Abstract
Background & objectives:
Group A streptococcal (GAS) pharyngitis, especially among children, leads to high prevalence of rheumatic fever (RF)/rheumatic heart disease (RHD) in India, as compared to the western world where invasive diseases are common. GAS encodes numerous virulence factors that cause diseases by exhibiting extraordinary biological diversity. Hence, we studied the virulence factors genes of GAS isolated from the throat of children with pharyngitis and also asymptomatic carriers.
Methods:
Fifty GAS isolates cultured from throats of north Indian children aged 5-15 yr with mild pharyngitis (20), severe pharyngitis (24) and asymptomatic pharyngeal carriers (6), during 2000-2003 along with reference M1 strain were emm typed and characterized for virulence factors genes by PCR. The presence of virulence factors was also checked for their association with emm type in pharyngitis.
Results:
Twenty emm types, six sequence types, and one non-typeable strain were found circulating in north India. The five most prevalent types were emm 74 (12%), 11 & StI129 (8% each) and emm 68 and NS292 (6% each). The spe B gene was found to be significantly higher (P=0.0007) in opacity factor (OF) negative isolates. emm 3, 11, 77, 86, 87, 109 and StI129 showed maximum virulence factors genes.
Interpretation & conclusions:
GAS isolates collected from throats of children from north India possess highly virulent antigens. This study also supports concept of isolate-associated virulence rather than type relatedness.
Keywords: emm types, GAS, virulence factors
Group A Streptococcus (GAS, Streptococcus pyogenes) shows an incredible history of changing disease pattern1 with numerous cell surface associated and secretary factors required for adherence and colonization of the host at various sites, destruction of the tissues for facilitating the spread, and other systemic effects causing autoimmune complications2. Proteomic analysis of GAS recently identified 79 proteins on its surface and 21 cytoplasmic proteins3. Streptococcal adherence to host pharyngeal epithelium is the basic step in colonization. Environmental conditions, cell density and growth phase are known to influence expression of virulence factors4,5.
Globally, GAS has emerged as a highly variable organism. High prevalence of rheumatic fever (RF)/rheumatic heart disease (RHD) in India in comparison to invasive diseases from developed nations and heterogeneity of GAS emm types have strengthened the need for looking into their virulence potential. The association of virulence factors, whether protein-binding or exotoxin with pharyngitis that ultimately leads to RF/RHD is poorly understood in Indian scenario. Therefore, an attempt was made to study the distribution of genes coding such factors to facilitate understanding of bacterial pathogenesis in GAS isolates from throat carriers and pharyngitis cases.
Material & Methods
A total of 50 clinical GAS isolates, collected from children aged 5-15 yr from Raipur Rani block of Panchkula, district Haryana and Government Medical College, Chandigarh (November 2000 to July 2003), identified by a Streptex Murex kit (Remel Europe Ltd, UK) and one M1 reference GAS strain were used in the present study. GAS isolates were categorized on the basis of severity of signs and symptoms of GAS pharyngitis into three groups: severe pharyngitis (n=24); mild pharyngitis (n=20) and no pharyngitis (n=6).
The opacity factor (OF) of these isolates was determined in a 96 well microplate6 and the results were read at 450 nm with an ELISA microplate reader (Tecan Austria, GmbH). Twenty seven GAS isolates (54%) were OF negative; 54 per cent in severe, 50 per cent in mild pharyngitis and 66 per cent in those without pharyngitis, i.e., in almost equal proportion among the three clinical categories.
Genomic DNA was isolated by Qiagen kit (Dneasy Tissue Kit, Qiagen GmbH, Germany) as per manufacturer’s instructions and preparations of OD260/OD280 >1.8 were used as template for PCR of different virulence factors. emm gene was identified using specific primers and following standardized PCR thermo cycling conditions7. PCR products were checked on 0.8 per cent gel, analyzed for yield and then purified by QIAGEN PCR purification kit. The emm gene sequencing was done in an ABI 377 Automated Sequencer as per manufacturer’s instructions (Applied Biosystems, USA) and then gene sequence was searched for homology at CDC website as described earlier7.
