Abstract
The genes encoding the structural components of the nitrate/nitrite assimilation system of the oceanic cyanobacterium Synechococcus sp. strain WH 8103 were cloned and characterized. The genes encoding nitrate reductase (narB) and nitrite reductase (nirA) are clustered on the chromosome but are organized in separate transcriptional units. Upstream of narB is a homologue of nrtP that encodes a nitrate/nitrite-bispecific permease rather than the components of an ABC-type nitrate transporter found in freshwater cyanobacteria. Unusually, neither nirA nor ntcA (encoding a positive transcription factor of genes subject to nitrogen control) were found to be tightly regulated by ammonium. Furthermore, transcription of glnA (encoding glutamine synthetase) is up-regulated in ammonium-grown cells, highlighting significant differences in nitrogen control in this cyanobacterium. Nitrogen depletion led to the transient up-regulation of ntcA, nirA, nrtP, narB, and glnA in what appears to be an NtcA-dependent manner. The NtcA-like promoters found upstream of nirA, nrtP, and narB all differ in sequence from the canonical NtcA promoter established for other cyanobacteria, and in the case of nirA, the NtcA-like promoter was functional only in cells deprived of combined nitrogen. The ecological implications of these findings are discussed in the context of the oligotrophic nature of oceanic surface waters in which Synechococcus spp. thrive.
Cyanobacteria of the genera Synechococcus and Prochlorococcus dominate the photosynthetic picoplanktonic biomass over vast tracts of the world's oceans (21, 27). Collectively, they account for a considerable fraction of marine primary production and are key players in the global carbon cycle. While they are often found as mixed populations, Prochlorococcus spp. are considerably more abundant than Synechococcus spp. in the highly stratified surface waters of the tropics and subtropics. Synechococcus spp., by contrast, are much more numerous in less oligotrophic waters and have a broader ecological range that extends to temperate and subpolar latitudes (20). Not least among the ecophysiological factors that may account for the differential oceanic distribution of these marine cyanobacteria are biogeochemically significant differences in their capacity to assimilate oxidized forms of combined nitrogen (18, 20).
The overwhelming majority of Synechococcus spp. can utilize both nitrate and nitrite as sole N sources, whereas most Prochlorococcus strains cannot grow on either (18). Exceptions in the former case are three recent isolates from the Red Sea that cannot utilize nitrate (6) and Synechococcus sp. strain MIT S9220, which can utilize nitrite only. This phenotype is also displayed by several low-light-adapted Prochlorococcus ecotypes but not by others or by any high-light-adapted strains examined to date. Only one Prochlorococcus isolate (PAC 1) examined to date appears to have the capacity to grow on nitrate (18). Given their differential distribution, this suggests that Prochlorococcus spp. are likely to be more efficient scavengers of ammonium and, perhaps, of other reduced forms of nitrogen (32) in the warm, nutrient-poor surface waters in which they dominate. By contrast, Synechococcus spp. seem to gain a greater foothold in somewhat less stable water columns where nitrate or nitrite concentrations are sufficient to meet or at least augment their N requirements. It is probable, therefore, that only Synechococcus spp. are likely to incorporate appreciable new nitrogen (nitrate N as opposed to remineralized forms of N such as ammonium and urea) into the oceanic food web.
Ammonium concentrations in oceanic surface waters dominated by Synechococcus spp. are frequently in the submicromolar range. Under these nutrient-depleted conditions, larger eukaryotic phytoplankton are capable of utilizing nitrate nitrogen in addition to ammonium (7). Whether this is true of open ocean cyanobacteria has not been investigated; although at nanomolar concentrations, the picoplankton size class shows a clear preference for ammonium (10). This is not unexpected, perhaps, since in such highly oligotrophic waters nitrogen uptake is likely to be dominated by the more abundant Prochlorococcus spp. rather than by Synechococcus spp. Whether nitrate/nitrite uptake by Synechococcus spp. is inhibited at low or very low concentrations of ammonium remains uncertain, although there is some field evidence to suggest that it is not (9). At much higher concentrations, however, ammonium does inhibit nitrate and nitrite uptake in the marine Synechococcus sp. strain WH 7803 (9, 14), presumably by repressing the genes involved in nitrite (and nitrate) assimilation.
Ammonium control of the genes involved in nitrate/nitrite assimilation in freshwater cyanobacteria has been studied extensively and involves the regulatory transcription factor NtcA (11). In Synechococcus sp. strain PCC 7942, ntcA is up-regulated in the absence of external ammonium and activates expression of the nirA operon that includes the genes encoding nitrite reductase (NiR; nirA), a nitrate/nitrite ABC-type transporter, and nitrate reductase (NR; narB). By contrast, while both nirA and narB are repressed by ammonium in Synechocystis sp. strain PCC 6803, the abundance of ntcA mRNA is similar in nitrate- and ammonium-grown cells (2). Full expression of nirA and narB is dependent on the enhancer NtcB, although ntcB mutants retain the capacity to grow on nitrate, albeit at substantially reduced rates compared to the wild type (1). NtcB is also implicated in enhancing expression of the nirA operon in Synechococcus sp. strain PCC 7942 (11), but unlike Synechocystis sp. strain PCC 6803 (1), positive regulation is absolutely dependent on the presence of nitrite supplied exogenously or generated endogenously via the reduction of nitrate.
Nitrogen assimilation in cyanobacteria occurs mainly via the glutamine synthetase (GS)/glutamate synthase pathway (11). The glnA gene (encoding GS) is ammonium repressible in Synechococcus sp. strain PCC 7942 and is up-regulated approximately sixfold in nitrate-grown cells (14). Ammonium has a less dramatic negative effect on glnA expression in Synechocystis sp. strain PCC 6803 (23), although like Synechococccus sp. strain PCC 7942, transcription is NtcA-regulated. Both of these nondiazotrophic cyanobacteria contain a second GS (encoded by glnN) that is specifically induced under nitrogen deprivation (23). In both cases, however, the NtcA-like promoters found upstream of glnN differ from the canonical GTAN8TAC-N22-TAN3T binding sequence found for other NtcA-activated genes (11).
The effect of N source and availability on expression of the nitrate/nitrite assimilation machinery of marine Synechococcus spp. has not been investigated in as much detail as that of freshwater cyanobacteria. However, ammonium control of ntcA expression in Synechococcus sp. strain WH 7803 is similar to freshwater Synechococcus spp. in that ammonium represses transcription substantially (13). Nitrate uptake is negligible in ammonium-grown cultures of this strain but is rapidly induced as ammonium is exhausted from the growth medium (8).
