Abstract
We developed a reliable and flexible green fluorescent protein (GFP)-based system for measuring gene expression in individual bacterial cells. Until now, most systems have relied upon plasmid-borne gfp gene fusions, risking problems associated with plasmid instability. We show that a recently developed GFP variant, GFP+, is suitable for assessing bacterial gene expression. Various gfp+ transcriptional fusions were constructed and integrated as single copies into the chromosome of Salmonella enterica serovar Typhimurium. A comparison of the expression levels of proU-lacZ and proU-gfp+ fusions showed that GFP+ reported proU activity in individual Salmonella cells as accurately as β-galactosidase reported activity for entire populations. The single-copy gfp+ fusions were ideal for monitoring up- and downregulation of Salmonella virulence genes. We discovered that in vitro induction of the SPI1gene prgH occurs only in a portion of the population and that the proportion varies with the growth phase. We determined the level of expression of the SPI2 gene ssaG in bacteria released from murine macrophages. Our results demonstrate for the first time that single-copy GFP+ fusions reliably report gene expression in simple and complex environments. This approach promises to allow accurate measurement of gene expression in individual bacteria during animal infection.
The increasing incidence of infectious diseases is driving renewed efforts to understand the interactions between bacterial pathogens and their hosts at a new level. This has prompted research to study the transcriptional response of bacteria during infection. The past decade has seen the development of a number of techniques for identifying in vivo-induced bacterial genes and for monitoring gene induction in complex environments. Genetic tools for visualization of gene expression in situ have also been devised. Fluorescent reporters have been developed for studying bacterial infection from both the host point of view and the bacterial point of view (44, 57, 60). The green fluorescent protein (GFP) from Aequoria victoria is an excellent tool for monitoring gene expression, because it is naturally fluorescent without any exogenous cofactor or substrate (9). Variants of GFP that have different spectral characteristics, folding properties, levels of chromophore brightness, or half-lives have broadened the spectrum of possible applications for GFP (4). Measurement of fluorescence by flow cytometry has revolutionized the use of fluorescent reporter genes in bacteria (27) and now allows observation of gene expression at the level of individual bacterial cells. Plasmid-borne gfp fusions have already been used to monitor gene expression in Salmonella strains (6, 61), as well as in other gram-negative or gram-positive bacteria, including Escherichia coli (42), Serratia liquefaciens (2), Erwinia herbicola (37), Staphylococcus aureus (45), Listeria monocytogenes (64), and Streptococcus gordonii (24). Such fusions have proven to be useful for identifying bacterial genes induced during infection of eukaryotic cells or animal hosts and have allowed some assessment of bacterial gene expression levels in such complex environments. However, the variation in plasmid copy number that can occur in individual bacterial cells makes plasmid-borne gene fusions unsuitable for measuring differences in gene expression between single bacteria (34, 35). Furthermore, plasmid-borne GFP fusions are useful only for reporting the induction of genes; any reduction in fluorescence may simply reflect plasmid loss, because it is not possible to guarantee that plasmids are maintained in every bacterial cell in plant or animal hosts. Indeed, high expression levels lead to accumulation of toxic levels of GFP in bacterial cells, which results in dramatic plasmid loss (63; I. Hautefort, J. M. Sidebotham, and J. C. D. Hinton, unpublished data). Hopkins et al. showed that a pBR322-derived plasmid-borne gfp fusion was unstable in wild-type Salmonella enterica serovar Typhimurium strain 14028 during infection of BALB/c mice, while it remained stable in a PhoPc mutant (29). More recently, it was reported that a plasmid-borne tac-gfp fusion was not stable in a wild-type Salmonella strain during murine infection (6). This study showed that the level of plasmid loss was related to promoter strength, meaning that the levels of expression of different promoters cannot be compared by using plasmid-borne gene fusions.
Our interest in determining the diversity of gene expression within bacterial populations in the host prompted us to develop an improved approach for construction of gfp transcriptional fusions that could be used reliably during infection. We reasoned that integration of single-copy gene fusions onto the bacterial chromosome was the best way to ensure genetic stability. Single-copy gfp gene fusions have been used to monitor the activity of the marRAB promoter in large populations of Salmonella serovar Typhimurium (46). This approach involves inactivation of the gene of interest, making it unsuitable for studying virulence genes during infection. Recently, the activity of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac hybrid promoter (65) was successfully monitored by using a single copy of gfpmut2 as a reporter in Bacillus subtilis (41). We performed preliminary experiments involving expression of single-copy gfpmut1 fusions in Salmonella; and we discovered that the GFPmut1 variant, which is even brighter than GFPmut2 (11), was only fluorescent enough to report particularly high levels of promoter activity in Salmonella during infection (Hautefort et al., unpublished). We overcame this problem by using the GFP+ variant (51), which carries the GFPuv mutations F99S, M153T, and V163A (12) along with the EGFP mutations F64L and S65T (11), resulting in better folding of the protein coupled with enhanced brightness. In this study we describe the use of GFP+-based single-copy gene fusions to measure Salmonella serovar Typhimurium gene induction in vitro and during infection of mammalian cells.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains used in this study are listed in Table 1. The Escherichia coli DH5α strain was used for gene cloning. All Salmonella serovar Typhimurium strains used in this study were derived from wild-type strain LT2 or SL1344.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 22 |
| S. enterica serovar Typhimurium strains | ||
| LT2 | Wild-type LT2A | 1 |
| SL1344 | Wild type | 28a |
| CH946 | LT2, proU1702::Mud1-8, Amprb | 7 |
| JH3008 | SL1344, promoterless gfp+, Cmrc | This study |
| JH3009 | SL1344, φ(ssaG′-gfp+)1, Cmrc | This study |
| JH3010 | SL1344, φ(prgH′-gfp+)1, Cmrc | This study |
| JH3016 | SL1344, φ(rpsM′-gfp+)1, Cmrc | This study |
| JH3017 | LT2, φ(proU′-gfp+)1, Cmrb | This study |
| JH3049 | LT2, promoterless gfp+, Cmrc | This study |
| Plasmids | ||
| pWH1012gfp+ | pBR322 derivative | 51 |
| pKD46 | pBAD18 derivative | 13 |
| pKD4 | pANTSγ derivative (Kmr) | 25 |
| pFPV25 | pBR322 derivative, ColE1 replicon | 61 |
| pFPV25.1 | pFPV25, φrpsM′-gfpmut3 | 61 |
| pZEP01 | pBR322 derivative (mob+/bla+), ColE1 replicon | This study |
| pZEP02 | φrpsM′-gfp+, pZEP01 derivative | This study |
| pZEP06 | pZEP01 derivative (Cmr) | This study |
| pZEP07 | pZEP06 derivative, t0T1 (Cmr) | This study |
| pZEP08 | pZEP07 derivative, (Cmr Kmr) | This study |
| pZEP09 | φssaG′-gfp+, pZEP08 derivative (Cmr Kms) | This study |
| pZEP10 | φprgH′-gfp+, pZEP08 derivative (Cmr Kms) | This study |
| pZEP16 | φrpsM′-gfp+, pZEP08 derivative (Cmr Kms) | This study |
Salmonella strain SL1344 was derived from strain 4/74, which was isolated from a calf bowel.
proU::Mud1-8 and the proU-gfp+ fusion are both inserted at position 2,956,849 in the LT2 genome (GenBank accession no. AE006468).
The gfp gene fusions are inserted at the putPA locus at positions 1,210,040 to 1,211,657 in the LT2 genome. φ indicates a transcriptional gene fusion.
