Abstract
The relationship between Bifidobacterium lactis and Bifidobacterium animalis was examined by comparative analysis of tuf and recA gene sequences and by restriction fragment length polymorphism analysis of their internal 16S-23S transcribed spacer region sequences. The bifidobacterial strains investigated could be divided into two distinct groups within a single species based on the tuf, recA, and 16S-23S spacer region sequence analysis. Therefore, all strains of B. lactis and B. animalis could be unified as the species B. animalis and divided into two subspecies, Bifidobacterium animalis subsp. lactis and Bifidobacterium animalis subsp. animalis.
Bifidobacterium lactis and Bifidobacterium animalis are closely related, representing a relatively small number of known strains. B. lactis includes strains employed as probiotics for dairy products and infant formula. Despite their high industrial importance, the taxonomy of B. lactis and B. animalis is still unclear. Historically, the species annotation of B. animalis (12, 15) was introduced first, followed by the isolation of a highly oxygen-tolerant Bifidobacterium organism from yogurt, which was classified in 1997 as the new species B. lactis (11). The definition criteria for B. lactis and B. animalis were based on phenotypic characteristics such as carbohydrate fermentation patterns and high tolerance of oxygen stress. However, due to the high levels of DNA relatedness and 16S ribosomal DNA (rDNA) sequence similarity between B. lactis and B. animalis, Cai et al. (1). proposed the rejection of the name B. lactis and suggested that B. lactis should be considered a junior subjective synonym of B. animalis. The nomenclature of the species B. lactis and B. animalis is still ambiguous, and a taxonomic decision by the International Committee on Systematic Bacteriology to maintain them as two separate species has been pending since 1999 (6).
Recently, we have demonstrated that analysis of 16S rDNA sequences, analysis of the 16S-23S spacer region, and enterobacterial repetitive intergenic consensus-PCR have powerful potential for tracing B. animalis or B. lactis (20-23). The tuf and recA genes have been proposed as useful markers in inferring bacterial phylogeny (2, 10, 18) and have recently been successfully used to differentiate species and subspecies within various bacterial genera (3, 7, 8, 17). Our results show that the sequence analysis of tuf and recA as well as restriction fragment length polymorphism (RFLP) analysis of the internal transcribed spacer (ITS) sequences are relatively simple and rapid methods by which B. lactis and B. animalis can be identified without resorting to the use of species-specific PCR primer sets.
tuf and recA sequence analysis.
Bacterial strains used in this study were either obtained from culture collections or isolated from human or animal fecal samples (Table 1) and grown as described previously (19). The DNA of all bifidobacterial strains was prepared as described by Ventura and Zink (19). The 940-bp tuf fragment sequences and the 690-bp recA sequences were amplified with the oligonucleotides tuf1 (5′-GAGTACGACTTCAACCAG-3′) (22) and tuf2 (5′-CAGGCGAGGATCTTGGT-3′) (22) and the oligonucleotides rec1 (5′-TCGAGGTGATTCCCACC-3′) and rec2 (5′-GAACCAAGAACCGGACTTC-3′), respectively. Each PCR mixture (50 μl) contained a reaction cocktail of 20 mM Tris-HCl at pH 8.0, 50 mM KCl, a 200 μM concentration of each deoxynucleoside triphosphate, 50 pmol of each primer, 1.5 mM MgCl2, and 1 U of Taq DNA polymerase (Gibco BRL, Paisley, United Kingdom). Each PCR cycling profile consisted of an initial denaturation step of 3 min at 95°C, followed by amplification for 30 cycles as follows: denaturation (30 s at 95°C), annealing (30 s at 52°C), and extension (2 min at 72°C). The PCR was completed with an elongation phase (10 min at 72°C). PCR fragments were purified with a PCR purification kit (Qiagen, West Sussex, United Kingdom) and were subsequently cloned in the pGEM-T Easy plasmid vector (Promega, Southampton, United Kingdom) following the supplier's instructions. Subsequently, the sequence of the inserted DNA fragment was determined by sequencing three randomly selected clones on both strands for each bacterial species to ensure that no sequencing errors were attributable to misincorporation by the Taq polymerase.