In the present study, we investigated the effect of nitrogen source and availability on the expression of genes involved in nitrogen assimilation in Synechococcus sp. strain WH 8103. Unlike strain WH 7803, which has a biliprotein complement that is characteristic of Synechococcus spp. found in shelf waters, strain WH 8103 is representative of open ocean strains. It is motile, produces phycoerythrins with high ratios of urobilin to erythrobilin chromophores, and is able to utilize urea as a nitrogen source (19, 21). Our results show that there are potentially ecologically important differences in N regulation in this marine cyanobacterium in comparison to freshwater strains and, in the case of ntcA at least, Synechococcus sp. strain WH 7803. These differences may reflect adaptations to life in a low-nutrient environment in which N concentrations are rarely, if ever, in great excess of demand.
MATERIALS AND METHODS
Culture conditions.
Stock cultures of Synechococcus sp. strain WH 8103 were incubated at 25°C under constant irradiance (20 μmol of photons m−2 s−1) in ASW medium (31). Experimental cultures were grown in modified ASW medium containing either 0.5 mM NaNO3 or 2 mM NH4Cl under the same conditions. In experiments in which NaNO2 (0.9 mM) was also supplied as the sole N source, cultures were incubated at a higher irradiance (60 μmol of photons m−2 s−1). Experimental cultures were maintained in the logarithmic phase of growth by regular dilution with fresh medium. For studies of the effect of nitrogen deprivation, cells from nitrate-grown cultures were collected by gentle filtration onto 90-mm-diameter, 0.6-μm-pore-size polycarbonate filters (Osmonics) and washed with several volumes of N-free ASW medium. The washed cells were resuspended to the original culture density in fresh N-free ASW medium and incubated under the same experimental conditions (continuous irradiance of 20 μmol of photons m−2 s−1; 25°C) for up to 24 h.
Isolation of genomic DNA.
Cell material from mid-logarithmic-phase cultures was harvested by centrifugation at 2,500 × g for 15 min at 4°C. Genomic DNA was extracted from the resulting cell pellet by a modified cetyltrimethylammonium bromide extraction procedure and further purified by cesium chloride density gradient centrifugation (3).
Amplification of narB, nirA, and ntcA gene fragments by PCR.
An ∼390-bp internal fragment of narB was amplified from genomic DNA with fully degenerate oligonucleotide primers (Table 1) targeting the conserved motifs TGQPNAM and MTNSER found in all cyanobacterial NarB peptide sequences entered in the GenBank database at the start of this study. The reaction conditions were as follows: 10 ng of template DNA, 12 pmol of each primer, 1 mM concentrations of each deoxynucleotide triphosphate, and 1 U of AmpliTaq DNA polymerase taken up in 50 μl of amplification buffer containing 1.5 mM Mg2+ (Perkin-Elmer). After denaturation at 95°C for 2 min, template DNA was amplified for 25 cycles (94°C for 1 min; 53°C for 1 min; 72°C for 1 min) followed by a final extension at 72°C for 10 min. PCR products were resolved on a 1.0% low-melting-point agarose gel, and fragments of the expected size were ligated into the T-tailed vector pCR2.1 and cloned in the Escherichia coli host strain INVα-F′ supplied with the vector (Invitrogen). Recombinant clones were grown overnight in Luria-Bertani broth plus 100 μg of ampicillin ml−1, plasmid DNA was purified with a commercial kit (Qiagen). The identity of the cloned fragment was verified by manual sequencing of both strands and by interrogating the GenBank database by using BlastP (National Center for Biotechnology Information [NCBI]) with the derived peptide sequence.
TABLE 1.
Oligonucleotide primers designed and used in this study
| Primer | Size of product (bp) | Primer use(s) | Sequence (5′-3′) | Temp (°C) |
|---|---|---|---|---|
| narF | 400 | Isolation of narB fragment | ACNGGYCARCCYAAYGCNATG | |
| narR | Isolation of narB fragment | CKHCKYTCNSWRTTNGTCAT | ||
| ntcAFor | 462 | Isolation of ntcA fragment, RT-PCR | GAACGCAACAAGACGATTTTCTT | 57.10 |
| ntcARev | Isolation of ntcA fragment, RT-PCR | GCTTCGGCAATGGCTTGATG | 59.40 | |
| glnAFor | 405 | RT-PCR | CACCGGTGAACCTCGTCTAT | 59.99 |
| glnARev | RT-PCR | TCGATCCAGTTGTCGATGAA | 60.20 | |
| narBFor | 352 | RT-PCR | GTTGGGTTACACCGAGCAGT | 60.04 |
| narBRev | RT-PCR | AGCACCAATGGGTAGGTGTC | 59.85 | |
| nirAFor | 288 | RT-PCR | TCGAAAACTCTCCACGCTCT | 60.13 |
| nirARev | RT-PCR | GGGAAACGCTGCAATAATGT | 59.97 | |
| nrtPFor | 244 | RT-PCR | CCTGACCTTTGTGGTCTGGT | 60.00 |
| nrtPRev | RT-PCR | GAACACCAGGATCGACGAGT | 60.12 | |
| rbcLFor | 193 | RT-PCR | CACCGACCTCGACTTCTACA | 58.88 |
| rbcLRev | RT-PCR | TGCGAGTTGATGTTCTCGTC | 59.99 | |
| nrtPtsp | NAa | Primer extension | GTGAGGTGAAGGGTTCGGTA | 59.40 |
| nirAext | NA | Primer extension | CCCGGGGACCTTTGTCGCTA | 63.50 |
| narBtsp | NA | Primer extension | TCACGGCTTGACCTTTCAC | 56.70 |
NA, not applicable.
A fragment of the coding sequence of nirA was obtained by serendipity during the course of an earlier investigation (J. T. Davies and M. Wyman, unpublished data). Genomic DNA was amplified with fully degenerate primers designed to target a 500-bp internal region of icfA (encoding carbonic anhydrase). PCR products of the expected size were isolated from a low-melting-point agarose gel and ligated into pCR2.1 as described for narB, and several of the resulting clones were sequenced. An NCBI BlastX search of the GenBank database revealed that the translated sequence of one of these clones (designated pCA500) shared significant similarity to the N terminus region of NirA from several other cyanobacteria.