E. coli and Salmonella strains were grown in Luria-Bertani (LB) medium (48) at 37°C unless stated otherwise. Cultures were shaken at 250 rpm. Chloramphenicol, ampicillin, and kanamycin were used at final concentrations of 12, 100, and 50 μg ml−1, respectively. For in vitro induction of proU expression, Salmonella serovar Typhimurium strains CH946, JH3017, and JH3049 were grown at 30°C in LB medium which lacked NaCl (LO medium). This medium was supplemented with 10 mM glucose (7) and sometimes with 0.06, 0.16, or 0.3 M NaCl. In vitro induction of the ssaG-gfp+ fusion involved growth of Salmonella serovar Typhimurium strains JH3008 and JH3009 in minimum medium at pH 5.8 (MM5.8) (30). MM5.8 is a low-salt, acidic medium based on medium N (40) and contains 38 mM glycerol, 0.1% Casamino Acids, and 100 mM BisTris/Tris-HCl (catalog no. B-6032; Sigma, St. Louis, Mo.) (pH 5.8). Induction of prgH-gfp+ in Salmonella serovar Typhimurium strain JH3010 was achieved by static growth in either LO medium or LO medium containing 0.3 M NaCl in 30-ml bottles filled to the top with medium to generate microaerophilic conditions.
Recombinant DNA techniques.
Plasmid and chromosomal DNA purification was performed by using protocols recommended by the suppliers (Sigma; Qiagen, Hilden, Germany). DNA was digested with restriction endonucleases or ligated with T4 DNA ligase under standard conditions recommended by the manufacturers (New England Biolabs, Beverly, Mass.; Roche, Basel, Switzerland; Promega, Madison, Wis.). Preparation of electrocompetent E. coli and Salmonella serovar Typhimurium cells and DNA transformation were performed as previously described (15).
Oligonucleotides and PCR.
All of the oligonucleotides used in this study are listed in Table 2 and were purchased from Sigma Genosys and MWG AG Biotech (Ebersberg, Germany). PCR amplification was performed in 96-well microtiter plates (MWG AG Biotech) by using a Primus HT thermocycler according to the recommendations of the manufacturer (MWG AG Biotech). For preparative PCR amplification, the PfuTurbo (Stratagene, La Jolla, Calif.) and BioXAct (Bioline, Canton, Mass.) proofreading polymerases were used. PCR products were gel purified by using a Qiagen gel purification kit. PCR screening reactions were performed by using HotStarTaq polymerase (Qiagen).
TABLE 2.
Oligonucleotides used
| Oligonucleotide | Sequencea |
|---|---|
| pFPV_F1 | 5′-GAATTCGAGCTCGGTACCCGG-3′ |
| pFPV_R1 | 5′-CGTATGTAGCATCACCTTCACC-3′ |
| GFP_F1 | 5′-GGTGAAGGTGATGCTACATACG-3′ |
| GFP_R1 | 5′-ATGCGATATCGCCACCTGACGTCTAAGAAACC-3′ |
| Cam_F3 | 5′-GCATGATATCCGTCATTTCTGCCATTCATCC-3′ |
| Cam_R3 | 5′-GCATGATATCGGGCTAGCCGGCCCGACGC-3′ |
| t0T1_F4 | 5′-ATGGGGTACCGGATCCGTCGACCTGCAGCC-3′ |
| t0T1_R4 | 5′-GCTCTAGAATAAGAATGCGGCCGCTCCCCGGGGGACCGAAACGCGCGAGGCAGC-3′ |
| Kan_F1 | 5′-CATGCGACGCTAGCAGCCCGGGCTGCGGCCGCACCAAGCGAACCGGAATTGCCAGC-3′ |
| Kan_R1 | 5′-GTCGCCATTCTAGAACGCTCAGAAGAACTCGTCAAGAAGG-3′ |
| PssaG_F2 | 5′-ACGTCCCGGGCGATTGCTAAAGCCGTCTCC-3′ |
| PssaG_R2 | 5′-CGATTCTAGACCATGTGGGAGAGCATATCC-3′ |
| PprgH_F1 | 5′-ACGTCCCGGGGATGACTATTACTTACAAAGG-3′ |
| PprgH_R1 | 5′-CGATTCTAGACGAACTATGTATGGCCCTGG-3′ |
| PrpsM_F3 | 5′-CATGCGACCCGGGGAAAGGCTACGGCCGTTAAT-3′ |
| PrpsM_R2 | 5′-GTCGCCATTCTAGACCAGCCAGGATGGCTTTAGAA-3′ |
| proUgfp+_F2 | 5′-TGAAATTATTACAGGACGAAGACCGTGAATATGGTTACGTCATTGAGCGTTAAGAAGGAGATATACATATGAG-3′ |
| proUgfp+_R2 | 5′-AATGCCGCTTTTAATGAGTCGATGGACACGACGCCCACGAATTTATTGCCTTATCACTTATTCAGGCGTA-3′ |
| T1_F1 | 5′-GCAGGTCACATTTAACGCGGTTGCACAAGTTGCAACATGGCCTGGGGTAATGACTCTCTAGC-3′ |
| Cam_R5 | 5′-GACCCGGATAGTAATTTTGCCCGGCCAGATGATAAATCGCGACGTCATTTCTGCCATTCATCC-3′ |
Sequences underlined indicate locations of restriction sites used for cloning.
Plasmids.
All of the plasmids used in this study are listed in Table 1. Plasmids pZEP01 and pZEP02 were constructed by PCR as follows. PCR amplification of part of the promoterless gfpmut3 gene or of the rpsM-gfpmut3 fusion was performed with primers pFPV_F1 and pFPV_R1 (Table 2) and plasmid templates pFPV25 and pFPV25.1 (61). The two resulting PCR products contained the common 5′ end DNA coding sequence of both the gfpmut3 and gfp+ genes, either with no promoter (from pFPV25; designated product 1) or fused to the rpsM promoter (from pFPV25.1; designated product 2). The gfp+-specific 3′ region, designated product 3, was amplified from the pWH1012gfp+ plasmid (51) with primers GFP_F1 and GFP_R1 (which contained an additional EcoRV site [Table 2]). Products 1 and 2 obtained from pFPV25 and pFPV25.1 and product 3 obtained from pWH1012gfp+ had a 118-bp homologous sequence located in their 3′ and 5′ ends, respectively. A crossover PCR approach was subsequently used (33). Left and right products 1 and 3 or products 2 and 3 were annealed at the overlapping region and amplified by PCR as single fragments with the outer primers pFPV_F1 and GFP_R1, generating ∼1,600-bp product 4 containing the new gfp+ reporter fused to the rpsM promoter and the corresponding 980-bp product 5 that lacked a promoter. PCR products 4 and 5 were digested with KpnI and EcoRV and cloned into the pFPV25 vector digested with KpnI and EcoRV to generate plasmids pZEP01 and pZEP02, respectively, in which the gfpmut3 gene was replaced by gfp+.
Plasmid pZEP07 was constructed as follows. A chloramphenicol resistance cassette was amplified by PCR from pACYC184 (10) by using primers Cam_F3 and Cam_R3 (Table 2). The resulting 1,004-bp product was digested with EcoRV and subsequently cloned into EcoRV-digested pZEP01 in the same orientation as the gfp+ gene, generating the pZEP06 plasmid. A 1,067-bp fragment, containing the strong t0 and T1 transcriptional terminators (49, 50), was amplified by PCR from the pQE9 plasmid (Qiagen) with primers t0T1_F4 and t0T1_R4 (Table 2), which added KpnI and XbaI restriction sites to the 5′ and 3′ ends of the product, respectively. After KpnI-XbaI digestion, the 1,067-bp fragment was cloned into KpnI-XbaI-digested pZEP06 to generate pZEP07.
