None of these phenomena were detected in the nonelicited plants, where bacterial cells appeared intact and proliferated in all tissues.
FIG. 3.
Transmission electron micrographs of ultrathin sections from cucumber leaves challenged with P. syringae pv. lachrymans at 96 h postchallenge. (A) Bacterial cell colonization of leaves sampled from nonelicited, challenged plants (T−Psl+). P. syringae pv. lachrymans progresses towards the inner leaf tissues mainly by intercellular (IS) growth. (B) Bacterial cell colonization of intercellular spaces in leaves of plants preelicited with T. asperellum 48 h prior to challenge (T+Psl+). Considerably fewer bacterial cells are observed. Bars, 2 μm.
FIG. 4.
Transmission electron micrographs of ultrathin sections from cucumber leaf tissues. (A) Xylem vessels (XV) heavily colonized with P. syringae pv. lachrymans in leaves of nonelicited plants (T−Psl+) at 96 h postchallenge with P. syringae pv. lachrymans. (B) Digestion of host cell wall by P. syringae pv. lachrymans (arrows) in leaves of nonelicited (T−Psl+) plants. (C) Intercellular spaces (IS) of T. asperellum-preelicited plants (T+Psl+) in which an amorphous matrix (AM) has accumulated. P. syringae pv. lachrymans cells trapped in this AM appear embedded, and their development is halted. (D) P. syringae pv. lachrymans cells (arrows) in leaves of T. asperellum-preelicited plants (T+Psl+) appear to be severely damaged, and their content is aggregated. Some of these cells accumulate dark-staining osmiophilic material. Bars, 2 μm (A and D) and 1 μm (B and C).
Gene expression.
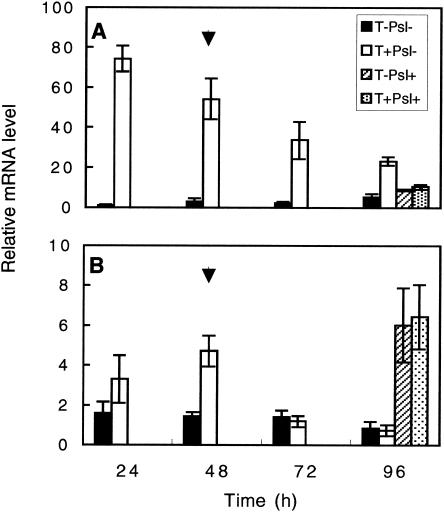
Increased expression levels of PAL1 were observed in the T. asperellum-preelicited roots (T+Psl−) relative to what was seen with nontreated roots (T−Psl−) throughout the examined period (Fig. 5A). Preelicited roots expressed a 70-fold-higher expression level of PAL1 at 24 h postinoculation (hpi), declining gradually to a fourfold difference at 96 hpi. Roots of plants challenged with P. syringae pv. lachrymans with or without preelicitation with Trichoderma expressed lower levels of PAL1. In the leaves, PAL1 expression levels were higher in the preelicited plants (T+Psl−) at 24 hpi and reached a maximum at 48 hpi (Fig. 5B). Although challenged leaves exhibited higher PAL1 expression than nonchallenged leaves, no significant difference in PAL1 expression was observed between T+Psl+ and T−Psl+ leaves (Fig. 5B).
FIG. 5.
Relative expression levels of Pal1 in the roots (A) and leaves (B) of cucumber plants. Expression was measured in both roots and leaves of cucumber at 24 to 96 h postelicitation of the root compartment with Trichoderma. At 48 h (arrow), challenge inoculation with P. syringae pv. lachrymans was performed, and mRNA levels postchallenge were recorded 48 h later. All experiments were repeated twice and showed similar results. Bars represent relative mRNA levels from one representative experiment ± SE.
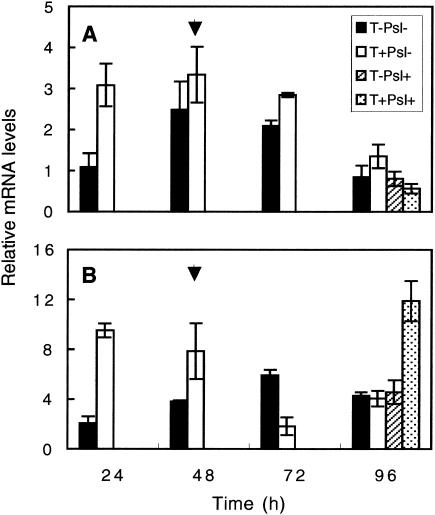
HPL expression levels in roots at 24 hpi were considerably higher in Trichoderma-pretreated plants than in controls (Fig. 6A). However, that difference declined during subsequent days. In the leaves at 24 to 48 hpi, plants preelicited with T. asperellum (T+Psl−) displayed higher HPL expression levels than controls (T−Psl−) (Fig. 6B). HPL expression levels in the leaves of all treatments at 96 hpi were similar, with the exception of the T+Psl+ plants, which expressed a threefold-higher expression level (Fig. 6B).