Primers ntcAFOR and ntcAREV were designed for the amplification of ntcA from Synechococcus sp. strain WH 8103 (Table 1) and correspond to amino acids 43 to 50 and 190 to 196 of NtcA in Synechococcus sp. strain WH 8102. At an annealing temperature of 55°C, the primers amplified a single fragment of the expected size of approximately 460 bp. This product was cloned in pCR2.1 TOPO, transformed into competent E. coli TOP10F′ cells (Invitrogen), and sequenced on both strands as described above to confirm its identity. This primer pair was further used for reverse transcription (RT)-PCR.
Isolation and characterization of narB and nirA.
The cloned narB and nirA fragments were labeled with digoxigenin-dUTP (Roche) by PCR with either the original gene-specific primers (narB) or a primer pair targeting pCR2.1 vector sequences flanking the insert DNA (nirA). Probe DNA was run out in low-melting-point agarose gels to remove unincorporated nucleotides and recovered with a commercial kit (Hybaid) prior to use in Southern hybridizations.
Single- and double-restriction digests of genomic DNA were resolved in analytical agarose gels and Southern blotted to nylon membranes (Roche). The blots were hybridized overnight at 68°C with each of the labeled probes and subsequently washed at high stringency in 0.1× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate, pH 7.5) and 0.1% sodium dodecyl sulfate (SDS) at 68°C for 30 min two times. Hybrids were detected with alkaline phosphatase-conjugated anti-digoxigenin in conjunction with the chemiluminescent substrate CDP-Star as recommended by the supplier (Roche). The resulting luminograms revealed that the narB probe hybridized to a 9.4-kb HindIII-SalI fragment, whereas nirA was localized to a 4.5-kb HindIII-SmaI fragment. Subgenomic libraries were constructed in pUC18 following digestion of genomic and vector DNA with each restriction enzyme pair, and the ligated fragments were transformed into competent cells of E. coli strain XL1-Blue.
The libraries were screened by colony hybridization with the hybridization and stringency wash conditions described above. Plasmid DNA was isolated from overnight cultures of positively hybridizing clones and restriction mapped prior to the construction of a series of subclones for sequencing. Automated sequencing was performed on an ABI prism 377 DNA sequencer with the Perkin-Elmer Big Dye PCR sequencing kit.
Experimental conditions and isolation of RNA.
Cell material was pelleted by centrifugation at 2,500 × g for 5 min at 4°C. Total RNA was isolated by a hot phenol extraction protocol and treated with DNase as described previously (29). For the preparation of RNA for RT-PCR, an equivalent concentration (1% wt/vol) of lithium dodecyl sulfate was included in the extraction buffer in preference to SDS. Following purification, RNA was dissolved in deionized water treated with diethyl pyrocarbonate and taken up in an equal volume of 4 M NaCl. RNA was precipitated overnight at 4°C and recovered the following day by centrifugation at 12,000 × g for 20 min.
The integrity of all RNA preparations was verified by electrophoresis through RNase-free nondenaturing gels stained with ethidium bromide. Total RNA concentrations were estimated spectrophotometrically and by densitometry by using the 16S rRNA band as an internal standard following electrophoresis. Prior to experimentation, RNA concentrations were normalized by adjusting the volume of RNA added to the RT-PCR mixture after quantitative Northern analysis of a dilution series of each RNA preparation with a digoxigenin-dUTP-labeled fragment of the 16S rRNA gene amplified from Synechococcus sp. strain WH 78703 as described previously (30).
Preparation of probe DNA and analysis of gene expression by Northern blotting.
Digoxigenin-labeled probe DNA was prepared by the PCR with a commercial kit (Roche), M13 primers, and the following templates: pCA500 (nirA), pNar1.1 (narB), pNB4.21 (nrtP), pNtc1.1 (ntcA), pGlnA1.1 (glnA), and pRBGL1.3 (rbcL). The plasmids pCA500 and pNar1.1 respectively harbor the nirA and narB PCR products isolated in this study. pNB4.21 harbors a HindII-XhoI fragment that includes the 5′ end of nrtP, whereas pNtc1.1 harbors the ntcA PCR product. The clones pGlnA1.1 and pRBGL1.1 contain gene internal fragments of Synechococcus sp. strain WH 8103 glnA and rbcL, respectively, and have been described previously (30).
For Northern blots, 10-μg aliquots of RNA were fractionated through 1.2% agarose gels containing 0.22 M formaldehyde and 0.1 ng of ethidium bromide ml−1 and buffered with 20 mM morpholinepropanesulfonic acid (MOPS), 5 mM sodium acetate, and 1 mM EDTA. Prior to electrophoresis, RNA was denatured at 68°C for 3 to 5 min in 4× gel running buffer containing 9 mM formaldehyde, 0.8 mM EDTA (pH 8.0), 30% formamide, and 20% glycerol. Gels were run in the gel buffer solution containing 0.25 M formaldehyde, and after a 20-min wash in 0.05 M NaOH, RNA was transferred to positively charged nylon membranes (Roche) in 10× SSC. Hybridization was carried out at 50°C in digoxigenin EasyHyb solution (Roche), and stringency washes were performed at 68°C in 0.05× SSC-0.1% SDS. Hybrids were detected by immunochemistry (as described above) and quantified by densitometry following exposure to Kodak Biomax MR film as described previously (30).
Analysis of gene expression by RT-PCR.
RT-PCR was performed with the Qiagen OneStep RT-PCR kit and primer pairs designed to amplify internal fragments of ntcA, rbcL, glnA, narB, nirA, and nrtP (Table 1). The nucleotide sequences of rbcL and glnA used to design the primers for these genes have been published previously (29) while those of the remaining genes were determined during this study. RT was carried out at 50°C for 30 min in a 50-μl reaction mixture containing 0.1 to 1 ng of RNA and 30 pmol of each primer. Equal quantities of RNA were used for each set of experiments. HotStar Taq DNA polymerase was activated by incubation at 95°C for 15 min, and cDNAs were amplified for 25 to 36 cycles (94°C for 1 min; 55°C for 1 min; 72°C for 1 min) prior to a final extension at 72°C for 10 min. The resulting products were resolved by electrophoresis through 2.5% agarose gels stained with ethidium bromide and quantified by using a gel documentation system (model GDS8000) and the GelWorks one-dimensional advanced software package (UVP). For each experiment, the exponential phase of the PCR was determined empirically by varying the amount of RNA added at the start of the reaction and the number of amplification cycles. The linear response range of the charge-coupled device camera used to capture the RT-PCR gel images was established by loading a twofold dilution series of products on the gel and selecting the appropriate loadings for each set of samples.