Plasmid pZEP08 was derived directly from pZEP07. A kanamycin resistance cassette was amplified from pKD4 (13) by using primers Kan_F1 and Kan_R1 (Table 2), which introduced an NheI site, a SmaI site, and a NotI site into the 5′ end and an XbaI site into the 3′ end of the resulting 993-bp fragment. The product was digested with NheI and XbaI and cloned into XbaI-digested pZEP07 to generate pZEP08.
Plasmids pZEP09, pZEP10, and pZEP16 containing the ssaG-gfp+, prgH-gfp+, and rpsM-gfp+ fusions were constructed as follows. The ssaG, prgH, and rpsM promoters were amplified from purified Salmonella LT2 chromosomal DNA with the following primer pairs: PssaG_F2 plus PssaG_R2, PprgH_F1 plus PprgH_R1, and PrpsM_F3 plus PrpsM_R2 (Table 2). All forward primers contained a 5′ SmaI site, and all reverse primers carried a 5′ XbaI site. Each PCR product was digested with SmaI-XbaI before gel purification and cloning into SmaI-XbaI-digested pZEP08, which resulted in deletion of the kanamycin cassette (Fig. 1A). The chloramphenicol-resistant (Cmr) and kanamycin-sensitive (Kms) E. coli transformants that harbored the new plasmid-borne fusions were identified.
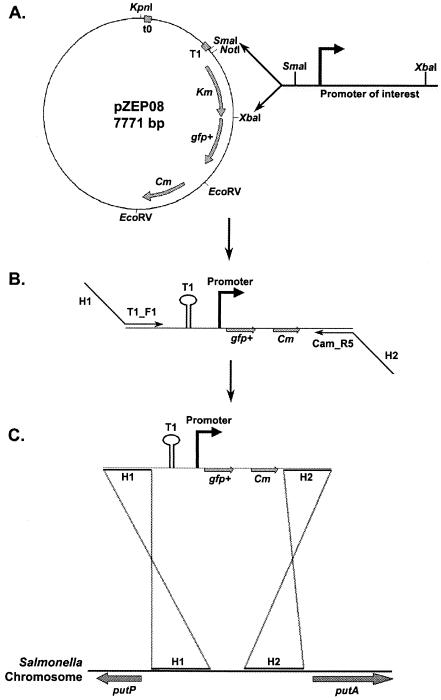
FIG. 1.
Strategy used for construction of single-copy gfp+ fusions. Promoters of interest were amplified by PCR from the Salmonella chromosome and inserted into the pZEP08 plasmid in place of the kanamycin resistance cassette (A). The resulting plasmid was then used as a template for PCR amplification of the fragment that contained the T1 terminator, the new gfp+ transcriptional fusion, and the chloramphenicol resistance cassette by using primers that had 40- to 50-nucleotide tails (H1 and H2) exhibiting perfect homology with the chromosomal site of insertion (i.e., putPA locus) (B). The linear PCR product containing the new fusion was moved to the chromosome of the recipient Salmonella strain by recombination by using the Lambda Red system (13) (C).
Chromosomal integration of single-copy fusions in Salmonella strains.
All gfp+ constructs were integrated into the chromosome of Salmonella strain LT2 or SL1344 by using the Lambda Red system as previously described (13, 43).
For construction of the single-copy proU-gfp+ fusion, a fragment containing the promoterless gfp+ gene and the chloramphenicol resistance cassette was amplified from the pZEP07 plasmid with primers proUgfp+_F2 and proUgfp+_R2 (Table 2). Each of these primers has a 5′ 50-nucleotide region that exhibits perfect homology with an internal part of the proV gene coding sequence (positions 2,956,849 to 2,956,948 on the Salmonella LT2 chromosome; GenBank accession no. AE006468).
Plasmids pZEP09, pZEP10, pZEP16, and pZEP07 were used as templates for PCR amplification of a fragment containing the T1 terminator, the ssaG, prgH, and rpsM-gfp+ fusions or the promoterless gfp+ gene, and the chloramphenicol resistance cassette by using primers T1_F1 and Cam_R5 (Table 2). Both of these primers had a 5′ 40- to 42-nucleotide region exhibiting perfect homology with the putPA locus of the Salmonella serovar Typhimurium SL1344 chromosome (Fig. 1) (H2 at positions 1,210,040 to 1,210,079 and H1 at positions 1,211,618 to 1,211,657 on the Salmonella LT2 chromosome). The PCR fragments were between 2 and 2.7 kb long.
For all single-copy fusions, between 500 ng and 1 μg of each linear PCR product was used for integrating fusions on the chromosome of Salmonella strain LT2 (proU fusion) or SL1344 (ssaG, prgH, and rpsM fusions) by the Lambda Red method (13). Between 10 and 15 transformants were obtained for each gene fusion. The loss of the pKD46 helper plasmid was monitored on LB medium plates at 37°C by using MAST ID Intralactam circles (MAST Diagnostics, Bootle Merseyside, United Kingdom) to screen for the absence of beta-lactamase in bacterial colonies. Putative constructs were verified by colony PCR by using specific primer pairs that annealed externally and internally with respect to the gfp+ fusions. The chromosomal regions containing the gfp+ fusions were sequenced on both DNA strands with specific primers by using an ABI 3700 sequencer and a Big Dye version 3 sequencing kit (ABI Prism).
β-Galactosidase assay.
The method used to measure β-galactosidase activity was adapted from the Miller method (36). Serial twofold dilutions of purified β-galactosidase (Sigma) were used at concentrations ranging from 100 to 1.56 mU/ml to establish a standard curve. Cells were permeabilized with chloroform-sodium dodecyl sulfate, and chlorophenol red β-d-galactopyranoside (Roche) was used as the substrate. Reactions were performed in 96-well microtiter plates, and the results were read with a Spectramax spectrophotometer (Molecular Devices, Sunnyvale, Calif.). The kinetics of substrate hydrolysis was determined for 20 min, and the Vmax was used to convert the data into milliunits per milliliter by using the SoftMaxPro 3.1.2 software (Molecular Devices) and the linear function formula y = (Ax + B)/OD600, where y is the β-galactosidase activity to be determined, x is the reading value, A is the slope of the reading curve, B is the y intercept of the line, and OD600 is the optical density at 600 nm of the culture resuspended in reaction buffer.
Fixation, immunostaining, and flow cytometric analysis.
For measurement of GFP in Salmonella, samples were immediately fixed for 1 min at room temperature in 4% (wt/vol) formalin (Sigma) freshly prepared in phosphate-buffered saline (PBS) (pH 7.4) (48). Fixed bacteria were subsequently washed, resuspended in PBS, and kept in the dark at 4°C until analysis. The PBS used in this study was filtered through a 0.22-μm-pore-size filter (Millipore, Billerica, Mass.) to reduce the background noise during flow cytometric analysis.
When appropriate, Salmonella cells were labeled with specific antibodies. A 1:200 final dilution of a rabbit anti-Salmonella lipopolysaccharide polyclonal primary antibody (catalog no. 2948-47-6; Biosciences Pharmingen, San Diego, Calif.) and a 1:40 final dilution of a goat R-phycoerythrin-conjugated anti-rabbit secondary antibody (catalog no. 4010-09; Southern Biotechnology Associates, Inc., Birmingham, Ala.) were used. Primary antibody staining and secondary antibody staining were performed in PBS containing 10% normal horse serum (Sigma) for 30 min at room temperature, followed by three washes in PBS.