The isolated DNA was used for amplifying nucleotide sequences corresponding to the regions of selected virulence factors: slo, ska, pbp, spe A, spe B, spe C, prtf, sfb, fbp-54, scp and sic by PCR. The PCR reaction mixture with specific primer of different virulence factors was prepared. All reagents were procured from Roche Molecular Biochemicals (Boehringer Manheim, Germany). PCR products were electrophoresed through 1.5 per cent agarose gel and separated bands were visualized under UV trans-illuminator after ethidium bromide staining (Sigma Chemicals Co, USA) and analyzed using Video Gel Documentation System (Imagemaster gel documentation system, M/S Bio-Rad Laboratories Pvt Ltd, Australia). EPI-6 (Epi Info Version 6) statcalc software was used for data analysis. Chi square test was applied and P<0.05 was considered to be statistically significant.
Results & Discussion
Significant diversity of emm types was observed. A total of 27 emm types; 20 known emm types, 6 sequence-types and a novel M non typeable strain were identified. The five most prevalent types were: emm 74 (12%), 11 and StI129 (8% each) and emm 68 and NS292 (6% each). Majority (65%, 13/20) of the isolates belonging to the five most prevalent types were OF negative; all the four isolates of emm11; 5 out of the 6 emm74 isolates and 3 out of the 4 StI129 isolates were OF negative, however, all the three isolates of emm 68 and two out of the three isolates of NS292 were OF positive. Amongst the less prevalent types emm 3, 28, 77, 49 and 74 were OF negative, and emm 2.1, 60, 68, 75, 81, 109 were OF positive (Table).
Table.
Distribution of virulence factor genes in group A Streptococcus emm types
| Sr. No. | emm type | OF | slo | ska | pbp | spe A | spe B | spe C | fbp-54 | sfb | prtf | scp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Presence of virulence factor genes | ||||||||||||
| Reference strain M1 | - | + | + | + | + | + | - | + | - | - | + | |
| Severe Pharyngitis | ||||||||||||
| 1 | AAB5113 | - | + | + | + | + | + | - | + | - | + | + |
| 2 | Sp11014/VT-15 | + | + | + | + | - | + | - | + | - | + | + |
| 3 | Sp11014/VT-15 | - | + | + | + | - | + | - | + | + | + | + |
| 4 | StI129 | + | + | + | + | - | + | + | + | + | + | + |
| 5 | 74 | - | + | + | + | - | + | - | + | + | + | + |
| 6 | 87 | + | + | + | + | + | + | - | + | + | + | + |
| 7 | 3 | - | + | + | + | + | + | - | + | + | + | + |
| 8 | NS292 | + | + | + | + | + | + | - | + | + | + | - |
| 9 | 74 | - | + | + | + | - | + | + | + | + | - | + |
| 10 | AAL28405 | - | + | + | + | + | + | + | + | - | + | + |
| 11 | 109 | + | + | + | + | - | + | - | + | + | + | + |
| 12 | 74 | - | + | + | - | - | + | - | + | + | + | - |
| 13 | 11 | - | + | + | + | - | - | - | + | + | + | + |
| 14 | 81 | + | + | + | + | - | + | - | + | - | + | + |
| 15 | 71 | - | + | + | + | - | + | - | + | + | + | + |
| 16 | 49 | + | + | + | + | + | + | - | + | - | + | + |
| 17 | 68 | + | - | + | + | - | + | + | + | + | + | + |