FIG. 6.
Relative expression levels of HPL in the roots (A) and leaves (B) of cucumber plants. Expression was measured in both roots and leaves of cucumber at 24 to 96 h postelicitation of the root compartment with Trichoderma. At 48 h (arrow), challenge inoculation with P. syringae pv. lachrymans was performed, and mRNA levels were recorded 48 h later. All experiments were repeated twice and showed similar results. Bars represent relative mRNA levels from one representative experiment ± SE.
Extraction and fractionation of phenolic compounds.
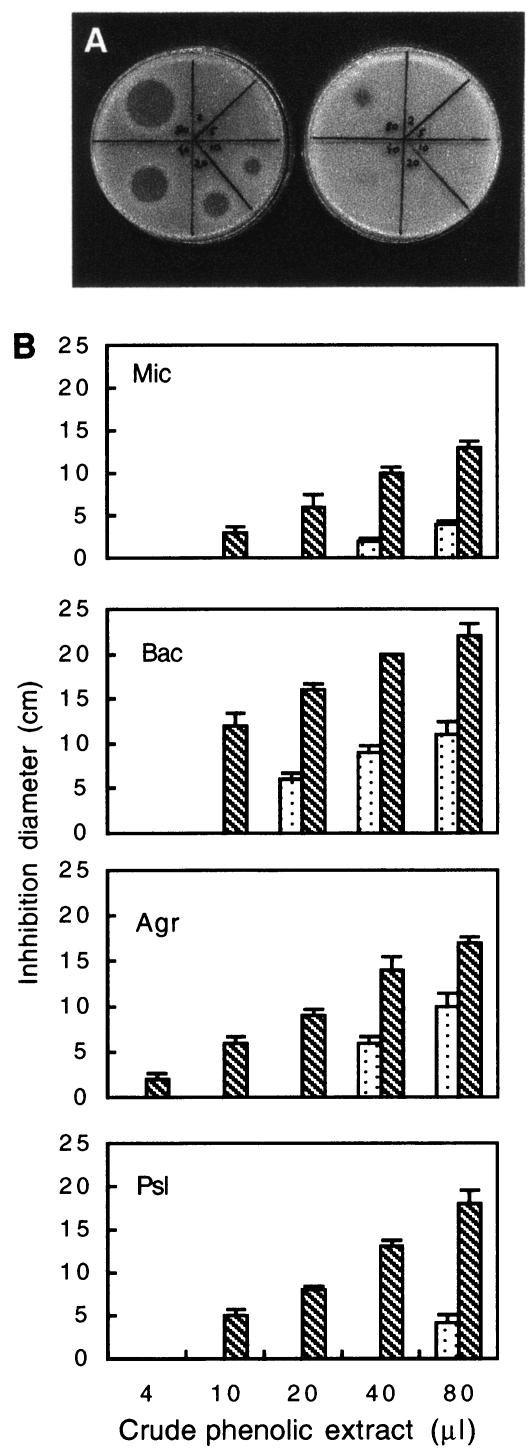
To further characterize the inhibition effect shown by plants preelicited with T. asperellum, leaves were extracted according to the method of Daayf et al. and Fawe et al. (5, 11) and the crude phenolic extract was assessed for antimicrobial activity. Since the P. syringae pv. lachrymans pathosystem was used throughout the study, the bioassay was designed to evaluate antibacterial activity of extractable phenolic fractions on P. syringae pv. lachrymans development. Each of the four treatments studied included a free-phenolic fraction, extracted prior to hydrolysis, and a conjugated-phenolic fraction consisting of the aglycones produced through acid hydrolysis. Similar basal antibacterial activities of both free and conjugated phenolics were found in the two unchallenged treatments, T−Psl− and T+Psl−. In the free-phenolic fraction, challenge inoculation with P. syringae pv. lachrymans resulted in an increase in the antibacterial activities of both the T−Psl+ and T+Psl+ plants, suggesting that P. syringae pv. lachrymans itself may trigger the accumulation of antimicrobial compounds in the leaves. Nevertheless, in the conjugated fraction containing the aglycones, challenge with P. syringae pv. lachrymans resulted in a remarkably higher antimicrobial activity of the preelicited challenged (T+Psl+) plants relative to that of nonelicited (T−Psl+) plants. A bioassay of the active conjugated fraction extracted from plants challenged with P. syringae pv. lachrymans exhibited a dose-dependent effect of T. asperellum preelicitation on the antibacterial potential of the extract. While 10 μl of the crude phenolics extracted from T+Psl+ plants was enough to produce a clear lytic zone on agar plates, indicating the presence of potent antibacterial compounds, as much as 80 μl of crude extract of the T−Psl+ plants was needed to produce inhibition (Fig. 7A).
FIG. 7.