For flow cytometric analysis, samples were diluted in 1 to 2 ml of PBS to obtain a maximum of approximately 106 particles per ml and were analyzed with a FACScalibur flow cytometer (Becton Dickinson, Franklin Lakes, N.J.) equipped with a 15-mW air-cooled argon ion laser as the excitation light source (488 nm). For analysis of bacterial cells released from macrophages, samples were gated for Salmonella-like particles by using the orange fluorescence of the anti-Salmonella labeling to identify bacterial cells and to exclude mammalian cell debris and background noise. Fluorescence compensation settings were determined in parallel under identical conditions by using the constitutively GFP+-expressing Salmonella strain JH3016 or the nonexpressing strain JH3008, with and without anti-Salmonella antibody labeling. All parameters were collected by using amplification gains set on LOG mode. Approximately 15,000 events identified as Salmonella cells were collected per sample. GFP fluorescence intensity values are presented below as medians for the populations after analysis with CellQuest 3.3 software (Becton Dickinson).
Macrophage infection by Salmonella strains.
Murine J774-A.1 macrophage-like cells (European Collection of Cell Cultures [ECACC] no. 91051511) were grown in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, Calif.) supplemented with 20% fetal bovine serum, 2 mM l-glutamine (Sigma), and 20 mM HEPES buffer (Sigma). For infection with Salmonella, 108 J774-A.1 cells were seeded in six-well plates (Becton Dickinson) at 37°C in the presence of 5% CO2 as described previously (17). Salmonella cells were grown overnight on LB agar plates, washed, and resuspended in sterile PBS. Complement opsonization of bacteria and macrophage infection were performed as described previously (18) by using a multiplicity of infection of 100 bacteria per macrophage. Contact with the macrophage monolayer was maximized by 5 min of centrifugation at 453 × g at room temperature. Infected J774-A.1 cells were immediately incubated for 1 h at 37°C in the presence of 5% CO2. Time zero of an experiment was defined as the beginning of this incubation. Salmonella cells that remained outside the macrophages were subsequently removed, immediately fixed, and used as a control in flow cytometry. The remaining extracellular bacteria were killed by addition of HEPES-buffered RPMI 1640 containing 10% fetal bovine serum and 30 μg of gentamicin per ml and incubation for an additional 1 h at 37°C in the presence of 5% CO2. The medium was then replaced by HEPES-buffered RPMI 1640 containing 5 μg of gentamicin per ml, and the preparation was incubated at 37°C in the presence of 5% CO2 until the end of the assay. At the end of the experiment, infected monolayers were washed with PBS and lysed under hypotonic conditions (17). Bacteria released from the intracellular environment were immediately fixed and kept at 4°C in PBS before anti-Salmonella labeling and flow cytometric analysis.
RESULTS AND DISCUSSION
Fixation does not impair fluorescence of GFP+.
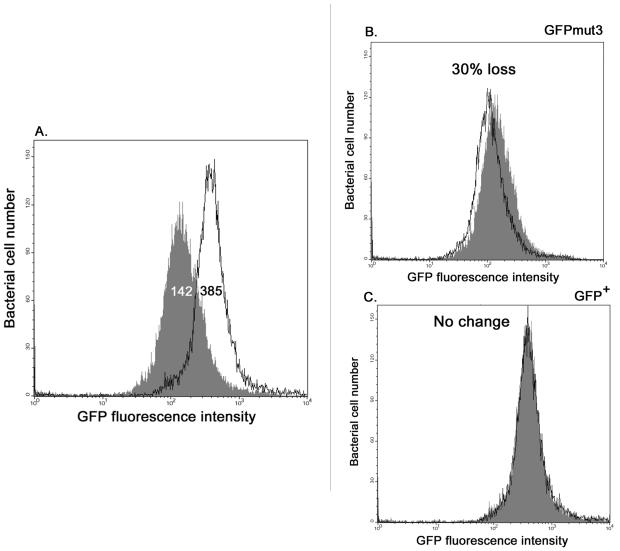
Visualization of fluorescence from single-copy transcriptional fusions requires a particularly bright version of GFP. Therefore, we compared the levels of fluorescence of the most promising GFP variants. These included the GFPmut3 variant (11) and the GFP+ variant, which was reported to be 130-fold brighter than the wild-type protein (51). The rpsM promoter was chosen because it was reported to be expressed at similar levels in various environments, including growth media and mammalian cells (62). We compared the rpsM-gfpmut3 plasmid-borne fusion pFPV25.1 with the pZEP02 plasmid (see Materials and Methods), which carried the gfp+ gene under control of the same transcriptional and translational signals as gfpmut3 in pFPV25.1. Both pZEP02 and pFPV25.1 were transformed into E. coli DH5α, and the levels of green fluorescence were determined by flow cytometry with and without formalin fixation.
Figure 2A shows that the plasmid-borne rpsM-gfp+ fusion is approximately three times brighter than the corresponding rpsM-gfpmut3 fusion. Use of fluorescent proteins to monitor bacterial gene expression by flow cytometry requires the use of chemical fixation to stop gene expression. It has previously been reported that formalin has less effect on GFP fluorescence than other fixatives have (4). Here, we compared the effects of formalin fixation on GFPmut3 fluorescence and GFP+ fluorescence, and we observed that the treatment reduced GFPmut3 fluorescence by about 30% compared to the fluorescence in unfixed E. coli (Fig. 2B). We discovered that GFP+ is the first GFP variant which is not adversely affected by fixation (Fig. 2C), probably due to the greater stability of the GFP+ protein. This confirms that the presence of the F64L, S65T, F99S, M153T, and V163A mutations results in increased fluorescence, making GFP+ a promising reporter for poorly expressed promoters in individual bacterial cells. We therefore constructed single-copy gfp+ fusions to monitor Salmonella gene expression.
FIG. 2.
Comparison of rpsM-gfpmut3 and rpsM-gfp+ expression in LB medium and after formalin fixation. E. coli strains harboring either pFPV25.1 (rpsM-gfpmut3) or pZEP02 (rpsM-gfp+) were grown overnight in LB broth containing ampicillin. Live or fixed (4% formalin) bacteria were immediately analyzed by flow cytometry. (A) Unfixed bacteria harboring either pFPV25.1 (shaded graph) or pZEP02 (solid line). (B and C) Fixed (solid line) and unfixed (shaded graph) bacteria harboring either pFPV25.1 expressing GFPmut3 (B) or pZEP02 expressing GFP+ (C). The values are the median values for fluorescence intensity for all individual bacteria in a population.
GFP+ is a reliable reporter of gene expression.
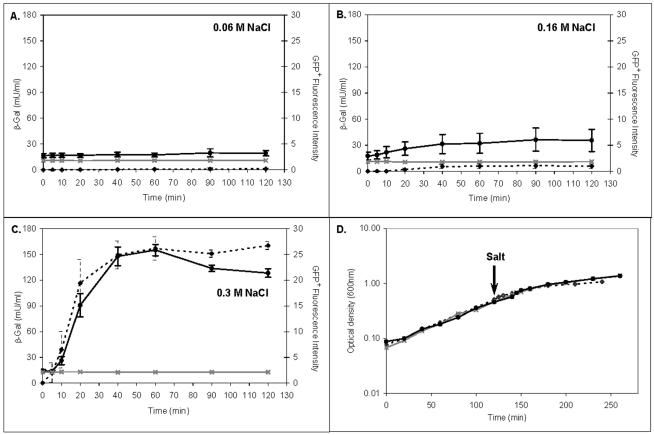
Since 1979, lacZ fusions have been used to obtain robust gene expression data for hundreds of bacterial genes (8, 26, 53). To assess the reliability of single-copy gfp+ fusions for monitoring gene expression, we compared a proU-gfp+ fusion with a well-characterized, salt-inducible proU-lacZ fusion in Salmonella (7). The proU operon encodes a betaine transport system that is involved in the adaptation of the bacteria to increases in environmental osmolarity (20). Cairney et al. reported the pattern of osmoregulation of proU expression in Salmonella serovar Typhimurium strain CH946, which carries a Mud1-8(lacZ) insertion in the proV gene (7, 56). We directly compared salt induction of the proU-lacZ fusion in CH946 with salt induction of a proU-gfp+ fusion that was constructed at exactly the same location in strain JH3017 (see Materials and Methods).