Primer extension analysis.
RNA was extracted from nitrate- and ammonium-grown cells and from N-deprived cells and further purified as described above. The reaction conditions were as follows: 1 ng of total RNA (denatured at 65°C for 5 min immediately prior to addition); 2 μl of 10× Omniscript RT buffer (Qiagen); 10 μM sequence-specific primer (Table 1) (nrtPtsp binds 30 bp downstream of the nrtP start codon; nirAtsp binds 59 bp upstream of the nirA start codon); 0.5 mM (each) dGTP, dTTP, and dCTP; 4 μM dATP; 12.5 μCi of [α-32P]dATP; 10 U of RNasin (Promega); and 8 U of Omniscript reverse transcriptase in a total volume of 20 μl. The reaction mix was incubated at 37°C for 2 h. Plasmids pNB4 (nrtP) and pSma3 (nirA) were used to generate sequencing ladders to run alongside the primer extension products. DNA sequencing reactions were performed with the same specific primers with the AmpliCycle sequencing kit (Applied Biosystems) and labeled with [α-32P]dATP according to the manufacturer's recommendations. The extension products were resolved on a 6% gel polymerized with Sequagel XR diluted in Sequagel ultra pure complete buffer reagent (National Diagnostics).
Primer extension reactions were also performed as described by Ausubel et al. (3) with the primers described above and a narB-specific primer, narBtsp (Table 1), which binds between positions 82 and 101 of the coding sequence. To ascertain whether nrtP and narB are coexpressed, an RT-PCR was carried out with primers nrtPFor and narBtsp. RT-PCR was performed with RNA from nitrate-grown cells, a DNA-only control, and the reaction conditions described above.
Nucleotide sequence accession numbers.
The nucleotide sequences of nrtP, narB, and nirA from Synechococcus sp. strain WH 8103 have been submitted to GenBank (accession numbers AY377536, AF127144, and AFO65403, respectively).
RESULTS
Characterization of the narB, nrtP, and nirA genes of Synechococcus sp. strain WH 8103.
The narB coding region is 2,229 bp in length and determines a polypeptide of 756 amino acid residues with a predicted molecular mass of 85.9 kDa. The deduced peptide sequence shares 55% identity with NarB from Anabaena sp. strain PCC 7120 and contains the two conserved domains that are characteristic of NRs. A molybdopterin oxidoreductase domain is located between amino acids 6 and 593 that shares 32.8% identity with the consensus sequence determined in the NCBI conserved domain database (data not shown). The second conserved region is toward the C terminus of the protein and is 37.8% identical to the 111-residue consensus sequence of molybdopterin dinucleotide binding domains (Fig. 1).
FIG. 1.
Alignments of conserved domains found in NarB, NirA, and NrtP from Synechococcus sp. strain WH8103. Conserved residues are highlighted in boldface type; those (cysteine) conserved residues that are involved in cofactor binding are underlined. X, any amino acid.
Upstream of narB and in the same orientation is a 1,542-bp open reading frame that encodes a protein with an estimated mass of 55.2 kDa (Fig. 2) that shares 91 and 61% identity with nrtP from Synechococcus sp. strain WH 7803 (accession number AAG45172) and Synechococcus sp. strain PCC 7002, respectively, and which encodes a nitrate/nitrite permease (24). In common with other NRT2-type nitrate-nitrite porters (NNP), the derived peptide sequence of the putative nrtP gene from Synechococcus sp. strain WH 8103 has an N-terminal domain that is virtually identical to the NNP consensus sequence (Fig. 1).
The nirA coding region is 1,539 bp and determines a polypeptide of 513 amino acid residues with a calculated molecular mass of 58.7 kDa. Surprisingly, the percent G+C content of the nirA gene (42%) is substantially lower than that of narB (64.4%), which more closely reflects the average percent G+C content (55 to 62%) of the MC-A group of marine Synechococcus spp. to which strain WH 8103 belongs (21). The deduced NirA peptide sequence is 48% identical to that of Plectonema boryanum and contains the two conserved domains found in both NiRs and sulfite reductases. A nitrite/sulfite reductase ferredoxin-like half domain between residues 60 to 130 shares 52.8% identity with the consensus while the second half domain between residues 312 to 377 is 40% identical (Fig. 1). Close to the C terminus (residues 393 to 456), a conserved domain found in all higher plant and cyanobacterial NiRs and sulfite reductases includes the four conserved cysteine residues involved in binding the 4Fe-4S prosthetic group (Fig. 1).
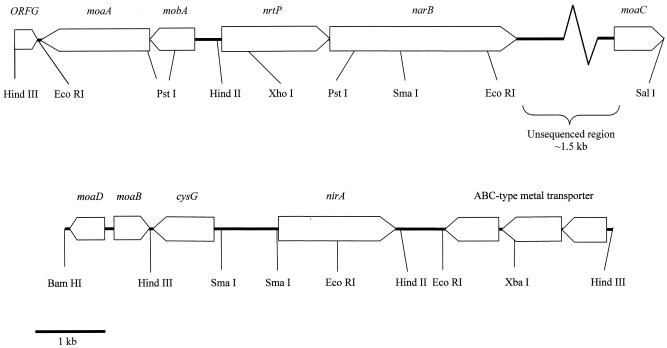
Restriction mapping and Southern blotting indicated that narB and nirA are quite closely linked on the chromosome (data not shown). Both genes are associated with a number of open reading frames whose gene products are predicted to be involved in molybdenum cofactor biosynthesis (Fig. 2). These include mobA and moaA, which are upstream of nrtP but in the opposite orientation, and cysG, moaB, and moaD upstream of nirA, in which moaB and moaD appear to be divergently transcribed. The overall gene arrangement is similar to that found in the recently sequenced genome of Synechococcus sp. strain WH 8102 (http://www.jgi.doe.gov/JGI_microbial/html/).
FIG. 2.
Map of the genomic regions harboring narB (top) and nirA (bottom) in Synechococcus sp. strain WH 8103.
Identification of putative NtcA-regulated promoters and primer extension analysis.
