Abstract
Background & objectives:
Bacillus cereus is one of the pathogens responsible for human diarrhoea, mainly due to consumption of contaminated food. The present study was undertaken to determine the occurrence of B. cereus among diarrhoeal patients and its phenotypic and genetic characteristics that determine the virulence and clonal features.
Methods:
Stool specimens were collected for two years from acute diarrhoeal patients attending the two referral hospitals in Kolkata. Presence of virulence genes in B. cereus was determined by PCR. Clonality was assessed by pulsed-field gel analysis (PFGE) by restriction digestion with SmaI and NotI enzymes. Enterotoxins were detected by haemolysin assay and using BCET-RPLA kit. Invasion assay was done on Hep-2 cell line. Antimicrobial susceptibility was tested by disc diffusion method.
Results:
B. cereus was identified in 54 (3.5%) of the 1536 diarrhoeal cases studied. Majority of the isolates were susceptible to many antibiotics but showed resistant to amoxyclav and cephalosporins. Six genes covering the two different enterotoxic complexes determining the pathogenicity of B. cereus have been characterized by PCR. The nhe genes were detected in a higher proportion than hbl. Except in two, clonal diversity was noticed among 21 B. cereus isolates. Haemolytic enterotoxin was detected in 76 per cent of the isolates. Majority of the isolates (67%) produced in vitro enterotoxin (BCET) confirming its involvement in the infection.
Interpretation & conclusions:
Though the presence of B. cereus was not high in patients with diarrhoea, several virulence factors confirm their association with diarrhoea. Distinct clonality was identified in majority of the isolates indicating their origin from different sources.
Keywords: B. cereus, BCET, diarrhoea, PFGE, virulence genes
Acute diarrhoea is an endemic disease in many parts of India especially, in Gangetic Bengal. Bacillus cereus is one of the pathogens responsible for human diarrhoea, and source of infestation is mainly due to consumption of contaminated food. B. cereus, a Gram-positive, rod shaped, endospore-forming, motile facultative anaerobic bacteria, can dominate in any given situation, because of its ubiquitous nature and ability to occur in a diversified range of foods1. Shortest mean incubation period (0.8 h) and onset of illness within 8 h make B. cereus more potent pathogen than the remaining enteric organisms2.
There are two types of B. cereus food poisoning syndromes caused by two independent toxins. The emetic toxin (<5kDa) is resistant to heat, proteolytic enzyme and low pH. This toxin causes nausea and vomiting within 1-5 h after the consumption of contaminated food. The diarrhoeal toxin is a 50kDa heat-labile protein, which is sensitive to proteolytic enzymes and expressed during the late exponential phase of growth. The onset of B. cereus mediated infection is about 8-16 h, lasts for 12-24 h, and mostly associated with abdominal pain, profuse watery diarrhoea and tenesmus than nausea and vomiting3. Two protein complexes from B. cereus isolates, haemolysin BL (HBL) and non-haemolytic enterotoxin (NHE) have been characterized. The haemolysin BL consists of a binding component B and two lytic components L1 and L 2 responsible for enterotoxigenicity of B. cereus4. The B and L (L1 and L2) components are encoded in the genes hblA, hblD and hblC, respectively5. These three components may be present in a different composition in B. cereus, and all the components together are necessary for the expression haemolysis to occur6. Non-haemolytic enterotoxin also consists of three different proteins, A, B and C with the corresponding encoding genes nheA, nheB and nheC, respectively7.
Systematic surveillance and molecular characterization of B. cereus isolated from acute diarrhoeal patients were not done in India. Hence, the present study was undertaken to determine the presence of B. cereus among patients with acute diarrhoea attending hospital and to characterize the isolates to understand the virulence features as well as clonal nature.