| 18 | 102.1 | - | + | + | + | - | + | + | + | - | - | + |
| 19 | 3 | - | + | + | + | + | + | - | + | + | - | + |
| 20 | 86 | + | + | + | + | + | + | - | + | + | + | + |
| 21 | 77 | - | + | + | + | + | + | - | + | + | + | + |
| 22 | 74 | + | + | + | + | + | - | - | - | - | + | + |
| 23 | StI129 | - | + | + | + | + | - | + | + | - | - | + |
| 24 | 60 | + | + | - | + | - | + | - | + | - | + | + |
| Mild Pharyngitis | ||||||||||||
| 25 | 68 | + | + | + | + | - | + | + | + | - | - | + |
| 26 | 28 | - | + | + | - | - | - | - | - | - | + | - |
| 27 | TP-c2135 | - | + | + | + | - | + | + | + | + | - | + |
| 28 | 89 | + | + | + | + | + | + | - | + | - | - | + |
| 29 | 75 | + | + | + | - | - | + | - | - | - | - | - |
| 30 | 74 | - | + | + | + | - | + | - | + | - | - | + |
| 31 | 11 | - | + | + | + | + | - | + | + | + | - | + |
| 32 | 77 | - | + | + | + | - | + | - | + | - | + | + |
| 33 | 11 | - | + | + | + | + | + | + | + | - | + | + |
| 34 | NS292 | + | + | + | + | - | + | - | + | - | + | + |
| 35 | 74 | - | + | + | + | + | + | - | + | + | - | + |
| 36 | 2.1 | + | + | + | + | - | + | + | + | + | - | + |
| 37 | 11 | - | + | + | + | - | + | - | + | + | + | + |
| 38 | 49 | - | - | + | + | + | + | - | + | - | - | + |
| 39 | 65 | + | + | + | + | - | + | - | + | + | + | + |
| 40 | 81 | + | + | + | + | - | + | - | + | + | + | + |
| 41 | 93 | - | + | + | - | + | + | - | + | - | + | + |
| 42 | 42 | + | + | + | + | + | + | + | + | - | + | + |
| 43 | SP10741/Allele75 | + | + | + | + | + | - | - | - | + | - | + |
| 44 | 71 | + | + | + | - | - | - | - | + | - | + | - |
| Asymptomatic | ||||||||||||
| 45 | 109 | + | + | + | + | + | + | - | + | + | + | + |
| 46 | 68 | + | + | + | + | - | + | - | - | - | + | + |
| 47 | stI129 | - | + | + | - | + | - | - | - | - | + | + |
| 48 | AAL28405 | - | + | + | - | - | - | - | - | - | - | - |
| 49 | NS292 | - | + | + | - | - | - | - | - | - | - | - |
| 50 | StI129 | - | + | + | - | - | - | - | - | - | - | - |
| Frequency of Clinical Isolates | ||||||||||||
| % | 54 | 96 | 98 | 82 | 42 | 78 | 24 | 82 | 50 | 72 | 84 | |
Percentage of isolates with following genotype = No. positive/No. tested (50) X 100. OF - opacity factor, slo - streptolysin, ska - streptokinase, pbp - plasminogen binding protein, spe A, spe B & spe C - streptococcus pyogenic exotoxin A, B , respectively, fbp-54 - fibronectin binding protein, sfb - streptococcal fibronectin binding protein, prtf - fibronectin binding protein F and scp- C5a peptidase. + presence, - absence
The amplification for slo, ska, sfb, fbp-54, prtf, spe A, spe B, spe C, scp and pbp genes when checked on agarose gel was found to be of <500, ~400, ~1400, 1500, <500, ~900, 1400, 1000, <1000 and 1200-1500 base pair in size, respectively. None of the isolates were positive for sic gene. When the OF production was compared with the prevalence of virulence genes, spe B was found to be statistically more common in OF negative isolates (p=0.0007), indicating its role in the pathogenesis.