TABLE 1.
Bacterial strains used in this study
| Species | Straina | Origin |
|---|---|---|
| B. lactis | DSM 10140T | Yogurt |
| NCC 363 | Human feces | |
| NCC 311 | Human feces | |
| NCC 239 | Human feces | |
| NCC 383 | Yogurt | |
| NCC 402 | Yogurt | |
| B. animalis | ATCC 25527T | Rat feces |
| ATCC 27672 | Rat feces | |
| ATCC 27673 | Sewage | |
| ATCC 27674 | Rabbit feces | |
| ATCC 27536 | Chicken feces |
T, type strain. NCC strains were isolated at our institute (Nestlè Culture Collection). All other strains were obtained from the American Type Culture Collection.
Nucleotide sequencing of both strands from cloned DNA was performed with a fluorescence-labeled primer cycle sequencing kit (Amersham Buchler, Braunschweig, Germany) following supplier's instructions. The primers used were tuf1, tuf2, rec1, and rec2 labeled with IRD800 (MWG Biotech, Germany). The tuf and recA sequences of all Bifidobacterium strains determined here as well as those available in the GenBank database were used for comparison. The partial nucleotide sequences of the tuf and recA genes from 11 Bifidobacterium strains belonging to B. lactis and B. animalis were determined, and phylogenetic trees based on these data as well as those retrieved from GenBank databases were constructed. Phylogenetic trees were constructed with the programs Clustal X, DNAML (maximum likelihood), and DNAPARS (parsimony) (PHYLIP [Phylogeny Inference Package], version 3.5c; J. Felsenstein, University of Washington, Seattle, Wash.).
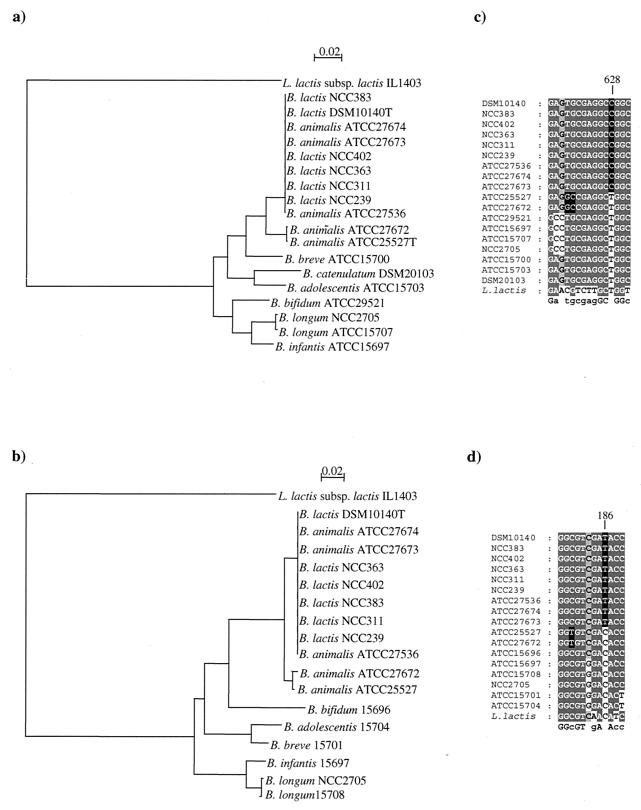
The topologies of the recA- and tuf-based trees were comparable (Fig. 1). In these trees, B. lactis and B. animalis strains were grouped into two clusters; the first one contained nine strains, including all B. lactis strains as well as B. animalis ATCC 27673, ATCC 27674, and ATCC 27536, while the second one contained only the type strain of B. animalis and B. animalis ATCC 27672. The phylogenetic distances among strains of the B. lactis are virtually zero (Fig. 1). The individuality of each of these strains was supported by various molecular typing tools, such as pulsed-field gel electrophoresis, random amplified polymorphic DNA PCR, and triplicate arbitrarily primed PCR (19; data not shown).
FIG. 1.