Material & Methods
Collection and processing of stool specimens:
Consecutive diarrhoeal stool specimens collected between October 2006 and September 2008 from outpatients attending the B.C. Roy Hospital and acute diarrhoeal patients admitted and enrolled in the active surveillance at the Infectious Disease Hospital, Kolkata, were included in the present study. Initial enrichment was made in Tryptone soy broth (TSB) (Hi-Media, Mumbai, India) supplemented with 500U/ml of polymyxin B at 37°C for 24-48 h followed by streaking on B. cereus selective agar (BCSA) (Hi-Media)8. The presumptive identification of B. cereus was made on the basis of colony characteristics (peacock blue coloured colonies with a surrounding zone of egg yolk precipitation). The other enteric bacterial pathogens were screened following standard protocols9. Diarrhoeagenic Escherichia coli were detected by multiplex PCR10.
Identification of B. cereus:
The presumptive cultures of B. cereus obtained from stool specimens were further tested for their motility, endospore formation, followed by species confirmation using biochemical tests with API-50 CHB kit (bioMerieux, La Balme Les Grottes, France).
Molecular characterization
Detection of virulence genes - PCR assay was made for the identification of haemolytic BL complex genes namely, hblC, hblD and hblA and non-haemolytic genes such as nheA, nheB and nheC. Overnight cultures of B. cereus from Luria Bertani (LB) agar (Difco, Detroit, MI, USA) were suspended in 200 µl of sterile distilled water and lysed by boiling for 10 min in a water bath and snap chilled on ice followed by centrifugation at 15000 × g for 5 min. The DNA containing supernatant was transferred into a new microcentrifuge tube and used immediately for PCR. Primers for the detection of the genes of the HBL and NHE complexes are shown in Table I. DNA extract (2.5 µl) was amplified with 0.6 U of Taq polymerase (GeNei, Bangalore, India) using published protocol11. PCR products were analyzed by 1.5 per cent agarose gel electrophoresis using 100 bp DNA ladder (NEB, Hitchin, Herts, UK) as molecular weight marker.
Table I.
List of primers used in the simplex PCR for the detection of virulence associated genes in B. cereus
| Gene | Primer (5’ →3’) | Amplicon (bp) |
|---|---|---|
| hblA | B component of haemolysin BL | |
| HBLA1:GTGCAGATGTTGATGCCGAT | 320 | |
| HBLA2:ATGCCACTGCGTGGACATAT | ||
| hblD | L1 component of haemolysin BL | |
| LIA:AATCAAGAGCTGTCACGAAT | 430 | |
| L1B:CACCAATTGACCATGCTAAT | ||
| hblC | L2 component of haemolysin BL | |
| L2A:AATGGTCATCGGAACTCTAT | 750 | |
| L2B:CTCGCTGTTCTGCTGTTAAT | ||
| nheA | A component of non-haemolytic ET | |
| nheA 344S: TACGCTAAGGAGGGGCA | 500 | |
| nheA 843A: GTTTTTATTGCTTCATCGGCT | ||
| nheB | B component of non-haemolytic ET | |
| nheB 1500S:CTATCAGCACTTATGGCAG | 770 | |
| nheB 2269A:ACTCCTAGCGGTGTTCC | ||
| nheC | C component of non-haemolytic ET | |
| nheC 2820S:CGGTAGTGATTTGCTGGG | 582 | |
| nheC 3401A:CAGCATTCGTACTTGCCAA |
Clonal analysis by pulsed-field gel electrophoresis (PFGE) - The genomic DNA of B. cereus was prepared using the protocol of Liu et al12 with modification. Since, the cell wall of B. cereus was difficult to lyse, an early log phase cultures were used to avoid sporulation. In brief, bacterial cells were grown in TSB for 3 h followed by centrifugation with Saline EDTA (SE) buffer (75 mM NaCl, 25 mM EDTA; pH 7.5) at 3000 g. The cell pellet was suspended in 0.5 ml SE buffer containing 1.5 mg lysozyme (Sigma, St. Louis, MO, USA) and 15 U lysostaphin (Sigma). This suspension was mixed with 0.5 ml 1.5 per cent low melting agarose and dispensed in plug molds. After the lysis and proteinase K treatment, bacterial cells embedded in agarose plugs were washed once for 1 h at room temperature in Tris-EDTA (TE) buffer [10 mM Tris-HCL (pH 7.5), 10 mM EDTA], once for 1 h at 37°C in TE buffer containing 1 mM phynylmethylsulphonylfluoride (Sigma) and thrice for 1 h each in TE buffer at 37°C. A slice from each plug (2.5 mm) was cut and incubated overnight with 20 U each of SmaI and NotI restriction endonuclease (NEB) with the appropriate buffers and the reaction conditions recommended by the manufacturer. After digestion, the plugs were loaded into 1 per cent PFGE agarose (BioRad, Hercules CA, USA) in 0.5 × TBE buffer (Tris, borate, EDTA). Electrophoresis was done in CHEF-Mapper system (BioRad, Hercules CA, USA) for 20 h at 14°C, with an electric field of 6 V/cm, and the pulse time increased from 5.3 to 34.9 sec for SmaI and 5 to 80 sec for NotI13. A bacteriophage lambda ladder (NEB) was used as the molecular weight marker. The reproducibility of the fingerprints was examined by repeated tests.