The presence of slo and ska was comparable to the previous studies8,9. Most of the GAS strains encode spe B while spe A and spe C occur less frequently. The strains associated with severe infections have been shown to produce spe A toxin9,10. Although there are few reports on the occurrence of scarlet fever and toxic shock syndrome from India, comparatively high frequency of spe A (42%) from non-invasive pharyngitis cases was found from worldwide9,11. In the past Nandi et al12 demonstrated low frequency of spe A gene (8.3%) within Indian isolates, indicating their less virulent nature, but the present study gives an indication that virulent strains are circulating within the Indian community. Such strains which have ability to produce specific exotoxins, in the absence of type specific immunity in a population, may lead to a community outbreak of streptococcal infections. spe B gene is assumed to be chromosomal encoded and highly conserved. Tyler et al9 showed spe B gene in all or majority of the GAS isolates, however all strains may not have it. Comparatively low frequency (78%) of spe B gene was found in our study than 93.3 per cent observed in the past from the same region12 and 99.3 per cent from Canada9 indicating that it might not have been retained during the course of evolutionary events that have also generated extensive genomic diversity in our population. The gene frequency for spe C (24%) was comparable with earlier study9,11. Almost all strains of GAS possess scp gene, similar to earlier report13. Fbp-54 gene has been reported in all clinical isolates, however its expression differs quantitatively14, in present study it was 82 per cent. Our frequency of prtf gene (72%) was similar to observations from Japan with prevalence of 77.3 per cent15. The sfb gene proportion was also comparable to published data with the frequency ranging between 50-70 per cent in all clinical isolates16. We could not find any GAS isolate positive for sic gene in contrast to closely related sic (crs) and distantly related sic (drs) genes encoding this protein reported recently from India and Japan15,17. It has been reported that all M1 and M57 strains have sic (crs); and all M12 and M55 strains have sic (drs) and hence express the respective proteins18. We could not isolate M1, 12, 55 and 57 types in our sample that could be well associated with the absence of sic gene from our GAS isolates.
The reference M1 strain possessed maximum virulence factors tested except spe C, sfb and prtf. The most commonly and least found genes were ska and spe C respectively. The most predominant emm types had almost all the virulence factors except sic: emm 11, 42 and AAL28405 lacked sfb gene; StI129 lacked spe A gene; and emm 87, 3, 86, 77 and 109 did not have spe C gene. AAL28405, emm 42 and 11 had genes for three exotoxins, and emm 11, 65, 81, 74, 87, 3, 109, 74, 11, 71, 68, 86, 77, NS292, StI129 and Sp11014/VT-15 possessed all the three fibronectin binding protein (FBP) genes. Hence emm 3, 11, 77, 86, 87, 109 and StI129 showed maximum number of virulence factors. The most prevalent five types (20 isolates) showed frequency of slo, ska, spe A, spe B, spe C, fbp-54, sfb, prtf, pbp and scp virulence factors genes in 95, 100, 35, 65, 35, 75, 50, 70, 80 and 80 per cent isolates respectively.
Varied genotypic combinations of the selected virulence factors were obtained in the GAS isolated from patients with throat infection of different severity (Table). Slo, ska, pbp, fbp54, prtf and scp encoding for major adherence/attachment factors were found in nearly 90 per cent of the severe pharyngitis cases whereas asymptomatic cases predominantly showed slo, ska and prtf. Significant association (P<0.05) was observed for the virulence factor genes in the three clinical pharyngitis categories (Fig.). The frequency of genes pbp, spe B and fbp-54 was significantly high in severe pharyngitis cases in comparison to asymptomatic cases (P<0.05). Significantly higher frequency of prtf (P=0.04) was seen in severe pharyngitis as compared to the ones isolated from mild pharyngitis cases. The frequency of fbp-54 was significantly high (P=0.01) in mild pharyngitis compared to asymptomatic cases.
Fig.