(A) Tryptic soy agar plates showing a dose-dependent bioassay (2 to 80 μl corresponding to 5 to 200 mg [fresh weight] of leaf tissue) of the aglycone fraction obtained by acid hydrolysis of the crude phenolics extract of cucumber leaves. The extraction was carried out 48 h postchallenge with P. syringae pv. lachrymans. Shown are results for plants elicited with T. asperellum (T+Psl+; left plate) and nonelicited plants (T−Psl+; right plate). The bioassay was executed with P. syringae pv. lachrymans as the test microorganism. (B) Bioassay comparing the antimicrobial capacity of 2 to 80 μl of the aglycone fraction obtained by acid hydrolysis of crude phenolics extract of cucumber leaves 48 h postchallenge with P. syringae pv. lachrymans. Shown are results for plants elicited with T. asperellum (T+Psl+; dotted columns) and nonelicited plants (T−Psl+; striped columns). The antimicrobial activity was assayed on P. syringae pv. lachrymans (Psl), Agrobacterium tumefaciens (Agr), Bacillus megaterium (Bac), and Micrococcus luteus (Mic) as the test microorganisms. Columns represent the mean inhibition diameter of two independent experiments ± SD.
Antimicrobial spectrum.
Dose-response bioassays were also conducted to explore the antimicrobial spectrum of the crude extract and HPLC fractions of the conjugated phenolics extracted from leaves of both T−Psl+ and T+Psl+ plants. The extracts were applied to two gram-positive bacteria (Micrococcus luteus and Bacillus megaterium) and two gram-negative bacteria (P. syringae pv. lachrymans and Agrobacterium tumefaciens). The T+Psl+ phenolic extract was far more potent in its antimicrobial potential than the T−Psl+ phenolic extract on all bacteria examined (Fig. 7B). Interestingly, the extract from the T−Psl+ treatment was less effective in its antimicrobial potential when applied on P. syringae pv. lachrymans than when applied on other bacterial types.
Quantification of total phenolics.
Total phenols were determined by using the Folin-Ciocalteu assay. Forty-eight hours after T. asperellum elicitation, a significant increase (35%) in the concentration of total phenols was found in both free and conjugated phenols (Table 1). However, this increase was not correlated with an elevation in antibacterial activity. Nonetheless, 48 h postchallenge with P. syringae pv. lachrymans, a marked increase was observed in the concentrations of total phenols of both free and conjugated fractions, with no significant difference between the T−Psl+ and T+Psl+ plants. Thus, it may be suggested that the marked rise in antibacterial activity of the T+Psl+ conjugated fraction described earlier (Fig. 7) indicates the production of specific substances with antimicrobial activity.
TABLE 1.
Concentration of total phenolics in cucumber leaves in response to T. asperellum elicitation of the root system and/or challenge inoculation of leaves with P. syringae pv. lachrymans
| Treatment group | Concn (μg of gallic acid equivalents/g of fresh leaf tissue)a
|
|
|---|---|---|
| Free phenolics | Conjugated phenolics | |
| T−Psl− | 72 ± 6b | 81 ± 9 |
| T+Psl− | 115 ± 7c | 121 ± 9 |
| T−Psl+ | 191 ± 18d | 160 ± 7 |
| T+Psl+ | 206 ± 31d | 174 ± 11 |
Total phenolics were determined in leaves 48 h after challenge inoculation with P. syringae pv. lachrymans. The crude extract was separated before and after acid hydrolysis to enable the analysis of free and conjugated phenolics, respectively. Values are means of two independent experiments ± SD for pooled samples of 40 g of fresh leaf tissue with three replicates.
Not significantly different from corresponding value for conjugated phenolics (P < 0.05, Student's t test) but different from those of T−Psl+ and t+Psl+.
Not significantly different from corresponding value for conjugated phenolics (and from values for T+Psl+ group) (P < 0.05, Student's t test) but different from those of T−Psl+.
Not significantly different from corresponding value for conjugated phenolics and from values for T−Psl+ group (P < 0.05, Student's t test) but different from those of T+Psl+.
Separation of phenolic compounds.