(a and b) Phylogenetic trees obtained with tuf gene sequences (a) and recA gene sequences (b). The scale indicates a genetic distances of 2%. (c and d) Alignment showing the sequence signatures for the tuf gene sequences (c) and recA gene sequences (d). Sequence signatures are shaded in black; residues shaded in light grey have ≤75% identity, while residues in dark grey have ≥76% identity.
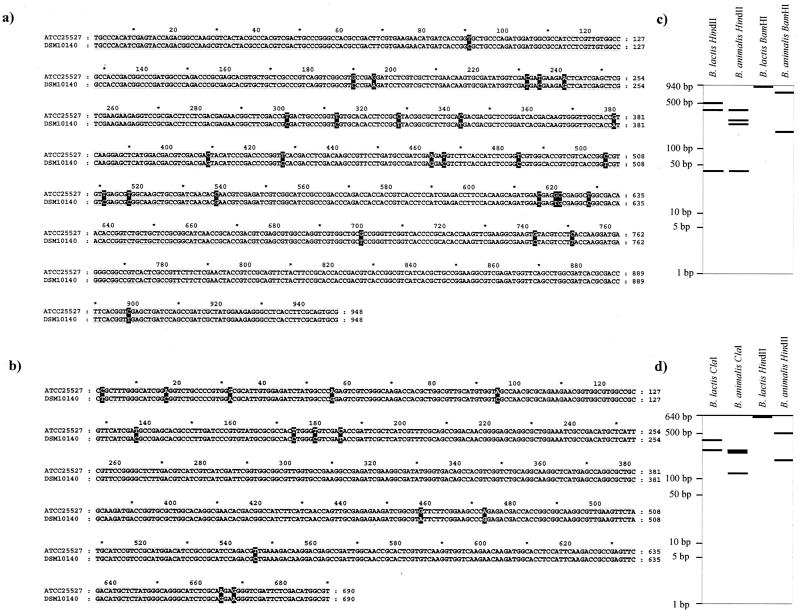
Twenty-seven nucleotide substitutions were observed between the tuf gene sequences of B. lactis DSM 10140 and B. animalis ATCC 25527, but only four contributed to an amino acid substitution. In parallel, only 13 synonymous nucleotide substitutions were noticed among the recA gene sequences in the same set of strains. Interestingly, many of the base differences observed between the tuf and recA sequences of the two taxa are guanine or cytosine in B. lactis and adenine and thymine in B. animalis. The spontaneous deamination of cytosine leading to a thymine is a frequent event, and it has already been described in relation to the species identification of Lactobacillus delbrueckii (4). Because of the similar G+C content of the organisms under consideration, sequence differences are likely due to real evolutionary divergence. Therefore, we focused our interest on the tuf and recA nucleotide sequences. When we aligned the tuf and the recA sequences of all B. lactis and B. animalis strains investigated, we noticed that these 27- and 13-nucleotide differences could be directly used to distinguish all B. lactis from B. animalis strains (Fig. 2). Some of these sequence signatures are typical of either B. lactis strains or B. animalis strains but are not exhibited in all other microorganism investigated here (Fig. 1b and d). These sequence signatures could be directly used for designing PCR-specific primers, or they could be a target for specific restriction enzymes, providing species-specific RFLP patterns. In fact, theoretical restriction profiles with different restriction enzymes were obtained from the tuf and recA sequences. The restriction enzymes HindII, BamHI, and ClaI were found to give the clearest and most reliable distinction in theoretical RFLP patterns in order to differentiate B. lactis and B. animalis strains (Fig. 2c and d).
FIG. 2.
(a and b) Pairwise alignment of the tuf sequences (a) and recA sequences (b) of B. lactis DSM 10140T and B. animalis ATCC 25527T. Nucleotides which are different between B. lactis strains and B. animalis ATCC 25527 are shaded in black. (c and d) Theoretical RFLP profiles from B. lactis-B. animalis tuf sequences (c) and from B. lactis-B. animalis recA sequences (d).