Enterotoxin assay
Haemolysin assay - Culture supernatant of individual isolates of B. cereus obtained from overnight culture grown in brain heart infusion broth (Difco, USA) with 0.1 per cent glucose (BHIG) at 37°C at 191 × g assessed for haemolytic activity by agar well plate assay using 5 per cent sheep blood agar (SBA) (bioMeriux, France). Twenty five microlitres of cell-free culture supernatants were added in 5 mm diameter well in the SBA plates and incubated at 30°C and monitored for haemolytic pattern14. The ATCC9139 B. cereus strain was used as positive control and an E. coli K12 used as negative control.
Enterotoxin assay (diarrhoeal type) -For the extraction of B. cereus enterotoxin, isolates from pure culture grown on BCSA plates were used after inoculation in 10 ml BHIG and incubation at 37°C for 18 h on a shaker (150 rpm). Culture supernatants obtained by centrifugation at 900 × g for 20 min at 4°C were used immediately for toxin assay using BCET-RPLA (reversed passive latex agglutination) kit (Oxoid, Hampshire, UK) as directed in the manual.
Tissue culture assay:
Free bacterial cells of B. cereus were harvested after 4 and 18 h growth and washed three times by centrifugation at 402 × g for 10 min each time with Dulbecco’s modified Eagles medium (DMEM, Gibco, Grand Island, NY, USA) and used immediately for HEp-2 monolayers grown in 5 per cent CO2 atmosphere at 37°C in DMEM supplemented with 10 per cent foetal calf serum (FCS) in 24-well tissue culture plates in triplicate using published protocol15. Shigella flexneri 2a and E. coli DH5α were used as positive and negative control strains, respectively for invasion assay in the HEp-2 cells.
Susceptibility to antimicrobials:
Antimicrobial susceptibility was determined by the disc agar diffusion method16. Single colonies of 24 h-old cultures were transferred to 5 ml of TSB (Difco) and incubated at 37°C for 6-8 h. A sterile cotton swab dipped into the TSB growth was applied evenly onto pre-dried Mueller-Hinton agar (Difco) plate. After drying for 15 min, the antimicrobial test discs (HiMedia) were placed aseptically and the plates were incubated at 37°C for 14-19 h. The zones were measured and sensitive, intermediate and resistance was categorized according to the standard methods16. ATCC E. coli strain 25922 was used as a quality control.
Results
Isolation and identification of B. cereus:
This study was conducted using 1536 stool specimens consecutively collected for two years (October 2006 through September 2008) from B. C. Roy Hospital and Infectious Disease Hospital in Kolkata. Fifty four stool specimens (3.5%) were found to be positive for B. cereus. Only a single B. cereus isolate was collected from each positive sample and all were confirmed using motility test, endospore formation and biochemical tests with API-50 CHB kit. In the Month-wise detection of B. cereus spreading over successive two years, higher incidence of B. cereus positive cases was recorded during October, December 2006, March, October and November 2007 (Table II).
Table II.