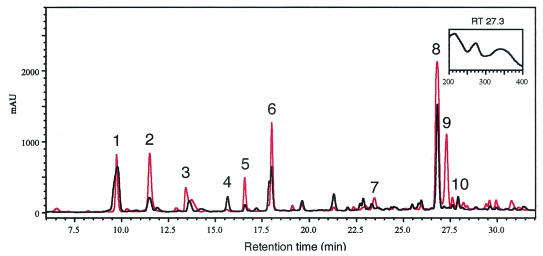
A comparative study of the profiles of the aglycone fractions (conjugated phenolics) in all treatments, using reverse-phase HPLC analysis, revealed different responses to T. asperellum elicitation with or without challenge inoculation with P. syringae pv. lachrymans (Fig. 8). Ten major peaks were observed, and changes in relative peak area and their absorbance spectra were recorded (Fig. 8) (Table 2). An increase was shown in the concentration of four phenolic compounds, while four compounds decreased in response to elicitation with T. asperellum (T+Psl−) relative to the control treatment (T−Psl−). However the increased concentration did not account for the increase in antimicrobial activity relative to the control. Challenge inoculation with P. syringae pv. lachrymans (T−Psl+) induced an increase in the concentration of five compounds, two of which showed antimicrobial activity in our system, while two compounds decreased relative to the control (T−Psl−). The increase in antimicrobial activity of the crude phenolics extracted from the leaves of T−Psl+ plants suggested that the pathogen itself triggers a defense response in the plant. In the T+Psl+ treatment, the level of six compounds increased, four of which were active against P. syringae pv. lachrymans in vitro. Two compounds decreased relative to controls. A compound designated peak 9 was recorded only in the preelicited and challenged plants (T+Psl+) (Fig. 8) (Table 2) and displayed a UV spectrum with λmax at 272 nm, which is typical of flavonoid compounds in cucumber (12). Other induced compounds, such as the compounds at retention times 11.5 and 18.0, displayed UV spectra typical of catechins, with peaks at 225 and 280 nm (37). Overall, a greater-than-sixfold increase in the antibacterial activity of the crude phenolics extract from leaves of T+Psl+ plants was observed relative to that in T−Psl+ plants.
FIG. 8.
Reverse-phase HPLC chromatogram at 272 nm. Shown are results for nonelicited, nonchallenged plants (T−Psl−) (black) and plants preelicited with T. asperellum 48 h prior to challenge with P. syringae pv. lachrymans (T+Psl+) (red). Numbers indicate analyzed peaks. The inset shows the UV spectrum of peak 9 observed only in the T+Psl+ treatment. This peak is typical of flavonoid compounds. AU, absorbance units.
TABLE 2.
Peak integration area calculated from HPLC chromatograms at 272 nm
| Peak no. | Retention time (min) | Absorbance units for treatmenta:
|
|||
|---|---|---|---|---|---|
| T−Psl− | T+Psl− | T−Psl+ | T+Psl+ | ||
| 1 | 9.7 | 11,832 | 8,839 | 10,313 | 6,199 |
| 2 | 11.5 | 2,383 | 3,847 | 8,388 | 9,071 |
| 3 | 13.5 | 1,825 | 3,143 | 2,506 | 3,940 |
| 4 | 15.6 | 1,836 | ND | ND | ND |
| 5 | 16.6 | 757 | 467 | 2,600 | 3,668 |
| 6 | 18.0 | 7,782 | 4,828 | 4,993 | 10,454 |
| 7 | 23.3 | 678 | 581 | 2,007 | 1,659 |
| 8 | 26.8 | 13,881 | 22,955 | 24,420 | 26,895 |
| 9 | 27.3 | ND | ND | ND | 11,293 |
| 10 | 27.9 | 1,183 | 5,096 | 1,179 | 769 |
Extracts were prepared from 40 g (fresh weight) of foliar material. HPLC runs were repeated twice (SD ≤ 1%). Peaks below detection level were specified as nondetectable (ND). Two independent experiments were performed and showed similar results. Presented data are from one representative experiment.
DISCUSSION
Attempts to exploit fungal antagonists such as Trichoderma spp. as potential biological control agents have recently led to the proposal that besides their recognized antifungal properties, such organisms could also act as elicitors of plant defense reactions, thereby promoting the expression of plant defense-related genes (1, 9, 20, 21, 48, 49). However, the exact mechanisms underlying induction of plant disease resistance by antagonistic fungi remain poorly understood. Several hypotheses have been put forward, but very few have been convincingly assessed through biochemical and cytological investigations of plant tissues challenged by these fungal agents.
The results of the present study demonstrate that cucumber plants grown in the presence of T. asperellum exhibit increased protection against P. syringae pv. lachrymans infection. These results are of particular relevance since they highlight the dual properties of T. asperellum, which is also capable of inducing systemic resistance against a foliar pathogen (Fig. 1). Moreover, our cytological and biochemical results demonstrate that the beneficial effect of T. asperellum in repressing Pseudomonas ingress in the leaf tissues does not rely on direct antifungal activity, as is the case for a number of plant growth-promoting rhizobacteria (43). Support for this concept came from the observations that Trichoderma propagules could not be isolated from the shoots and that antagonistic activity between Trichoderma and Pseudomonas did not occur in vitro (data not shown).
While cucumber has been used frequently as a model for systemic induced resistance (7, 8, 18, 39), little has been reported on the relative importance of phytoalexins in the overall defense response of cucumber. Nevertheless, it has been proposed that lignification (18) and to a lesser extent peroxidases and chitinases (7) are among the potential defense mechanisms employed by this plant. In a recent study, Yedidia et al. provided evidence that T. asperellum (T-203) may induce a transient systemic increase in the activity of these enzymes (48). Indeed, chitinases were shown to inhibit bacterial proliferation to some extent; however, since Trichoderma was absent from the site of challenge with the pathogen, this increase could not explain the observed reduction in disease symptoms (Fig. 1).