The LT2 strain JH3049 carrying a promoterless gfp+ gene was used as a negative control. Expression of proU-lacZ was assessed by measuring the β-galactosidase activity of the entire population, and proU-gfp+ expression was monitored by flow cytometry of fixed bacteria (Fig. 3). Figure 3A shows that in the presence of 0.06 M NaCl, neither the proU-lacZ fusion nor the proU-gfp+ fusion was induced. When 0.16 M NaCl was added, a low level of induction was observed for both proU-lacZ and proU-gfp+ 20 min after the salt was added; this level of induction stabilized after 40 min and remained very low until the end of the assay (Fig. 3B). Figure 3C shows that both proU-lacZ and proU-gfp+ were highly induced by 0.3 M NaCl and exhibited similar expression patterns through time. Induction of both proU-lacZ and proU-gfp+ was detected just 10 min after salt was added. The slight difference in expression observed for the two fusions at 20 min probably reflected the greater variation in the β-galactosidase measurements for that time point. For both reporter systems maximum induction of the proU promoter was obtained 60 min after addition of 0.3 M NaCl. Subsequently, proU expression slowed, and both reporters exhibited constant or slightly reduced expression in the stationary phase (Fig. 3D).
FIG. 3.
Osmotic induction of proU. Induction of proU-lacZ and proU-gfp+ expression was tested as follows. Salmonella strains CH946 (proU-lacZ; β-galactosidase [β-Gal] activity indicated by the dashed line), JH3017 (proU-gfp+; GFP fluorescence indicated by the solid black line), and JH3049 (promoterless gfp+; GFP fluorescence indicated by the solid grey line) were grown in LO medium containing glucose at 30°C to an optical density at 600 nm of 0.5. NaCl was then added to a final concentration of 0.06 M (A), 0.16 M (B), or 0.3 M (C). Samples were collected at 0, 5, 10, 20, 40, 60, 90, and 120 min after addition of the salt. The optical density of each culture was measured at 600 nm (panel D shows the growth curves obtained before and after addition of 0.3 M NaCl). The arrow indicates when the salt was added to each mid-log-phase culture. The median β-galactosidase activities for three independent experiments are shown (see Materials and Methods). The GFP+ fluorescence intensities are the median values for the intensities of all individual bacteria in a population for seven independent experiments. The error bars indicate the standard deviations.
In summary, these results confirmed the previously described osmoregulation of proU (7) and demonstrated that GFP+ reports proU activity as accurately as β-galactosidase does. These data agree with a previous study involving plasmid-borne fusions (51). However, insertion of a reporter gene into a gene of interest, as described above, necessarily generates a mutation that could have a polar effect. To maintain an intact copy of each wild-type promoter and to avoid production of virulence mutants while allowing direct comparison of many promoters in the same chromosomal context, we developed a system for insertion of single-copy transcriptional fusions at a different chromosomal location.
Construction of single-copy gfp+ chromosomal fusions in Salmonella serovar Typhimurium.
Rapid construction of single-copy GFP+ fusions in Salmonella involved the pZEP08 plasmid (see Materials and Methods), which carried transcriptional terminator T1, a promoterless gfp+ gene, a chloramphenicol resistance cassette, and a kanamycin resistance cassette (Fig. 1A). Fragments containing the promoter regions of genes of interest were amplified by PCR and cloned upstream of the gfp+ gene. The whole constructs were amplified by PCR (Fig. 1B) and integrated onto the Salmonella chromosome (13). A similar approach was used to generate the negative control construct; this approach involved amplification of the corresponding fragment from parental plasmid pZEP07, which carried the promoterless gfp+ gene located directly downstream of the T1 terminator, preventing production of GFP+. The gene fusions were integrated at the putPA locus (Fig. 1B and C). We have shown that interruption of putPA does not affect the ability of Salmonella to colonize the spleen and liver in the BALB/c mouse model (Hautefort, Proença, and Hinton, unpublished data).
The rpsM, ssaG, and prgH gene fusions were integrated as a single copy into the putPA locus on the Salmonella chromosome. In parallel, the promoterless gfp+ gene derived from pZEP07 was inserted at exactly the same position. Colony PCR and subsequent DNA sequencing (see Materials and Methods) were used to verify that all colonies carried the correct fusion. The approach summarized in Fig. 1 allowed successful construction and integration of gfp+ fusions, and it has proved to be a rapid and flexible method for generating single-copy chromosomal gfp+ fusions in Salmonella.
Single-copy gfp+ fusion allows detection of in vitro induction of virulence gene expression.
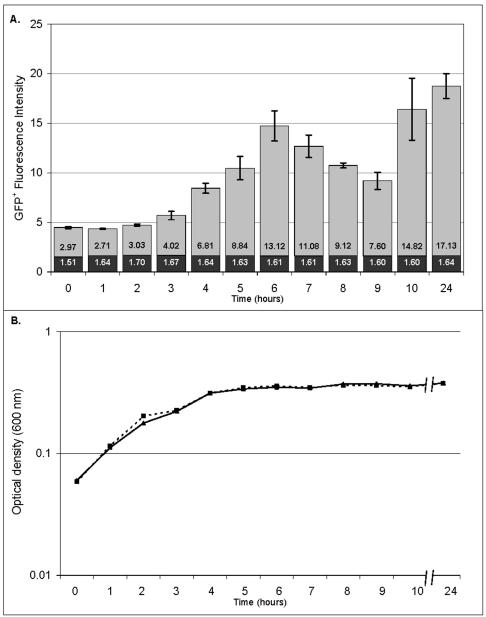
Understanding the host-pathogen interaction requires monitoring of virulence gene expression during infection of mammalian cells and animal models, as well as in vitro. We first verified the single-copy ssaG-gfp+ fusion by monitoring expression during growth of Salmonella serovar Typhimurium under inducing conditions in vitro. The ssaG gene encodes a component of the SPI2 type III secretion system, which is highly induced during macrophage infection (18, 62), when Salmonella faces an acidic pH combined with low levels of phosphate and magnesium (14, 47). We chose an acidic minimal medium (MM5.8) (30) to reproduce some of these conditions. The negative control Salmonella strain JH3008 (promoterless gfp+) did not express GFP+ and had a constant fluorescence intensity of about 1.6 (Fig. 4). This confirmed that the transcriptional terminator T1 included in our constructs efficiently prevented transcriptional readthrough. Induction of ssaG from strain JH3009 was detected after 3 h of growth, corresponding to the mid-exponential growth phase. Expression of ssaG-gfp+ increased eightfold at 6 h, when the bacteria entered the stationary growth phase. This is in agreement with the previously described pattern of expression of SPI2 transcriptional fusions to the luciferase gene, as monitored in acidic medium (3). Figure 4 shows that ssaG-gfp+ was then switched off, since GFP+ fluorescence decreased twofold between 7 and 9 h. A second induction of ssaG-gfp+ expression was shown by the doubling of fluorescence intensity between 9 and 10 h, which was maintained at 24 h. Monitoring of ssaG-gfp+ expression showed that increases as well as decreases in GFP+ fluorescence intensity could be measured through time (Fig. 4), confirming that single-copy fusions are a valuable tool for looking at growth phase-dependent gene expression and for performing time course experiments.