A putative NtcA-regulated promoter was identified 131 bp upstream of the nrtP translational start site with the structure GTAGCAATTCCAAC-N22-TTAACT, which closely matches the GTANCARNWRNTAC-N22-TAN3T consensus binding site established for NtcA-regulated promoters (11). The −10 region and 3′ flanking nucleotides share a primary structure (TTAACTGCCCGAG) similar to the NtcA-regulated transcriptional start point (tsp [underlined]) of the nirA operon of Synechococcus sp. strain PCC 7942 (15). A putative E. coli-like σ70 promoter with the structure GTAGCA-N22-TTN3T is also present, in which the −35 box overlaps the putative NtcA binding site, and is identical to the σ70-like promoter found upstream of ntcA in Synechococcus sp. strain PCC 7942 (15).
No recognizable promoters were identified in the 78-bp intergenic region between nrtP and narB, suggesting either that these genes are coexpressed from a common promoter upstream of nrtP or that the narB promoter lies within the coding region of nrtP. A sequence (GTGCTGTGACCTAC) that resembles an NtcA binding site is centered 197 bp upstream of the stop codon of nrtP but lacks a recognizable −10 box. As was the case with nrtP, no extension products that corresponded to initiation from a recognizable NtcA-regulated or σ70-like promoter were identified for narB. Unlike the DNA control, however, RT-PCR with ntrP and narB primers failed to yield a product. It seems likely, therefore, that narB is transcribed independently from nrtP from an as yet unidentified promoter (data not shown).
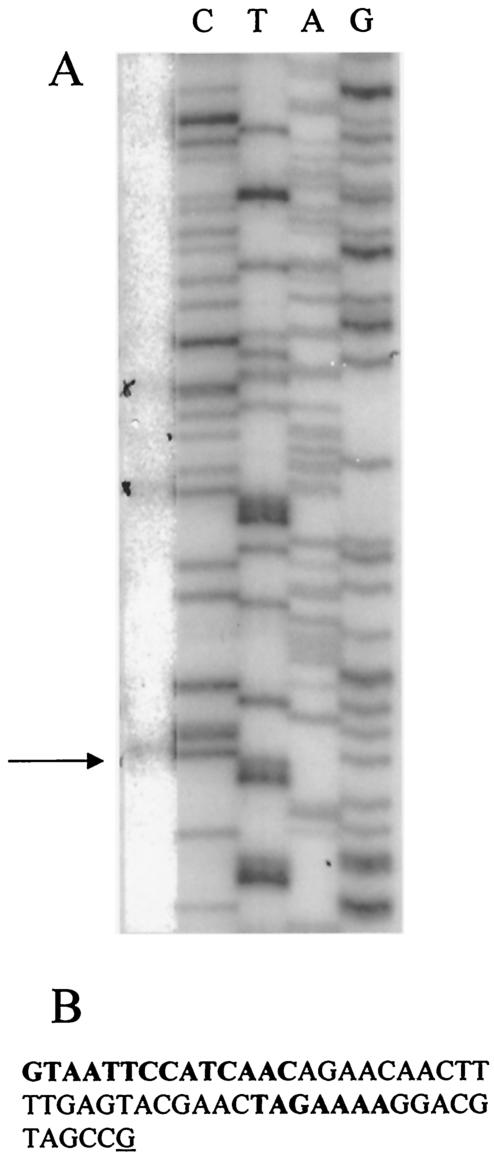
A further putative NtcA binding site was identified centered in a noncoding region 353 bp upstream of the start codon of nirA with the architecture GTAN8AAC-N22-TAGAAA. Like nrtP, a second putative E. coli-like σ70 promoter that has the structure TTTACA-N24-TAN3T is located at −248 with respect to the first initiation codon of nirA. Primer extension analysis revealed several possible transcriptional start points, including one identified only in RNA isolated from N-deprived cells that is flanked by the putative NtcA binding site and that initiates at a guanidine residue located 12 bp downstream of the −10 region (Fig. 3).
FIG. 3.
Primer extension analysis of the promoter region of nirA. (A) The left hand lane shows the extension product (arrow) obtained from RNA isolated from N-starved cells while the other lanes contain a sequencing ladder produced by using the same primer. (B) The nucleotide sequence upstream of the nirA transcriptional start point (base underlined). The nucleotide bases in boldface type are the putative NtcA binding site and associated −10 region and are separated by 22 nucleotides.
The influence of nitrogen source and nitrogen deprivation on transcriptional regulation of the nitrate/nitrite assimilatory system.
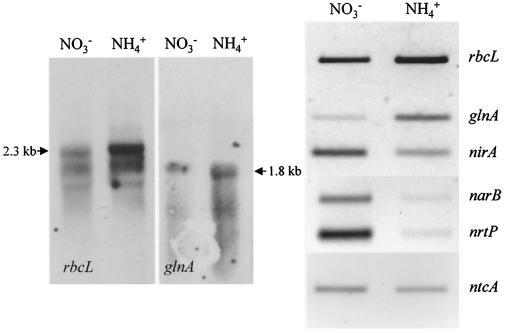
Northern analyses of rbcL and glnA revealed that both genes are substantially up-regulated in ammonium-grown cells (Fig. 4). The major rbcL transcript (2.3 kb) and that of glnA (1.8 kb) are at least two- to threefold-more abundant in comparison to nitrate-grown cells. Northern blots probed with nrtP, narB, or nirA all produced smeared hybridization signals that were difficult to quantify (data not shown) and which suggested that the mRNAs originating from these three genes are turned over rapidly. Therefore, a quantitative RT-PCR protocol was developed to investigate the expression of these genes and also that of rbcL, glnA, and ntcA.
FIG. 4.
Influence of nitrogen source on the expression of genes involved in carbon and nitrogen assimilation in Synechococcus sp. strain WH 8103 grown under low irradiance. The results of the Northern analyses are shown on the left side of the figure while the panels on the right show the results of the RT-PCR experiments conducted with equal quantities (1 ng) of RNA from cells grown with either nitrate or ammonium as the sole N source.
A very pronounced negative effect of ammonium on the abundance of narB and nrtP mRNAs was observed in low-light-grown cells; the abundance of both transcripts was ≤10% of that found in nitrate-grown cells (Fig. 4). By comparison, ammonium had a much smaller inhibitory effect on the abundance of nirA and ntcA mRNAs which were present at about half the concentrations found in nitrate-grown cells. Consistent with the results of the Northern analysis, RT-PCR confirmed that rbcL and glnA were both up-regulated in ammonium-grown cultures.