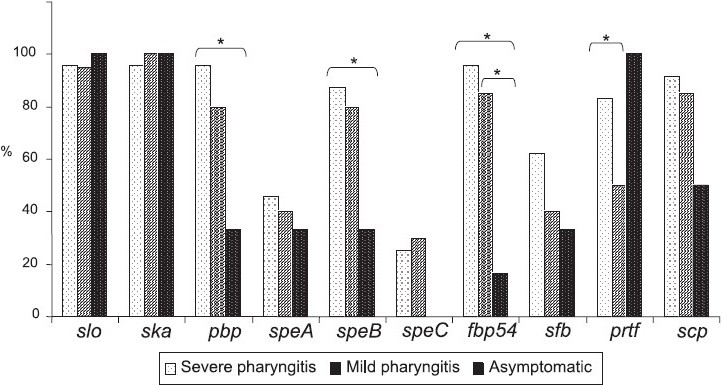
Virulence factors genes and their association with severity of pharyngitis. *Significant (P<0.05).
Since slo, ska, spe A, sfb and scp were present in all the throat isolates studied, their presence or absence may not be associated with increased virulence. Similar to our study for FBP genes particularly prtf, no significant difference in their distribution in pharyngitis isolates and asymptomatic carriers was observed earlier19. All the GAS isolates from asymptomatic cases carried the prtf gene, almost similar to the previous findings. High proportion of fbp-54 was described in GAS from asymptomatic carriers, however, it was quite low in our study (Fig.). Hence, GAS might recruit different FBPs genes for different purposes. Individual contributions of spe genes that elicit potent inflammatory responses is currently unclear, but is reported in invasive GAS diseases20. High frequency of spe A and its production associated with severity has been predicted10, a case different than slo, spe B and spe C9. The frequency of spe A gene (43%) in pharyngitis or RF/RHD GAS isolates in the present study was higher as compared to reports from India and elsewhere (6 to 25%)9,12; but quite similar to the reports from severe infections (42.9%)12, which is a matter of concern. spe B (84%) in GAS isolated from pharyngitis was in agreement with the previous findings (86.5%) from the same region12. Its frequency was less (33%) in asymptomatic cases, justifying its role in severe streptococcal diseases. Spe C was present in 25 per cent GAS isolates from severe pharyngitis cases and was absent in asymptomatic cases as compared to 55.4 and 65.8 per cent reported earlier11. No association of asymptomatic GAS with spe C gene or its production in vitro has been reported. The fact that almost all GAS types possess scp gene is a reason to believe that specific cleavage of C5a chemotaxin contributes to the virulence of these streptococci13.
On comparing OF negative and positive isolates for throat infection severity in terms of their virulence factors, significant differences (P<0.05) were recorded for pbp, spe B, fbp-54, sfb, scp in OF negative; and prtf in OF positive isolates respectively, suggesting heterogeneous distribution of virulence factors in OF negative GAS isolates.
This study demonstrated different emm types in GAS isolates, with varied genotypic potential. GAS isolates collected from northern India possessed highly virulent antigens. This study also supports concept of isolate-associated virulence rather than virulence broadly related to a given serotype.
Acknowledgments
We acknowledge the financial support by Indian Council of Medical Research (ICMR): Jai Vigyan Mission Mode Project on “Community Control of RF/RHD in India” to the first author (VD).