Several lines of evidence, from TEM of ultrathin sections, monitoring of P. syringae pv. lachrymans populations, and in vitro bioassays of phenolic leaf extracts, confirmed the systemic inhibition of bacterial proliferation in the leaves. A reduction of 2 orders of magnitude in the proliferation rates of bacterial cells in the preelicited plants (T+Psl+) was observed relative to those in nonelicited (T−Psl+) plants (Fig. 2). This reduction correlated temporally with the TEM results, demonstrating that there were considerably fewer bacteria in the preelicited plants (Fig. 3B). Apparently, the surviving bacterial cells were restricted in space and their ingress into the inner leaf tissue was blocked. The accumulation of osmiophilic material inside P. syringae pv. lachrymans cells was associated with the presence of phenolic compounds known to stain densely upon reaction of O-dihydroxy groups with osmium tetroxide (38). Some bacterial cells embedded in the amorphous matrix were altered and their cell content was aggregated (Fig. 4C and D). Similar responses have been observed during invasion by Xanthomonas campestris of cotton, cabbage, and Pelargonium spp., in which collapsed bacterial cells were observed mainly in phenol-containing areas, indicating that these compounds probably have a bactericidal effect in planta (3). These results suggest that antimicrobial compounds are involved in the inhibition of bacterial growth in cucumber leaves.
Phytoalexins were only recently isolated from long English cucumber, a member of the Cucurbitaceae (5, 6, 11, 12, 33, 40). These previous studies convincingly demonstrated de novo synthesis of a number of phytoalexins, thus establishing the role of antimicrobial compounds in the defense response of cucumber (5, 6, 11).
Previous studies have also demonstrated that both preformed and neosynthesized conjugates play a role in the accumulation of phytoalexins following infection with a pathogen (11, 13, 14). Accordingly, nearly all elements of plant chemical defense and the bulk of antimicrobial activity were found in the aglycone form (5, 6, 11). Our results confirmed that the production of glycosylated phytoalexins in cucumber requires acid hydrolysis of the extract as a prerequisite for the expression of antimicrobial activity. The increased antimicrobial activity of the aglycone fraction of both T−Psl+ and T+Psl+ treatment groups suggests that P. syringae pv. lachrymans itself triggers the production of antimicrobial compounds. Such potential of a bacterial pathogen to induce systemic defense responses in plants has been shown previously (19). However, in the present study, when plants were elicited with T. asperellum prior to challenge with Psl (T+Psl+), a prominent increase in antimicrobial potential was observed (Fig. 7). This increase was apparent in both the MIC required for bacterial inhibition and the spectrum of antimicrobial activity of the T+Psl+ plants relative to that of the T−Psl+ plants. The spectrum of antimicrobial activity of the T+Psl+ phenolics was broader and was proven to be effective against gram-positive and gram-negative bacteria (Fig. 7), the yeast Saccharomyces cerevisiae, and fungi such as Fusarium oxysporum, Botrytis cinerea, and Penicillium italicum (data not shown). A wide spectrum of activity is characteristic of the ISR afforded by rhizobacteria and fungi (41, 43). To our knowledge, although phytoalexin accumulation is a well-established mechanism in systemic acquired resistance (17, 19), the role of these compounds in ISR triggered by selected strains of PGPR and PGPF is still ambiguous. The enhanced antimicrobial activity of the T+Psl+ plants and the broad spectrum of that activity suggest that a prior application of T. asperellum may enhance the plant's capacity to defend itself against subsequent infection through the production of secondary metabolites with antimicrobial activity.
PAL has been shown to regulate the production of several secondary compounds, including phytoalexins (16). Steady-state mRNA levels of Pal1 have been shown to increase in plant tissues during resistance to pathogen infection (15, 44). Application of Trichoderma to the root system resulted in a significant elevation of Pal1 mRNA levels both locally and systemically (Fig. 5). Thus, Pal1 activation peaked at 48 h postelicitation with T. asperellum, coincident with the observed phytoalexin accumulation in the leaves (Fig. 5) (Table 1). A similar time course for Pal1 expression was observed by Martinez et al. (28), using active cellulase from T. longibrachiatum as an inducer. Daayf et al. (6) showed that the first 48 h postchallenge are the most important for synthesis of the phytoalexin para-coumaric acid methyl ester in Milsana-elicited cucumber plants. Ninety-six hours post-challenge inoculation with P. syringae pv. lachrymans, Pal1 expression reached its maximal level, which was not further elevated in response to preelicitation of the roots with Trichoderma. However, the increase in P. syringae pv. lachrymans-induced Pal1 expression and the related accumulation of phenolic compounds were not complemented by an elevated level of antimicrobial capacity. Such an increase in antimicrobial potential was recorded only in the preelicited and challenged plants (T+Psl+), suggesting the involvement of other enzymes along the biosynthetic pathway.