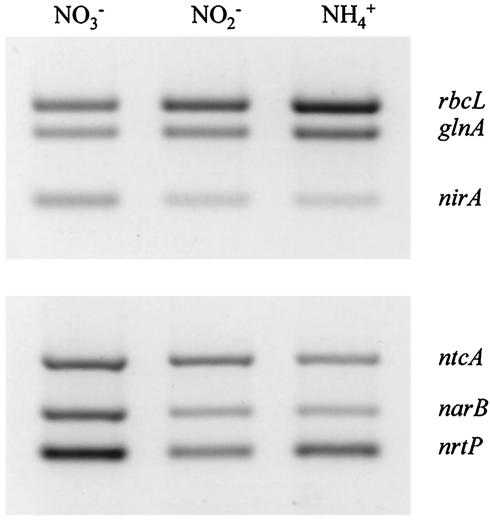
A similar effect of ammonium on the abundance of glnA and rbcL mRNAs was also observed in high-light-grown cells (Fig. 5). Likewise, nirA mRNA was at ∼50% of the concentrations found in nitrate-grown cells, but surprisingly, ammonium had a far less pronounced negative effect on the transcription of nrtP and narB under this light regimen. The abundance of nrtP mRNA was ∼60% of that found in nitrate-grown cells while that of narB was just under 40% (cf. ≤10% for both transcripts under low light). With the exception of nrtP, which was down-regulated somewhat, the overall abundance of transcripts of the other five genes investigated in nitrite-grown cells was intermediate between that found in ammonium- and nitrate-grown cells.
FIG. 5.
Influence of nitrogen source on the expression of genes involved in carbon and nitrogen assimilation in Synechococcus sp. strain WH 8103 grown under high irradiance. The panels show the results of RT-PCR experiments conducted with equal quantities (1 ng) of RNA from cells grown with either nitrate, nitrite, or ammonium as the sole N source.
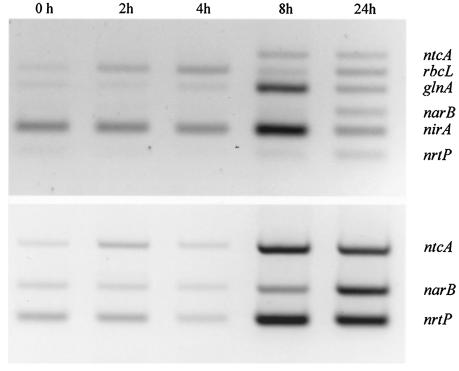
The influence of nitrogen-deprivation on gene expression was also investigated by RT-PCR. Apart from a steady increase in the abundance of rbcL mRNA, little change in the overall pattern of gene expression was detected during the first 4 h of N deprivation (Fig. 6). The abundance of ntcA, glnA, nirA, narB, and nrtP transcripts all increased over the next 4 h, whereas conversely, rbcL expression declined somewhat. After 8 h the abundance of glnA and nirA mRNAs had increased ∼10- and ∼3-fold, respectively, whereas ntcA and nrtP transcripts had increased ∼10- and ∼5-fold, respectively, in comparison to nitrate grown cells. Expression of ntcA and nrtP mRNAs was still elevated after 24 h, at which time point the abundance of narB mRNA reached the highest concentration observed, increasing a further 2.5-fold over that found at 8 h and ∼10-fold in comparison to unstarved cells.
FIG. 6.
Influence of N deprivation on the expression of genes involved in carbon and nitrogen assimilation in Synechococcus sp. strain WH 8103. RNA was extracted from cells at the times indicated, up to 24 h after transfer to N-free medium. The abundance of mRNAs was determined by RT-PCR with 30 (top panel) or 36 (bottom panel) cycles of the PCR.
DISCUSSION
Isolation and characterization of nirA, narB, and nrtP from Synechococcus sp. strain WH 8103.
The genes involved in nitrate and nitrite assimilation in Synechococcus sp. strain WH 8103 were isolated and characterized during the present study. The identities of nirA, narB, and nrtP were inferred by sequence similarities to known genes and by interrogation of the NCBI conserved domain database. Both nirA and narB contain the conserved domains found in other NiRs and NRs, and their peptide sequences share significant identity with those from other cyanobacteria. Together with nrtP, which is >90% identical to the homologue present in Synechococcus sp. strain WH 7803, these genes constitute the structural components of the nitrate/nitrite assimilation apparatus of this marine cyanobacterium.
Like the euryhaline Synechococcus sp. strain PCC 7002 (24) and the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH 9601 (26), the narB gene of Synechococcus sp. strain WH 8103 is closely linked with an open reading frame homologous to nrtP. This gene encodes a nitrate/nitrite permease rather than the component genes (nrtABCD) of an ABC-type nitrate/nitrite transporter found upstream of narB in most freshwater cyanobacteria. The presence of nrtP in Synechococcus sp. strain WH 8103 lends further support to the prediction that there are significant differences between the nitrate/nitrite transporters present in marine and freshwater cyanobacteria (24). A copy of nrtP is also found upstream of narB in the recently completed genome of the closely related isolate, Synechococcus sp. strain WH 8102. Intriguingly, components of an ABC-type transporter with significant similarity to nrtABCD are also present in the genome of Synechococcus sp. strain WH 8102 (http://genome.ornl.gov/microbial/syn_wh/embl/syn_wh_P_cogs.html). Whether these genes are functional remains to be investigated, but in Synechococcus sp. strain PCC 7002 at least, NrtP has been shown to be the primary bispecific transporter (24).
In Synechococcus sp. strain PCC 7002, nrtP and narB occur in the same orientation (24) as that found for Synechococcus sp. strain WH 8103. Unlike Synechococcus sp. strain WH 8103, however, both genes have NtcA-like promoters in their upstream regions in this euryhaline strain and Northern analyses confirmed that nrtP and narB are transcribed as monocistronic mRNAs (24). The lack of a recognizable promoter in the much shorter intergenic region (78 bp) located between nrtP and narB in Synechococcus sp. strain WH 8103 suggested that these genes might be coexpressed from a common promoter upstream of nrtP. Despite a number of attempts to map the 5′ end of nrtP and narB mRNAs by primer extension, it was not possible to verify this experimentally. The failure to amplify the intergenic region between these genes by RT-PCR, however, strongly suggests that these genes are transcribed independently.