Per cent positivity of B. cereus isolated from acute diarrhoeal patients during 2006-2008
| Month & year | No. of sample | No. of positive sample (%) |
|---|---|---|
| Oct 2006 | 66 | 6 (9.1) |
| Nov 2006 | 99 | 5 (5) |
| Dec 2006 | 39 | 5 (12.8) |
| Jan 2007 | 61 | 3 (4.9) |
| Feb 2007 | 51 | 0 |
| Mar 2007 | 58 | 4 (6.9) |
| Apr 2007 | 70 | 3 (4.3) |
| May 2007 | 79 | 3 (3.8) |
| Jun 2007 | 80 | 4 (5) |
| Jul 2007 | 65 | 0 |
| Aug 2007 | 53 | 0 |
| Sep 2007 | 47 | 0 |
| Oct 2007 | 53 | 4 (7.5) |
| Nov 2007 | 90 | 6 (6.7) |
| Dec 2007 | 60 | 1 (1.7) |
| Jan 2008 | 74 | 2 (2.7) |
| Feb 2008 | 56 | 1 (1.8) |
| Mar 2008 | 53 | 2 (3.8) |
| Apr 2008 | 59 | 1 (1.7) |
| May 2008 | 66 | 2 (3) |
| Jun 2008 | 51 | 1 (2) |
| Jul 2008 | 80 | 1 (1.3) |
| Aug 2008 | 68 | 0 |
| Sep 2008 | 58 | 0 |
B. cereus was identified more often in male patients (54%) than females (46%). When age-wise distribution of confirmed B. cereus related acute diarrhoeal patients was considered, there was no association between different age groups and only 0.8 per cent of the affected individuals were above 60 yr of age (Table III).
Table III.
Prevalence of B. cereus in different age groups of diarrhoeal patients
| Age groups (yr) |
|||||
|---|---|---|---|---|---|
| <15 | 15 - <30 | 30 - <45 | 45 - >60 | >60 | |
| No. of samples | 599 | 338 | 286 | 192 | 121 |
| No. of positive samples (%) | 23 (3.8) | 13 (3.8) | 12 (4.2) | 5 (2.6) | 1 (0.8) |
Of the 54 isolates, 42 (78%) were found as sole pathogen associated with diarrhoea. The mixed infections were checked for Vibrio cholerae, V. parahaemolyticus, diarrhoeagenic E. coli, Salmonella and Shigella. Polymicrobial aetiology was detected in the remaining 12 (22%) patients.
Detection of virulence genes:
Representative amplicons of hblA, hblC and hblD encoding the enterotoxin HBL complex and nheA, nheB and nheC encoding the nonhaemolytic enterotoxin of NHE complex, are shown in Fig. 1. The three genes, hblA, hblC and hblD were detected in 28 isolates (51.9%). Eight (14.8%) isolates possessed two of the three hbl genes and three (5.6%) had only one gene coding the HBL complex. Fifteen (27.8%) B. cereus isolates had none of HBL complex. All the three genes nheA, nheB and nheC, were detected in 48 (88.9%) of 54 B. cereus isolates. Five (9.3%) isolates harboured two nhe genes, whereas only one (1.9%) isolate lacked all three genes of NHE complex (Table IV). The nonhaemolytic enterotoxin (NHE) genes nheA, nheB, and nheC (98, 96 and 91%, respectively) were frequently detected than haemolytic enterotoxin (HBL) genes, hblA, hblC and hblD (65, 59 and 67%, respectively).
Fig. 1.
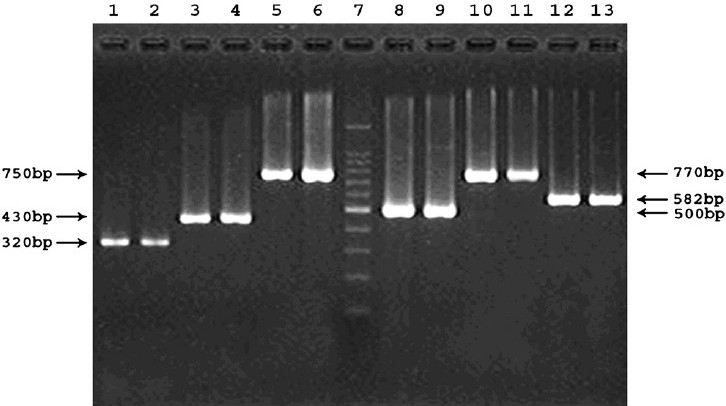
Representative PCR products showing amplicons of six virulence genes in B. cereus isolates. Lanes 1 to 2, hblA positive isolates M20833 and M20890; 3 to 4, hblD positive isolates M20833 and IDH00060; 5 to 6, hblC positive isolates M20890 and M21325; 7, molecular size marker 100 bp ladder; 8 to 9, nheA positive isolates M20833 and IDH00195; 10 to 11, nheB positive isolates IDH00278 and IDH00339; 12 to 13, nheC positive isolates M20890 and M21325.