HPL and lox1 are defense enzymes belonging to a distinct pathogen-induced metabolic pathway. This pathway, also known as the lipoxygenase pathway, leads to the production of signaling molecules, induction of defense genes (2, 29), and phytoalexin accumulation (4, 50). To the best of our knowledge, this is the first report that T. asperellum treatment may induce HPL transcript accumulation in cucumber both locally and systemically. The increased expression of HPL in the Trichoderma-preelicited plants was further elevated by challenge inoculation with P. syringae pv. lachrymans. This suggests a role for HPL in the potentiation of the plant to respond more rapidly and effectively to a subsequent pathogenic attack. In addition, lox1, which produces the substrates for HPL and jasmonic acid, was also induced in cucumber 1 h after root elicitation with Trichoderma (data not shown). Jasmonic acid and other structural derivatives function as signals involved in plant defense to wounding and pathogens and have also been shown to induce phytoalexin and antimicrobial phenolic accumulation (46).
Indeed, an increase of more than 30% in total phenolic concentration was observed following T. asperellum elicitation (T+Psl−) relative to what was seen with control plants (T−Psl−) (Table 1). However, this increase did not account for an increase in antimicrobial activity in vitro. Nonetheless, challenge inoculation with P. syringae pv. lachrymans elevated the level of total phenolics for both treatments, T−Psl+ and T+Psl+, with no significant difference between them (Table 1). Thus, it is suggested that the prominent increase in antimicrobial activity of the T+Psl+ treatment is the result of an increase in specific phenolic compounds (Table 2), while the accumulation of other phenolic compounds is involved in the in vivo inhibition of the pathogen. The existence of different patterns of phenolic compound accumulation was supported here by the HPLC profile analysis (Fig. 8) (Table 2).
Further separation of the phenolic extract from the T+Psl+ plants resulted in an increase in the levels and antimicrobial activities of several compounds relative to the T−Psl+ treatment. Daayf et al. and Fawe et al. (5, 11) identified two phenolic compounds, rhamnetin and p-coumaric methyl ester, in cucumber leaves following elicitation with Milsana and subsequent challenge with powdery mildew. Using specific standards, we showed that none of these compounds were present in our system.
In conclusion, pretreatment of the roots with T. asperellum was shown to systemically inhibit proliferation of P. syringae pv. lachrymans in the leaves. This reduction appeared to be associated with transcript accumulation of biosynthetic defense-related genes and a prominent accumulation of phenolic compounds showing substantial antimicrobial activity. Future efforts should focus on characterizing the chemically complex nature of these compounds and the study of their production and accumulation in greenhouse and field experiments.
Acknowledgments
This research was partially supported by Binational Agricultural Research and Development Fund grant IS-2880-97.
We thank J. Kloepper for generously providing the strain of P. syringae pv. lachrymans used throughout this study.
REFERENCES
- 1.Ahmed, S. A., C. P. Sanchez, and M. E. Candela. 2000. Evaluation of induction of systemic resistance in pepper plants (Capsicum annum) to Phytophtora capsici using Trichoderma harzianum and its relation with capsidiol accumulation. Eur. J. Plant Pathol. 106:817-824. [Google Scholar]
- 2.Bate, N. J., and S. J. Rothstein. 1998. C6-volatiles derived from lipoxygenase pathway induce a subset of defense related genes. Plant J. 16:561-569. [DOI] [PubMed] [Google Scholar]
- 3.Benhamou, N., and M. Nicole. 1999. Cell biology of plant immunization against microbial infection: the potential of induced resistance in controlling plant diseases. Plant Physiol. Biochem. 37:703-719. [Google Scholar]
- 4.Croft, K. P. C., F. Juttner, and A. J. Slusarenko. 1993. Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv. phaseolicola. Plant Physiol. 101:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daayf, F., R. Bel-Rhlid, and R. R. Bélanger. 1997. Methyl ester of p-coumaric acid: a phytoalexin like compound from long English cucumber leaves. J. Chem. Ecol. 23:1517-1527. [Google Scholar]
- 6.Daayf, F., A. Schmitt, and R. R. Bélanger. 1997. Evidence of phytoalexins in cucumber leaves infected with powdery mildew following treatment with leaf extracts of Reynoutria sachalinensis. Plant Physiol. 113:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalisay, R. F., and J. A. Kuc'. 1995. Persistence of induced resistance and enhanced peroxidase and chitinase activities in cucumber plants. Physiol. Mol. Plant Pathol. 47:315-327. [Google Scholar]