Although the transporter is different, the overall arrangement of the genes encoding the nitrate/nitrite assimilation system in Trichodesmium sp. strain WH 9601 (nirA, nrtP, and narB) resembles that of Synechococcus sp. strain PCC 7942 (26). The nirA gene is in a separate transcriptional unit from narB in Synechococcus sp. strain WH 8103, however, and the comparatively minor negative effect of ammonium on nirA transcription when compared to its freshwater counterpart indicates that unlike Synechococcus sp. strain PCC 7942, nirA and narB are differentially regulated in this marine cyanobacterium. Mapping of the 5′ end of nirA mRNA identified a transcriptional start site (tsp) 353 bp upstream of the initiation codon that has an NtcA-like promoter in the upstream flanking sequence. This tsp was only detected in N-starved cells, however, in which nirA is transiently up-regulated. This suggests that this promoter is active only under nitrogen deprivation and that it differs from the constitutive promoter active in unstarved cells.
The NtcA-binding site upstream of nirA and its 5′ and 3′ flanking sequences has a structure AATCGTAATTCCATCAACAGAA that deviates in a number of nucleotide positions (matches underlined) from the NtcA consensus promoter and therefore is unlikely to bind NtcA with the same efficiency (11). Nevertheless, this NtcA-like promoter should be functional at the elevated NtcA concentrations that are likely to be present under N deprivation (25), since ntcA is itself substantially up-regulated. The effect of prolonged N deprivation on the overall abundance of the nirA message was not as dramatic as that observed for nrtP, narB, or glnA; however, this might be because expression of nirA is comparatively high in unstarved cells irrespective of the N source supplied for growth.
Intriguingly, the moles percent G+C content of the coding region of nirA (42%) is considerably lower than that typical of the genomes of the MC-A group of Synechococcus spp. (mol% G+C, 55 to 62) (22). Phylogenetic analysis of nirA, including the draft nirA peptide sequence submitted to GenBank during an earlier part of this study, revealed that the Synechococcus sp. strain WH 8103 gene grouped more closely with higher plant and green algal sequences rather than with typical cyanobacterial NiRs (26). While the overall topology of the trees varied slightly after several sequencing ambiguities had been corrected post-primary sequence submission, our own phylogenetic analyses confirmed the relatedness with eukaryotic sequences (4). Horizontal gene transfer appears to have occurred comparatively recently in marine Synechococcus spp. (28), but whether the nirA gene has been inherited in this manner or is an example of convergent evolution is a matter of conjecture.
Effect of nitrogen source and availability on gene expression.
In addition to the comparative lack of ammonium control in regulating transcription of nirA, a further remarkable feature of nitrogen metabolism in Synechococcus sp. strain WH 8103 is the up-regulation of glnA in ammonium-grown cells. In both Synechococcus sp. strain PCC 7942 and Synechocystis sp. strain PCC 6803, glnA is substantially down-regulated by ammonium, although to a lesser extent in the latter case (23). Why Synechococcus sp. strain WH 8103 differs in this regard is not clear. The growth rate of Synechococcus sp. strain WH 8103 utilizing ammonium is a little higher than that with nitrate, but it seems unlikely that the observed increase in glnA mRNA abundance could be due entirely to global effects on transcription. Interestingly, GS activity in Prochlorococcus marinus strain PCC 9511 is not enhanced when switched from ammonium to nitrogen-free medium (5), indicating that unlike other cyanobacteria (17) GS is not subject to N control in this cyanobacterium. Like Synechococcus sp. strain WH 8103, transcription of ntcA is up-regulated during N-deprivation, whereas amt1, encoding a high-affinity ammonium transporter, is not (12).
One possible explanation for the observed up-regulation of glnA by ammonium in Synechococcus sp. strain WH 8103 is that NR and NiR compete with RubisCO for photosynthetically generated reductant in nitrate-grown cells. In comparison to ammonium-grown cells, this may limit the supply of 2-oxoglutarate that is essential for the incorporation of nitrogen through the GS/glutamate synthase pathway (11). Since at the transcriptional level at least, glnA expression is not regulated by the N source, enhanced transcription may be a response to the greater availability of C skeletons for glutamate synthesis in ammonium-grown cells. The lack of competition for reducing power to support CO2 fixation may also explain why rbcL is also up-regulated by ammonium. In natural populations of Synechococcus spp., the steady accumulation of photosynthate during the first few hours of sunlight and the effect this has on the cell C-N balance has been put forward as an explanation for the observed increase in the abundance of glnA mRNA during the latter half of the of the diel cycle (29).
The lack of repression of glnA and the relatively high level of expression of nirA and ntcA in ammonium-grown cells point to some unique features of nitrogen control in this marine cyanobacterium. All of the genes involved in N assimilation that were investigated are responsive to nitrogen deprivation and in most cases are up-regulated substantially in what appears to be an NtcA-dependent manner. Both nrtP and narB are also tightly regulated by ammonium (and particularly so under low irradiance), but this is clearly not the case for either glnA, nirA, or indeed, ntcA itself. The lesser degree of ammonium inhibition of nrtP and narB transcription at higher irradiance is of interest and suggests that the expression of these genes might be redox-responsive, as has been shown for glnA and ntcA in Synechocystis sp. strain PCC 6803 (2, 22).
None of the genes sequenced in this study have a canonical NtcA-type promoter, but the putative promoter located upstream of nrtP is likely to bind NtcA far more efficiently than that identified for nirA (11). This variability in the degree of conservation in the putative NtcA-like promoters for nrtP and nirA might help explain why the response to ammonium differs. The nrtP promoter should compete for NtcA much more effectively than that for nirA at the considerably lower concentrations of this transcription factor likely to be found in N-replete cells. Perhaps surprisingly, a search of the genome sequence of the closely related Synechococcus sp. strain WH 8102 revealed only one putative promoter and its associated −10 box (GTAATAACTACTAC-N22-TAATTT) that matches the NtcA consensus sequence. Intriguingly, this was found upstream of ntcA.
Concluding remarks.