The synonymous distances calculated by the method of Nei et al. (13) from the nucleotide substitution ratios at synonymous positions in tuf and recA were examined for all possible combinations of these 22 Bifidobacterium gene sequences. A significant correlation between the synonymous distances in the tuf genes and those in the recA genes were observed, with a correlation coefficient of 0.99. This result was not unexpected, because it has been demonstrated before that a synonymous substitution rate is constant for many chromosomal genes in various organisms and can thus serve indeed as a molecular clock of their evolution (9). The evolutionary distances calculated for tuf, recA, ITS, and 16S rDNA sequences for strains of B. lactis and B. animalis (Table 2) clearly underline the inadequacy of the 16S rDNA sequences for distinguishing these closely related taxa. From these data, it appears that the Knuc values (average extent of sequence change at any position in two homologous sequences [5]) calculated for 16S rDNA sequences are at least threefold lower than those calculated for the other taxonomic molecular markers (tuf and recA gene sequences) and sevenfold lower than those of the ITS. Hence, tuf, recA, and ITS sequences provide different complementary phylogenetic information. While tuf and recA sequences are excellent tools for inferring interspecific relationships, 16S-23S rDNA spacer sequence comparisons provide information concerning intraspecific links (e.g., characterization of strains).
TABLE 2.
Knuc values for tuf, recA, ITS, and 16S rDNA sequences of B. lactis DSM 10140 with respect to B. lactis and B. animalis strains
PCR-RFLP analysis of the 16S-23S ITS region.
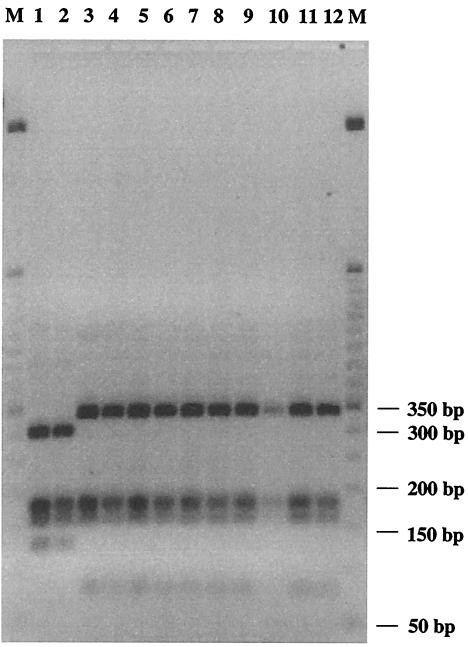
Theoretical restriction profiles for the ITS region of B. lactis and B. animalis with different enzymes were obtained by using the Webcutter online analysis tool (http://users.unimi.it/∼camelot/tools/cut2.html). The enzyme Sau3AI gave the clearest and most reliable theoretical PCR-RFLP pattern, in order to differentiate between the B. animalis and B. lactis type strains. PCR was used to amplify the 16S-23S ITS region of all B. lactis and B. animalis strains with the primers 16S-for (5′-GCTAGTAATCGCGGATCAG-3′) (19) and 23Si (5′-CATTCGGACACCCTGGGATC-3′) (19). Before a restriction digestion of all PCR products was done, amplicons were purified with the QIAquick PCR purification kit (Qiagen, Valencia, Calif.). In order to achieve complete digestion, reactions were carried out for 3 h at 37°C in a 10-μl volume of incubation buffer containing 2 U of Sau3AI (Boehringer-Mannheim, Mannheim, Germany) and 1 μg of the purified PCR product. The restriction products were loaded on 3% (wt/vol) agarose gel (NuSieve; FMC Bioproducts, Rockland, Maine) and separated at 7 V/cm, followed by ethidium bromide staining (UV at 260 nm). PCR-amplified 16S-23S rDNA ITS sequences of all B. lactis and B. animalis strains were digested with Sau3AI, leading to polymorphic patterns and hence allowing direct species-level identification (Fig. 3). The clustering from these patterns consistently grouped all restriction fragment patterns into two distinct subsets. One shared the restriction pattern of the type strain of B. lactis and includes B. lactis DSM 10140, B. animalis ATCC 27536, B. animalis ATCC 27673, B. animalis ATCC 27674, and all other B. lactis isolates, and the other includes the type strain of B. animalis, ATCC 25527, and B. animalis ATCC 27672. In all experiments, we obtained clearly distinguishable and always reproducible RFLP patterns of the 16S-23S ITS region, confirming all theoretically expected restriction profiles.