- 8.Dalisay, R. F., and J. A. Kuc'. 1995. Persistence of reduced penetration by Colletotrichum lagenarium into cucumber leaves with induced systemic resistance and its relation to enhanced peroxidase and chitinase activities. Physiol. Mol. Plant Pathol. 47:329-338. [Google Scholar]
- 9.De Meyer, G., J. Bigirimana, Y. Elad, and M. Höfte. 1998. Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea. Eur. J. Plant Pathol. 104:279-286. [Google Scholar]
- 10.Elad, Y., I. Chet, and Y. Henis. 1982. Degradation of plant pathogenic fungi by Trichoderma harzianum. Can. J. Microbiol. 28:719-725. [Google Scholar]
- 11.Fawe, A., M. Abou-Zaid, J. G. Menzies, and R. R. Bélanger. 1998. Silicon-mediated accumulation of flavonoid phytoalexins in cucumber. Phytopathology 88:396-401. [DOI] [PubMed] [Google Scholar]
- 12.Fofana, B., D. J. McNally, C. Labbé, R. Boulanger, N. Benhamou, A. Seguin, and R. R. Bélanger. 2002. Milsana-induced resistance in powdery mildew-infected cucumber plants correlated with induction of chalcone synthase and chalcone isomerase. Physiol. Mol. Plant Pathol. 61:121-132. [Google Scholar]
- 13.Graham, T. L., J. E. Kim, and M. Y. Graham. 1990. Role of constitutive isoflavone conjugates in the accumulation of glyceollin in soybean infected with Phytophthora megasperma. Mol. Plant-Microbe Interact. 3:157-166. [Google Scholar]
- 14.Grayer, R. J., and J. B. Harborne. 1994. A survey of antifungal compounds from higher plants, 1982-1993. Phytochemistry 37:19-42. [DOI] [PubMed] [Google Scholar]
- 15.Guo, A., G. Salih, and D. F. Klessig. 2000. Activation of a diverse set of genes during the tobacco resistance response to TMV is independent of salicylic acid; induction of a subset is also ethylene independent. Plant J. 21:409-418. [DOI] [PubMed] [Google Scholar]
- 16.Hahlbrock, K., and D. Scheel. 1989. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40:347-369. [Google Scholar]
- 17.Hammerschmidt, R. 1999. Phytoalexins: what have we learned after 60 years? Annu. Rev. Phytopathol. 37:285-306. [DOI] [PubMed] [Google Scholar]
- 18.Hammerschmidt, R., and J. Kuc'. 1982. Lignification as a mechanism for induced systemic resistance in cucumber. Physiol. Plant Pathol. 20:61-71. [Google Scholar]
- 19.Hammerschmidt, R., and J. Smith Becker. 1997. Acquired resistance to disease in plants. Hortic. Rev. 18:247-280. [Google Scholar]
- 20.Howell, C. R., L. E. Hanson, R. D. Stipanovic, and L. S. Puckhaber. 2000. Induction of terpenoid synthesis in cotton roots and control of Rhizoctonia solani by seed treatment with Trichoderma virens. Phytopathology 90:248-252. [DOI] [PubMed] [Google Scholar]
- 21.Howell, C. R., and R. D. Stipanovic. 1995. Mechanisms in the biocontrol of Rhizoctonia solani-induced cotton seedling disease by Gliocladium virens: antibiosis. Phytopathology 85:469-472. [Google Scholar]
- 22.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of phycocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 23.Kloepper, J. W., S. Tuzun, and J. A. Kuc'. 1992. Proposed definitions related to induced resistance. Biocontrol Sci. Technol. 2:349-351. [Google Scholar]
- 24.Kuc', J. 1987. Plant immunization and its applicability for disease control, p. 255-274. In I. Chet (ed.), Innovative approaches to plant disease control. John Wiley & Sons, New York, N.Y.
- 25.Kullnig, C., T. Krupica, S. L. Woo, R. L. Mach, M. Rey, T. Benitez, M. Lorito, and C. P. Kubicek. 2001. Confusion abounds over identities of Trichoderma biocontrol isolates. Mycol. Res. 105:770-772. [Google Scholar]
- 26.Liu, L., J. W. Kloepper, and S. Tuzun. 1992. Induction of systemic resistance against cucumber mosaic virus by seed inoculation with selected rhizobacterial strains. Phytopathology 82:1109. [Google Scholar]
- 27.Liu, L., W. Kloepper, and S. Tuzun. 1995. Induction of systemic resistance in cucumber against Fusarium wilt by plant growth promoting rhizobacteria. Phytopathology 85:695-698. [Google Scholar]
- 28.Martinez, C., F. Blanc, E. Le Claire, O. Besnard, M. Nicole, and J. C. Baccou. 2001. Salicylic acid and ethylene pathways are differentially activated in melon cotyledons by active or heat-denatured cellulase from Trichoderma longibrachiatum. Plant Physiol. 127:334-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui, K., C. Ujita, S. Fujimoto, J. Wilkinson, V. Knauf, T. Kajiwara, and I. Feussner. 2000. Fatty acid 9- and 13-hydroperoxide lyases from cucumber. FEBS Lett. 481:183-188. [DOI] [PubMed] [Google Scholar]