FIG. 4.
Virulence gene induction in vitro. Salmonella serovar Typhimurium strains JH3009 (ssaG-gfp+) and JH3008 (promoterless gfp+) were grown overnight in LB broth. Both strains were subsequently diluted 50-fold in MM5.8 and were grown for 24 h. Time zero corresponded to the beginning of incubation in MM5.8. Samples were collected every hour from time zero until 10 h and once after 24 h, immediately fixed in 4% formalin, and analyzed by flow cytometry. (A) Fluorescence intensity of Salmonella strain JH3009 (grey bars) determined in triplicate. The fluorescence intensity of the negative control JH3008 strain is also indicated for each time point (solid bars). The values in the bars are the median fluorescence intensities of all individual bacteria in the populations. The error bars indicate the standard deviations. (B) Corresponding growth curve. Dashed line, strain JH3008; solid line, strain JH3009.
Single-copy gfp+ fusion reveals variation in gene expression between individual bacterial cells.
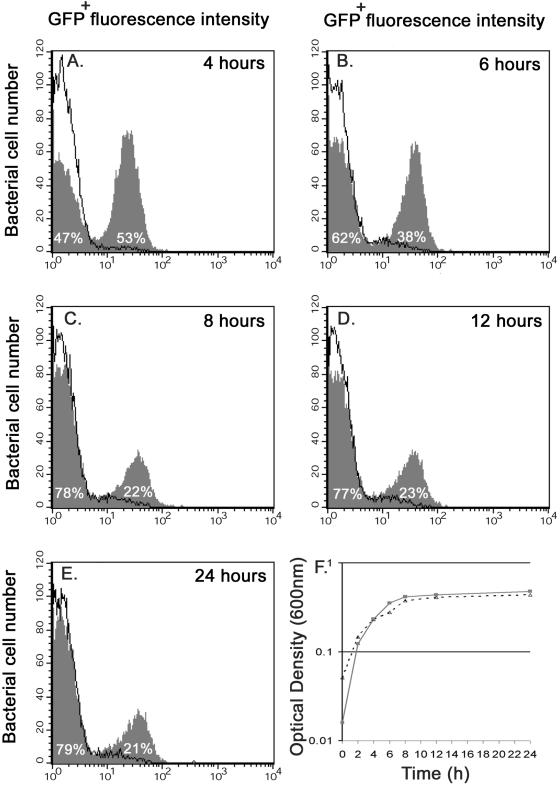
The combination of single-copy gfp+ fusions and the ability to measure fluorescence in individual bacterial cells offers a powerful system for searching for different levels of gene expression in bacteria within a population. We monitored expression of the prgH-gfp+ single-copy fusion in Salmonella strain JH3010. The prgH gene encodes a basal component of the needle complex of the SPI1 type III secretion machinery. SPI1 genes are induced by the high osmolarity, low oxygen levels, and short-chain fatty acids thought to be present in the ileum of the digestive tract (19, 31). Figure 5 shows that the prgH-gfp+ fusion was induced 12-fold after 4 h of growth only in the presence of salt (LO medium containing 0.3 M NaCl). The salt-dependent induction of prgH-gfp+ increased to 18-fold at 6 h and remained at 17-fold at 8, 12, and 24 h. This suggests that prgH is induced soon after salt addition (<4 h, mid-exponential phase) and that expression of this gene is dramatically reduced after 6 h (from the late exponential phase to the stationary phase) since no more GFP+ accumulates in each bacterial cell. Concomitantly, the experiment revealed that there was significant differential gene expression within the bacterial population (Fig. 5). The sequential flow cytometric analysis showed for the first time that prgH-gfp+ is not induced in every bacterial cell. Only 53% of the population showed prgH-gfp+ induction at 4 h, and the percentage decreased to approximately 22% from 8 h until the end of the experiment. To ensure that the single-copy prgH-gfp+ fusion was stable on the chromosome and had not been lost from any of the cells, we screened the bacteria from the 24-h culture for Cmr resistance. All of the 200 Salmonella colonies tested were Cmr, showing that every bacterial cell still carried prgH-gfp+. This observation of differential expression of an SPI1 gene is completely novel, and the effect of this expression on the ability of Salmonella to succeed during infection merits further investigation.
FIG. 5.
Differential expression of a promoter within a genetically identical population. Salmonella serovar Typhimurium strain JH3010 (carrying prgH-gfp+) was grown overnight in LO medium containing no salt. A culture containing 104 bacteria per ml (final concentration) was then grown in either LO medium (solid line) or LO medium containing 0.3 M NaCl (shaded graph). Samples were collected after 4 h (A), 6 h (B), 8 h (C), 12 h (D), or 24 h (E) of growth with or without salt (F) (dotted line, LO medium; solid line, LO medium containing 0.3 M NaCl), fixed in 4% formalin, and analyzed by flow cytometry. The level of induction was calculated by comparing the fluorescence for the most fluorescent peak at each time point with the fluorescence at the same time obtained when strain JH3010 was grown in LO medium. The percentage of the population in each fluorescence peak is indicated on the graph. To ensure that the differential expression of the prgH-gfp+ fusion was genuine, the experiment was repeated 25 times, and the data from a single representative experiment are shown.
Monitoring virulence gene expression in complex environments.
To determine the utility of single-copy gfp+ fusions in complex environments, we monitored GFP+ expression in bacteria following infection of mammalian cells. We detected and quantified ssaG-gfp+ expression in individual bacteria released from infected J774-A.1 macrophage-like cells. Data that were generated from a plasmid-borne ssaG-gfpmut3 fusion (originally referred to as ssaH) showed that ssaG was highly expressed 6 h after macrophage infection (62). We infected J774-A.1 murine macrophages with opsonized strains JH3009, JH3016, and JH3008 carrying single-copy ssaG-gfp+ and rpsM-gfp+ fusions and a promoterless gfp+ gene, respectively. The rpsM gene had previously been reported to be expressed at similarly high levels in LB medium and in macrophages (61), suggesting it would be an appropriate positive control.