FIG. 3.
Restriction patterns of PCR-amplified fragments of 16S-23S rDNA digested with Sau3AI. Lane 1, B. animalis ATCC 25527; lane 2, B. animalis ATCC 27672; lane 3, B. lactis DSM 10140; lane 4, B. animalis ATCC 27674; lane 5, B. animalis ATCC 27673; lane 6, B. animalis ATCC 27536; lane 7, B. lactis NCC 363; lane 8, B. lactis NCC 311; lane 9, B. lactis NCC 402; lane 10, B. lactis NCC 311; lane 11, B. lactis NCC 383; lane 12, B. lactis NCC 239; lanes M, 50-bp DNA ladder (Gibco BRL).
Conclusions.
Our study shows that comparison of tuf and recA sequences as well as RFLP analysis of the ITS sequences of B. lactis and B. animalis strains provides additional and much further detailed practical means to discriminate these closely related taxa. The analysis of tuf and recA gene sequences and classical molecular species identification tools (rDNA sequencing and species-specific oligonucleotide probes) data should therefore be incorporated in a modern polyphasic approach for bifidobacterial taxonomy. Our results suggest that tuf and recA gene sequencing and RFLP analysis of the ITS sequences have specific advantages over other molecular tools for tracing B. lactis-B. animalis strains (19). In fact, tuf and recA are relatively short sequences that can easily be sequenced on both polynucleotide strands. In addition, it has been shown that bifidobacterial genomes carry only one tuf and one recA gene (16), which might be advantageous because the interpretation and power of rRNA-based data (e.g., the use of a single gene or operon) in molecular taxonomy appear sometimes to be questionable (24). Finally, the application of tuf and recA gene sequencing and RFLP analysis of the 16S-23S spacer sequences for our species-specific identification allows simultaneous handling and comparison of many isolates, in contrast to a required repeated PCR amplification with species-specific primers. Establishing PCR primers based on tuf and recA sequences for quantitative detection of B. lactis and B. animalis is a task for the future, and research into real-time quantitative detection avoiding the use of multicopy genes (e.g., ribosomal genes) is rapidly proceeding (14).
In the present study, genotypic analysis carried out on B. lactis and B. animalis strains targeting the tuf and recA genes showed a consistent similarity between these two taxa. However, taxonomic trees based on tuf and recA indicate a separate branching of B. lactis and B. animalis strains. Recently, a reorganization of the B. animalis species with the rejection of few strains (ATCC 27673, ATCC 27674, and ATCC 27536) from this species and a reclassification of them as B. lactis have been proposed (19). This hypothesis is also validated by the tuf and recA gene analyses, which suggested a revision of various strains, actually designed to the B. animalis species. This study provides a clear image of the genetic variability within the B. lactis-B. animalis taxa. In fact, the use of genes encoding proteins (e.g., tuf and recA) instead of ribosomal genes (19) shows that the taxon B. lactis is in fact highly homogeneous. In contrast, the analysis of the tuf gene in all investigated B. animalis strains depicted a significant variability. This might be due to the fact that the taxon B. animalis-B. lactis was originally a single group that has diverged only recently as a result of different environmental conditions (e.g., growth in different ecological niches). Therefore, based on the distinct phenotypic characteristics (12, 19) of all B. lactis strains and further detailed molecular evidence (tuf and recA sequences as well as their ITS restriction patterns), we proposed that B. lactis and B. animalis be unified into one species and that this species should be divided into two subspecies, Bifidobacterium animalis subsp. animalis and Bifidobacterium animalis subsp. lactis.
Nucleotide sequence accession numbers.
The accession numbers for the tuf and recA sequences are AY370912 to AY370929 and AY372028 to AY372031.