- 30.Meera, M. S., M. B. Shivanna, K. Kageyama, and M. Hyakumachi. 1994. Plant growth promoting fungi from Zoysiagrass rhizosphere as potential inducers of systemic resistance in cucumbers. Phytopathology 84:1399-1406. [Google Scholar]
- 31.Meera, M. S., M. B. Shivanna, K. Kageyama, and M. Hyakumachi. 1995. Responses of cucumber to induction of systemic resistance against anthracnose by plant growth promoting fungi. Eur. J. Plant Pathol. 101:421-430. [Google Scholar]
- 32.Okon, Y., I. Chet, and Y. Henis. 1973. Effects of lactose, ethanol and cycloheximide on the translation pattern of radioactive compounds and on Sclerotium rolfsii. Gen. Microbiol. 74:251-258. [Google Scholar]
- 33.Ongena, M., F. Daayf, P. Jacques, P. Thonart, N. Benhamou, T. C. Paulitz, and R. R. Bélanger. 2000. Systemic induction of phytoalexins in cucumber in response to treatments with fluorescent pseudomonas. Plant Pathol. 49:523-530. [Google Scholar]
- 34.Ongena, M., F. Daayf, P. Jacques, P. Thonart, N. Benhamou, T. C. Paulitz, P. Cornelis, N. Koedam, and R. R. Bélanger. 1999. Protection of cucumber against Pythium root rot by fluorescent pseudomonas: predominant role of induced resistance over siderophores and antibiosis. Plant Pathol. 48:66-76. [Google Scholar]
- 35.Pieterse, C. M. J., S. C. M. van Wees, E. Hoffland, J. A. van Pelt, and L. C. van Loon. 1996. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8:1225-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pieterse, C. M. J., S. C. M. van Wees, J. A. van Pelt, A. Trijssenaar, Y. A. M. Van't Westende, E. M. Bolink, and L. C. van Loon. 1996. Systemic resistance in Arabidopsis thaliana induced by biocontrol bacteria. Meded. Fac. Landbouwkd. Toegep. Biol. Wet. Univ. Gent 61:209-220. [Google Scholar]
- 37.Sakakibara, H., Y. Honda, S. Nakagawa, H. Ashida, and K. Kanazawa. 2003. Simultaneous determination of all polyphenols in vegetables, fruits, and teas. J. Agric. Food Chem. 51:571-581. [DOI] [PubMed] [Google Scholar]
- 38.Scalet, M., E. Crivaletto, and F. Mallardi. 1989. Demonstration of phenolic compounds in plant tissue by an osmium-iodide post fixation procedure. Stain Technol. 64:273-290. [DOI] [PubMed] [Google Scholar]
- 39.Siegrist, J., W. Jeblick, and H. Kauss. 1994. Defense responses in infected and elicited cucumber (Cucumis sativus L.) hypocotyl segments exhibiting acquired resistance. Plant Physiol. 105:1365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stange, R., Jr., J. J. Sims, S. L. Midland, and R. E. McDonald. 1999. Isolation of a phytoalexin, trans-p-coumarylaldehyde, from Cucurbita maxima, Cucurbitacea. Phytochemistry 52:41-43. [Google Scholar]
- 41.Tuzun, S., and E. Bent. 1999. The role of hydrolytic enzymes in multigenic and microbially induced resistance in plants, p. 95-116. In A. A. Agrawal, S. Tuzun, and E. Bent (ed.), Induced plant defenses against pathogens and herbivores: biochemistry, ecology, and agriculture. APS Press, St. Paul, Minn.
- 42.Tuzun, S., and J. W. Kloepper. 1994. Induced systemic resistance by plant growth promoting rhizobacteria, p. 104-109. In M. H. Ryder, P. M. Stephens, and G. D. Bowen (ed.) Improving plant productivity with rhizosphere bacteria. CSIRO, Adelaide, South Australia, Australia.
- 43.van Loon, L. C., P. A. H. M. Bakker, and C. M. J. Pieterse. 1998. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36:453-483. [DOI] [PubMed] [Google Scholar]
- 44.van Wees, S. C., M. Luijendijk, I. Smoorenburg, L. C. van Loon, and C. M. Pieterse. 1999. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41:537-549. [DOI] [PubMed] [Google Scholar]
- 45.van Wees, S. C., C. M. Pieterse, A. Trijssenaar, Y. A. van't Westende, F. Hartog, and L. C. van Loon. 1997. Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol. Plant-Microbe. Interact. 10:716-724. [DOI] [PubMed] [Google Scholar]
- 46.Wasternack, C., and B. Parthier. 1997. Jasmonate-signal plant gene expression. Trends Plant Sci. 2:302-307. [Google Scholar]
- 47.Wei, G., J. W. Kloepper, and S. Tuzun. 1992. Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth promoting rhizobacteria. Phytopathology 81:1508-1512. [Google Scholar]
- 48.Yedidia, I., N. Benhamou, and I. Chet. 1999. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yedidia, I., N. Benhamou, Y. Kapulnik, and I. Chet. 2000. Induction and accumulation of pathogenesis related protein activity during the early stages of root colonization by Trichoderma harzianum. Plant Physiol. Biochem. 38:863-873. [Google Scholar]
- 50.Zeringue, H. J. J. 1992. Effects of C6-C10 alkenals and alkanals on eliciting a defense response in the developing cotton ball. Phytochemistry 31:2305-2308. [Google Scholar]