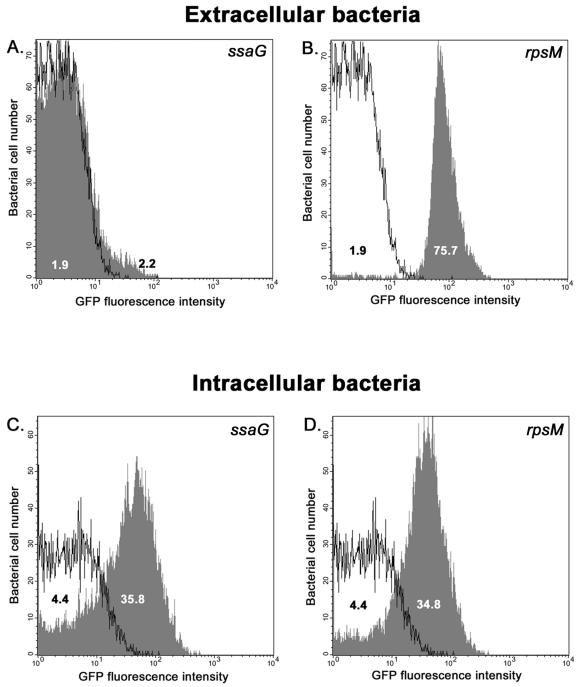
For flow cytometric analysis, detection of multiple fluorescent colors requires adjustment of settings to ensure that each fluorescent signal does not spill over into a second signal and to avoid false-positive data. This adjustment, referred to as compensation, required the use of comparable positive and negative control strains, JH3016 and JH3008. The analysis was performed with bacteria that either were released from mammalian cells after 6 h or remained outside the macrophages following the initial incubation (see Materials and Methods). To be able to detect all bacteria, we used an anti-Salmonella antibody to distinguish Salmonella cells from host cell debris. This crucial part of the protocol allowed us to observe all bacteria that either expressed or did not express GFP+. Figure 6A shows that extracellular JH3009 Salmonella cells did not express the ssaG fusion and exhibited levels of fluorescence similar to those of the control JH3008 extracellular bacteria. As expected, the JH3016 strain expressed the rpsM-gfp+ fusion outside the macrophages with 40-fold more GFP+ fluorescence than the negative control (Fig. 6B). Figure 6C clearly shows the novel finding that the single-copy ssaG-gfp+ fusion was induced eightfold in all bacterial cells when Salmonella was internalized within the macrophages. This increase in ssaG expression is consistent with the increase observed at the RNA level by DNA microarray analysis (18) and confirms that reporter genes can accurately reflect the level of bacterial gene transcription within mammalian cells. No differential expression of ssaG-gfp+ was observed within the intracellular bacterial population. Interestingly, the fluorescence of strain JH3016, which expressed the rpsM-gfp+ fusion, decreased fivefold 6 h after phagocytosis and was only eightfold higher than the fluorescence of the negative control strain JH3008 (Fig. 6D), suggesting that a high level of expression of the small S13 ribosomal protein is no longer required once the bacteria are inside a Salmonella-containing vacuole. This is consistent with the threefold decrease in rpsM expression observed at the RNA level (18). Figures 6C and D also show that the intrinsic green autofluorescence of Salmonella cells increased within macrophages. Indeed, the relative fluorescence intensity of JH3008 (promoterless gfp+) doubled from the extracellular location to the intraphagosomal location (Fig. 6B and D). This observation shows that it is not sufficient simply to compare extracellular bacteria with intracellular bacteria, ignoring the variation in the level of bacterial autofluorescence with cellular location. A simplistic comparison would have suggested that ssaG-gfp+ expression from strain JH3009 was induced more than 16-fold inside macrophages compared to the level of expression in extracellular bacteria. Because the level of autofluorescence of the promoterless control strain JH3008 more than doubled intracellularly, the true level of ssaG induction was eightfold. This observation shows that choosing an appropriate negative control is crucial when gfp+ fusions are used to study in vivo gene expression. The results presented in Fig. 6 validate the use of our system for monitoring induction of virulence gene expression in the complex environment of infected mammalian cells.
FIG. 6.
Single-copy SPI2 gene fusion accurately reports gene expression in mammalian cells. J774-A.1 murine macrophages were infected with Salmonella serovar Typhimurium strains JH3008, JH3009, and JH3016 harboring a promoterless gfp+ gene, an ssaG-gfp+ fusion, and an rpsM-gfp+ fusion, respectively. Six hours after infection, intracellular bacteria were released under hypotonic conditions and immediately fixed in 4% formalin. Salmonella cells were then labeled with a specific antibody, and their fluorescence was measured by flow cytometry. The results shown represent the GFP+ fluorescence intensity of extracellular or intracellular bacteria identified as Salmonella cells by antibody labeling. Each panel shows an overlay of the GFP+ fluorescence of either JH3009 or JH3016 (shaded graph) with the GFP+ fluorescence of JH3008 (solid line), which was used as negative control. (A and B) Levels of expression of ssaG-gfp+ or rpsM-gfp+ in extracellular Salmonella cells. (C and D) Fluorescence in bacteria of ssaG-gfp+ or rpsM-gfp+ released from inside macrophages at 6 h postinfection. The values indicate the median GFP fluorescence intensity of all individual bacteria in a population.
This study confirmed that single-copy GFP+ fusions allow low levels of gene expression to be quantified in individual bacterial cells as accurately as has been possible in entire bacterial populations with lacZ fusions. For the first time, the combination of a single-copy transcriptional fusion with flow cytometric analysis revealed different levels of expression of a virulence promoter.
The ability to record the level of variation in the expression of a particular gene in genetically identical populations is particularly important because of the unexplained phenotypic heterogeneity that has been reported previously for bacterial populations (5, 32, 55). Flow cytometry and cell sorting have been used to measure the variation in several phenotypic parameters in bacterial cells (39). Heterogeneity has only begun to be studied at the level of gene expression in the last decade; Mulec et al. (38) showed that induction of a plasmid-borne cka-gfp fusion occurred in only 3% of E. coli cells, and Siegele and Hu (52) observed variations in the level of expression of an araBAD-GFP transcriptional fusion in different bacterial cells. Other techniques, such as in situ PCR, have been used to detect qualitative differences in mRNA levels between individual bacterial cells (59). However, the study of variations in gene expression within bacterial populations has been hampered by the paucity of techniques to measure the levels of promoter activity within individual bacteria. Recently, a robust mathematical modeling study based on single-copy gfp fusions in Bacillus subtilis showed that phenotypic variations commonly observed between bacterial cells of a genetically identical population are strongly linked to translational rather than transcriptional efficiency (41). This confirms that transcriptional fusions can be relied upon to show real variations in gene expression rather than phenotypic noise. The single-copy gfp fusions described here permit the study of virulence gene expression in individual bacterial cells during infection of mammalian cells.
In the last two decades, the technology for construction of single-copy reporter gene fusions has been in constant development and has relied upon the site-specific recombination systems of various phages, transposons, and suicide vectors, as summarized by Slauch and Silhavy in 1991 (54) and by Hand and Silhavy more recently (23). In a large number of studies the workers have successfully used these methods to study and dissect regulatory pathways (21, 58). However, many of the approaches have been problematic, because integration has been restricted to one specific site on the chromosome or there has been genetic instability. The recently developed Lambda Red system has revolutionized recombinant genetics in enteric bacteria. This system provides scientists with an excellent tool for stable insertion of DNA fragments anywhere in the bacterial chromosome (13). This system has recently been used for construction of single-copy lacZ fusions in the chromosome of Salmonella serovar Typhimurium (16). Ellermeier et al. used FLP/FRT-mediated site-specific recombination events to incorporate a promoterless lacZ gene at the site of a mutated gene of interest, which had previously been knocked out by using the Lambda Red system. However, this approach initially involves creation of a gene knockout. Because no wild-type copy of the gene remains, the resulting strain might show attenuated virulence or phenotypes might be affected. Our system is also based on the Lambda Red recombination method but has the advantage of leaving an intact copy of the gene of interest in its original site, as well as the benefit of generating single-copy gfp+ gene fusions in a chromosomal location that is known to have no apparent effect on Salmonella virulence. The insertion locus can also be varied without a requirement for a supplementary cloning step, which makes this tool an adaptable system that is applicable to a large number of gram-negative bacteria. The approach described here promises to provide answers to key biological questions concerning the pattern of bacterial gene expression within populations, both in vitro and during the process of infection itself.
Acknowledgments
We thank Sofia Eriksson for sharing her expertise with Salmonella infection of macrophages, Martin Goldberg for useful technical advice, and Roy Bongaerts for critical evaluation of the manuscript. We are grateful to Chris Higgins and Julie Sidebotham for their input at the early stages of this work. We thank Mikael Niederweis for providing us with the pWH1012gfp+ plasmid.