Footnotes
Conflict of Interest: None
References
- 1.Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heath A, Rita VJ, Barg NL, Engleberg NC. A two component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, slo and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Severin A, Nickbarg E, Wooters J, Quazi SA, Matsuka YV, Murphy E, et al. Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J Bacteriol. 2007;189:1514–22. doi: 10.1128/JB.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaussee MA, Dmitriev AV, Callegari EA, Chaussee MS. Growth phase-associated changes in the transcriptome and proteome of Streptococcus pyogenes. Arch Microbiol. 2008;189:27–41. doi: 10.1007/s00203-007-0290-1. [DOI] [PubMed] [Google Scholar]
- 5.McIver KS, Heath AS, Scott JR. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange and iron limitation of emm transcription. Infect Immun. 1995;63:4540–2. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DR, Kaplan EL. Micro technique for serum opacity factor characterization of group A Streptococcus adaptable to the use of human sera. J Clin Microbiol. 1988;26:2025–30. doi: 10.1128/jcm.26.10.2025-2030.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beall B, Facklam RR, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–8. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malke H. Polymorphism of the streptokinase gene: implications for the pathogenesis of post-streptococcal glomerulonephritis. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:246–57. doi: 10.1016/s0934-8840(11)80842-x. [DOI] [PubMed] [Google Scholar]
- 9.Tyler SD, Johnson WM, Huang JC, Ashton FE, Wang G, Low DE, et al. Streptococcal erythrogenic toxin genes: Detection by polymerase chain reaction and association with disease in strains isolated in Canada from 1940 to 1991. J Clin Microbiol. 1992;30:3127–31. doi: 10.1128/jcm.30.12.3127-3131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inagaki Y, Konda T, Murayama S, Yamai S, Matsushima A, Gyobu Y, et al. Serotyping of Streptococcus pyogenes isolated from common and severe invasive infections in Japan, 1990-95: Implication of the T3 serotype strain expansion in TSLS. Epidemiol Infect. 1997;119:41–8. doi: 10.1017/s0950268897007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creti R, Gherardi G, Imperi M, Von Hunolstein C, Baldassarri L, Pataracchia M, et al. Association of group A streptococcal emm types with virulence traits and macrolide-resistance genes is independent of the source of isolation. J Med Microbiol. 2005;54:913–7. doi: 10.1099/jmm.0.46035-0. [DOI] [PubMed] [Google Scholar]
- 12.Nandi S, Chakraborti A, Bakshi DK, Rani A, Kumar R, Ganguly NK. Association of pyrogenic exotoxin genes with pharyngitis and rheumatic fever and rheumatic heart disease among Indian isolates of Streptococcus pyogenes. Lett Appl Microbiol. 2002;35:237–41. doi: 10.1046/j.1472-765x.2002.01176.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen CC, Cleary PP. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J Biol Chem. 1990;265:3161–7. [PubMed] [Google Scholar]
- 14.Haynes W. Virulence factors of the group A streptococci and genes that regulate their expression. Front Biosci. 2004;9:3399–433. doi: 10.2741/1491. [DOI] [PubMed] [Google Scholar]
- 15.Ma X, Kikuta H, Ishiguro H, Yoshioka M, Ebihara T, Murai T, et al. Association of the prtF1 gene (encoding fibronectin binding protein F1) and the sic gene (encoding the streptococcal inhibitor of complement) with emm types of group A streptococci isolated from Japanese children with pharyngitis. J Clin Microbiol. 2002;40:3835–7. doi: 10.1128/JCM.40.10.3835-3837.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodfellow AM, Hibble M, Talay SR, Kreikemeyer B, Currie BJ, Sriprakash KS, et al. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the Northern Territory, Australia. J Clin Microbiol. 2000;38:389–92. doi: 10.1128/jcm.38.1.389-392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sagar V, Kumar R, Ganguly NK, Chakraborti A. Comparative analysis of emm type pattern of group A Streptococcus throat and skin isolates from India and their association with closely related SIC, a streptococcal virulence factor. BMC Microbiol. 2008;8:150–8. doi: 10.1186/1471-2180-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sriprakash KS, Hartas J, White A. Antibodies to streptococcal inhibitor of complement function and M peptides in a post-streptococcal glomerulonephritis endemic region of Australia. J Med Microbiol. 2002;51:589–94. doi: 10.1099/0022-1317-51-7-589. [DOI] [PubMed] [Google Scholar]
- 19.Musumeci R, Bue CL, Milazzo I, Nicoletti G, Serra A, Speciale A, et al. Internalization-associated proteins among Streptococcus pyogenes isolated from asymptomatic carriers and children with pharyngitis. Clin Infect Dis. 2003;37:173–9. doi: 10.1086/375589. [DOI] [PubMed] [Google Scholar]
- 20.Chatellier S, Thendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, et al. Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and non-severe invasive diseases. Infect Immun. 2000;68:3523–34. doi: 10.1128/iai.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