REFERENCES
- 1.Cai, Y., M. Matsumoto, and Y. Benno. 2000. Bifidobacterium lactis Meile et al. 1997 is a subjective synonym of Bifidobacterium animalis (Mitsuoka 1969) Scardovi and Trovatelli 1974. Microbiol. Immunol. 44:815-820. [DOI] [PubMed] [Google Scholar]
- 2.Chavagnat, F., M. Haueter, J. Jimeno, and M. G. Casey. 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol. Lett. 217:177-183. [DOI] [PubMed] [Google Scholar]
- 3.Felis, G. E., F. Dellaglio, L. Mizzi, and S. Torriani. 2001. Comparative sequence analysis of recA gene fragment brings new evidence for a change in the taxonomy of the Lactobacillus casei group. Int. J. Syst. Evol. Microbiol. 51:2113-2117. [DOI] [PubMed] [Google Scholar]
- 4.Germond, J. E., L. Lapierre, M. Delley, B. Mollet, G. E. Felis, and F. Dellaglio. 2003. Evolution of the bacterial species Lactobacillus delbrueckii: a partial genomic study with reflections on the prokaryotic species concept. Mol. Biol. Evol. 20:93-104. [DOI] [PubMed] [Google Scholar]
- 5.Hori, H., and S. Osawa. 1979. Evolutionary changes in 5S RNA secondary structure and phylogenetic tree of 54S RNA species. Proc. Natl. Acad. Sci. USA 76:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Committee on Systematic Bacteriology. 2001. Minutes of the meetings, 22 and 23 September 1999, Veldhoven, The Netherlands. Int. J. Syst. Evol. Microbiol. 51:259-261. [Google Scholar]
- 7.Ke, D., F. J. Picard, F. Martineau, C. Menard, P. H Roy, M. Ouellette, and M. G. Bergeron. 1999. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 37:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kullen, M., J., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence, J. G., D. L. Hartl, and H. Ochaman. 1991. Molecular considerations in the evolution of bacterial genes. J. Mol. Evol. 33:241-250. [DOI] [PubMed] [Google Scholar]
- 10.Ludwig, W., J. Neumaier, N. Klugbauer, E. Brockmann, C. Roller, S. Jilg, K. Reetz, I. Schachtner, A. Ludvigsen, M. Bachleitner, U. Fischer, and K. H. Schleifer. 1993. Phylogenetic relationships of bacteria based on comparative sequence analysis of elongation factor Tu and ATP-synthase beta subunit genes. Antonie Leeuwenhoek 64:285-305. [DOI] [PubMed] [Google Scholar]
- 11.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Senger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:57-64. [Google Scholar]
- 12.Mitsuoka, T. 1969. Comparative studies on bifidobacteria isolated from the alimentary tract of man and animals (including descriptions of Bifidobacterium thermophilum nov. spec. and Bifidobacterium pseudolongum nov. spec.). Zentralbl. Bakteriol. 210:52-64. [PubMed] [Google Scholar]
- 13.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 14.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scardovi, V., and L. D. Trovatelli. 1974. Bifidobacterium animalis (Mitsuoka) comb. nov. and the “minimum” and “subtile” groups of new bifidobacteria found in sewage. Int. J. Syst. Bacteriol. 24:21-28. [Google Scholar]
- 16.Shell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, N. H., E. C. Holmes, G. M. Donovan, G. A. Carpenter, and B. G. Spratt. 1999. Networks and groups within the genus Neisseria: analysis of argF, recA, rho, and 16S rRNA sequences from human Neisseria species. Mol. Biol. Evol. 16:773-783. [DOI] [PubMed] [Google Scholar]
- 18.Torriani, S., G. E. Felis, and F. Dellaglio. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67:3450-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 21.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ventura, M., V. Meylan, and R. Zink. 2003. Identification and tracing of Bifidobacterium species by enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura, M. C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization and loci of the tuf genes in Lactobacillus and Bifidobacterium and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed]
- 24.Waterhouse, R. N., and L. A. Glover. 1993. Differences in the hybridization pattern of Bacillus subtilis genes coding for rRNA depend on the method of DNA preparation. Appl. Environ. Microbiol. 59:919-921. [DOI] [PMC free article] [PubMed] [Google Scholar]