This work was originally supported by Wellcome Trust Programme grant 045490. Isabelle Hautefort was initially supported by a Training and Mobility of Researchers fellowship from the European Union (contract number ERBFMRXCT9), and the work was subsequently supported by the BBSRC.
REFERENCES
- 1.Ames, B. N., and H. J. Whitfield, Jr. 1966. Frameshift mutagenesis in Salmonella. Cold Spring Harbor Symp. Quant. Biol. 31:221-225. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 4.Bongaerts, R. J., I. Hautefort, J. M. Sidebotham, and J. C. Hinton. 2002. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol. 358:43-66. [DOI] [PubMed] [Google Scholar]
- 5.Booth, I. R. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19-30. [DOI] [PubMed] [Google Scholar]
- 6.Bumann, D. 2002. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol. Microbiol. 43:1269-1283. [DOI] [PubMed] [Google Scholar]
- 7.Cairney, J., I. R. Booth, and C. F. Higgins. 1985. Osmoregulation of gene expression in Salmonella typhimurium: proU encodes an osmotically induced betaine transport system. J. Bacteriol. 164:1224-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 10.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 15.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson, S., J. Bjorkman, S. Borg, A. Syk, S. Pettersson, D. I. Andersson, and M. Rhen. 2000. Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell. Microbiol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 19.Galan, J. E., and R. D. Curtiss. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gowrishankar, J., and D. Manna. 1996. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica 97:363-378. [DOI] [PubMed] [Google Scholar]
- 21.Haldimann, A., L. L. Daniels, and B. L. Wanner. 1998. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of Escherichia coli phosphate regulon. J. Bacteriol. 180:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Hand, N. J., and T. J. Silhavy. 2000. A practical guide to the construction and use of lac fusions in Escherichia coli. Methods Enzymol. 326:11-35. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 25.Hasan, N., M. Koob, and W. Szybalski. 1994. Escherichia coli genome targeting. I. cre-lox-mediated in vitro generation of ori-plasmids and their in vivo chromosomal integration and retrieval. Gene 150:51-56. [DOI] [PubMed] [Google Scholar]
- 26.Hautefort, I., and J. C. Hinton. 2000. Measurement of bacterial gene expression in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hautefort, I., and J. C. D. Hinton. 2002. Molecular methods for monitoring bacterial gene expression during infection. Methods Microbiol. 31:55-90. [Google Scholar]
- 28.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 29.Hopkins, S. A., F. Niedergang, I. E. Corthesy-Theulaz, and J. P. Kraehenbuhl. 2000. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer's patch dendritic cells. Cell. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 30.Kox, L. F. F., M. M. S. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 32.Levin, M. D., C. J. Morton-Firth, W. N. Abouhamad, R. B. Bourret, and D. Bray. 1998. Origins of individual swimming behavior in bacteria. Biophys. J. 74:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Link, A., D. Phillips, and G. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Løbner-Olesen, A. 1999. Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J. 18:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and the AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Miller, W. G., M. T. Brandl, B. Quinones, and S. E. Lindow. 2001. Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Appl. Environ. Microbiol. 67:1308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulec, J., Z. Podlesek, P. Mrak, A. Kopitar, A. Ihan, and D. Zgur-Bertok. 2003. A cka-gfp transcriptional fusion reveals that the colicin K activity gene is induced in only 3 percent of the population. J. Bacteriol. 185:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nebe von-Caron, G., P. J. Stephens, C. J. Hewitt, J. R. Powell, and R. A. Badley. 2000. Analysis of bacterial function by multi-colour fluoresence flow cytometry and single cell sorting. J. Microbiol. Methods 42:97-114. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, D. L., and E. P. Kennedy. 1971. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J. Biol. Chem. 246:3042-3049. [PubMed] [Google Scholar]
- 41.Ozbudak, E. M., M. Thattai, I. Kurtser, A. D. Grossman, and A. van Oudenaarden. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31:69-73. [DOI] [PubMed] [Google Scholar]
- 42.Patkar, A., N. Vijayasankaran, D. W. Urry, and F. Srienc. 2002. Flow cytometry as a useful tool for process development: rapid evaluation of expression systems. J. Biotechnol. 93:217-229. [DOI] [PubMed] [Google Scholar]
- 43.Poteete, A. R. 2001. What makes the bacteriophage Lambda Red system useful for genetic engineering: molecular mechanism and biological function. FEMS Microbiol. Lett. 201:9-14. [DOI] [PubMed] [Google Scholar]
- 44.Qazi, S. N., C. E. Rees, K. H. Mellits, and P. J. Hill. 2001. Development of gfp vectors for expression in Listeria monocytogenes and other low G+C gram positive bacteria. Microb. Ecol. 41:301-309. [DOI] [PubMed] [Google Scholar]
- 45.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Randall, L. P., and M. J. Woodward. 2001. Multiple antibiotic resistance (mar) locus in Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 67:1190-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathman, M., M. D. Sjaastad, and S. Falkow. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337-1346. [DOI] [PubMed] [Google Scholar]
- 50.Scholtissek, S., and F. Grosse. 1987. A cloning cartridge of lambda t(o) terminator. Nucleic Acids Res. 15:3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scholz, O., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267:1565-1570. [DOI] [PubMed] [Google Scholar]
- 52.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silhavy, T. J., and J. R. Beckwith. 1985. Uses of lac fusions for the study of biological problems. Microbiol. Rev. 49:398-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slauch, J. M., and T. J. Silhavy. 1991. Genetic fusions as experimental tools. Methods Enzymol. 204:213-248. [DOI] [PubMed] [Google Scholar]
- 55.Spudich, J. L., and D. E. Koshland, Jr. 1976. Non-genetic individuality: chance in the single cell. Nature 262:467-471. [DOI] [PubMed] [Google Scholar]
- 56.Stirling, D. A., C. S. Hulton, L. Waddell, S. F. Park, G. S. Stewart, I. R. Booth, and C. F. Higgins. 1989. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol. Microbiol. 3:1025-1038. [DOI] [PubMed] [Google Scholar]
- 57.Terskikh, A., A. Fradkov, G. Ermakova, A. Zaraisky, P. Tan, A. V. Kajava, X. Zhao, S. Lukyanov, M. Matz, S. Kim, I. Weissman, and P. Siebert. 2000. “Fluorescent timer”: protein that changes color with time. Science 290:1585-1588. [DOI] [PubMed] [Google Scholar]
- 58.Tinker, J. K., L. S. Hancox, and S. Clegg. 2001. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tolker-Nielsen, T., K. Holmstrom, L. Boe, and S. Molin. 1998. Non-genetic population heterogeneity studied by in situ polymerase chain reaction. Mol. Microbiol. 27:1099-1105. [DOI] [PubMed] [Google Scholar]
- 60.Unge, A., and J. Jansson. 2001. Monitoring population size, activity, and distribution of gfp-luxAB-tagged Pseudomonas fluorescens SBW25 during colonization of wheat. Microb. Ecol. 41:290-300. [DOI] [PubMed] [Google Scholar]
- 61.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 62.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 63.Wendland, M., and D. Bumann. 2002. Optimization of GFP levels for analyzing Salmonella gene expression during an infection. FEBS Lett. 521:105-108. [DOI] [PubMed] [Google Scholar]
- 64.Wilson, R. L., A. R. Tvinnereim, B. D. Jones, and J. T. Harty. 2001. Identification of Listeria monocytogenes in vivo-induced genes by fluorescence-activated cell sorting. Infect. Immun. 69:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yansura, D. G., and D. J. Henner. 1984. Use of Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 81:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]