The present study has shown that, unusually, nirA is expressed at comparatively high levels in the presence of ammonium in Synechococcus sp. strain WH 8103 and that there is at least the potential for coutilization of both ammonium and nitrite. What remains to be established is whether the nirA message is translated in ammonium-grown cells and whether they do indeed maintain a capacity to utilize nitrite. We are investigating this currently, and since we have shown that nrtP is down-regulated by ammonium, one part of this investigation will examine whether there is a second nitrite-specific active transport system, as has been shown for Synechococcus sp. strain PCC 7942 (16). In view of the low N concentrations and abundant energy supply available in well-illuminated oceanic surface waters, the potential competitive benefits of retaining a capacity to coutilize different forms of combined N may offset the obvious biosynthetic and maintenance costs involved. The majority of Prochlorococcus spp. appear to have adopted a quite different strategy by losing the capacity to utilize oxidized forms of combined N and reducing maintenance costs by compacting the genome such that it is now the smallest of any known photosynthetic prokaryote.
Acknowledgments
This research was supported by a studentship to C.B. (ref. GT04/97/280/MAS) from the Natural Environment Research Council of the United Kingdom.
REFERENCES
- 1.Aichi, M., N. Takatani, and T. Omata. 2001. Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfonso, M., I. Perewoska, and D. Kirilovsky. 2001. Redox control of ntcA gene expression in Synechocystis sp. PCC 6803. Nitrogen availability and electron transport regulate the levels of NtcA protein. Plant Physiol. 125:969-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 4.Bird, C. 2001. Molecular cloning and characterisation of genes involved in nitrate/nitrite assimilation in the marine cyanobacterium, Synechococcus sp. strain WH 8103. Ph.D. thesis. University of Stirling, Stirling, United Kingdom.
- 5.El Alaoui, S., J. Diez, L. Humanes, F. Toribio, F. Partensky, and J. M. Garcia-Fernandez. 2001. In vivo regulation of glutamine synthetase activity in the marine chlorophyll b-containing cyanobacterium Prochlorococcus sp. strain PCC 9511 (Oxyphotobacteria). Appl. Environ. Microbiol. 67:2202-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glibert, P. M., J. C. Goldman, and E. J. Carpenter. 1982. Seasonal variations in the utilisation of ammonium and nitrate by phytoplankton in Vineyard Sound, Massachusetts, USA. Mar. Biol. 70:237-249. [Google Scholar]
- 8.Glibert, P. M., and R. T. Ray. 1990. Different patterns of growth and nitrogen uptake in two clones of marine Synechococcus spp. Mar. Biol. 107:273-280. [Google Scholar]
- 9.Glover, H. E., B. B. Prézelin, L. Campbell, M. Wyman, and C. Garside. 1988. A nitrate dependent Synechococcus bloom in surface Sargasso Sea water. Nature 331:161-163. [Google Scholar]
- 10.Harrison, W. G., L. R. Harris, and B. D. Irwin. 1996. The kinetics of nitrogen utilization in the oceanic mixed layer: nitrate and ammonium interactions at nanomolar concentrations. Limnol. Oceanogr. 41:16-32. [Google Scholar]
- 11.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindell, D., D. Erdner, D. Marie, O. Prasil, M. Koblizek, F. Le Gall, R. Rippka, F. Partensky, D. J. Scanlan, A. F. Post. 2002. Nitrogen stress response of Prochlorococcus strain PCC 9511 (Oxyphotobacteria) involves contrasting regulation of ntcA and amt1. J. Phycol. 38:1113-1124. [Google Scholar]
- 13.Lindell, D., E. Padan, and A. F. Post. 1998. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luque, I., A. Contreras, G. Zabulon, A. Herrero, and J. Houmard. 2002. Expression of the glutamyl-tRNA synthetase gene from the cyanobacterium Synechococcus sp. PCC 7942 depends on nitrogen availability and the global regulator NtcA. Mol. Microbiol. 46:1157-1167. [DOI] [PubMed] [Google Scholar]
- 15.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda, S., M. Okamura, M. Kobayashi, and T. Omata. 1998. Nitrite-specific active transport system of the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180:6761-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merida, A., P. Candau, and F. J. Florencio. 1991. Regulation of glutamine synthetase activity in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 by the nitrogen source: effect of ammonium. J. Bacteriol. 173:4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilisation of different nitrogen sources by marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47:989-996. [Google Scholar]
- 19.Olson, R. J., S. W. Chisholm, E. R. Zettler, and E. V. Armbrust. 1988. Analysis of Synechococcus pigment types in the sea using single and dual beam flow cytometry. Deep Sea Res. 35:425-440. [Google Scholar]
- 20.Partensky, F., J. Blanchot, and D. Vaulot. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. 19:457-476. [Google Scholar]
- 21.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyes, J. C., and F. J. Florencio. 1995. Electron-transport controls transcription of the glutamine-synthetase gene (glnA) from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 27:789-799. [DOI] [PubMed] [Google Scholar]
- 23.Reyes, J. C., I. Muro-Pastor, and F. J. Florencio. 1997. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differentially regulated in response to nitrogen availability. J. Bacteriol. 179:2678-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakamoto, T., K. Inoue-Sakamoto, and D. Bryant. 1999. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 181:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaquez-Bermudez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71-74. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Q., H. Li, and A. Post. 2000. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J. Bacteriol. 182:1764-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214:71-120. [Google Scholar]
- 28.Watson, G. M. F., and F. R. Tabita. 1996. Regulation, unique gene organisation, and unusual primary structure of carbon fixation genes from a marine phycoerythrin-containing cyanobacterium. Plant Mol. Biol. 32:1103-1115. [DOI] [PubMed] [Google Scholar]
- 29.Wyman, M. 1999. Diel rhythms in ribulose-1, 5-bisphosphate carboxylase/oxygenase and glutamine synthetase gene expression in a natural population of marine picoplanktonic cyanobacteria (Synechococcus spp.). Appl. Environ. Microbiol. 65:3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyman, M., J. T. Davies, D. W. Crawford, and D. A. Purdie. 2000. Molecular and physiological responses of two classes of marine phytoplankton (diatoms and prymnesiophytes) during the development of nutrient-stimulated blooms. Appl. Environ. Microbiol. 66:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyman, M., R. P. F. Gregory, and N. G. Carr. 1985. Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science 230:818-820. [DOI] [PubMed] [Google Scholar]
- 32.Zubkov, M. V., B. M. Fuchs, G. A. Tarran, P. H. Burkill, and R. Amann. 2003. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl. Environ. Microbiol. 69:1299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]