Table IV.
Results of PCR, haemolysin assay and enterotoxin production of B. cereus
| Isolate No | hblA | hblD | hblC | nheA | nheB | nheC | Haemolysis | BCET (ng/ml) |
|---|---|---|---|---|---|---|---|---|
| VTE2563 | + | + | + | + | + | + | + | 16 |
| L19524 | - | - | - | + | + | + | - | - |
| L19632 | + | + | + | + | + | + | + | 128 |
| L20144 | - | - | - | + | + | + | - | - |
| L20166 | - | - | - | + | + | + | - | - |
| L20190 | - | - | - | + | + | + | - | - |
| L21161 | + | + | + | + | + | + | + | 128 |
| L21500 | + | + | + | + | + | + | + | 128 |
| L21519 | + | + | + | + | + | + | + | 64 |
| VTE2582 | + | + | + | + | + | + | + | 32 |
| VTE2584 | + | + | + | + | + | + | + | 16 |
| L22622 | - | + | + | + | + | + | + | 32 |
| VTE2591 | - | - | - | + | + | + | + | - |
| L22959 | - | - | - | + | + | + | - | - |
| VTE2593 | + | + | + | + | + | + | + | 128 |
| L23188 | - | - | - | + | + | + | + | - |
| M78 | + | + | + | + | + | + | + | 64 |
| M103 | - | - | - | + | + | + | - | - |
| M293 | + | + | + | + | + | + | + | 16 |
| M3102 | + | + | + | + | + | + | + | 8 |
| M3139 | - | - | - | + | + | + | - | - |
| M3551 | + | + | + | + | + | + | + | 8 |
| VTE2613 | + | + | + | + | + | + | + | 32 |
| VTE2620 | + | + | + | + | + | + | + | 16 |
| M4698 | + | + | + | + | + | + | + | 32 |
| M5007 | + | + | + | + | + | + | + | 128 |
| M7351 | + | + | + | + | + | + | + | 64 |
| M7826 | + | + | + | + | + | + | + | 16 |
| M7985 | + | + | + | + | - | + | + | 32 |
| M8263 | + | + | + | + | + | + | + | 128 |
| M8278 | + | + | + | + | + | + | + | 64 |
| M8432 | + | + | + | + | + | + | + | 16 |
| VTE2665 | - | - | - | + | + | - | - | - |
| M20833 | + | + | + | + | + | + | + | ≥256 |
| M20885 | + | - | + | + | + | + | + | ≥256 |
| M20890 | + | - | + | + | + | + | + | 128 |
| M21325 | - | - | + | + | + | + | + | ≥256 |
| M21518 | + | - | - | + | + | - | + | - |
| M21558 | + | - | - | + | + | - | + | - |
| VTE2742 | - | + | + | + | + | + | + | ≥256 |
| IDH48 | - | - | - | + | + | + | - | - |
| IDH60 | + | + | - | + | + | + | + | - |
| IDH80 | + | - | + | + | + | + | + | ≥256 |
| IDH106 | - | - | - | + | + | - | - | - |
| IDH195 | + | - | + | + | + | + | + | >256 |
| IDH232 | - | - | - | - | - | - | - | - |
| IDH278 | - | + | + | + | + | + | + | 64 |
| IDH339 | - | - | - | + | + | + | - | - |
| IDH348 | + | + | + | + | + | + | + | 128 |
| IDH413 | + | + | + | + | + | + | + | 64 |
| IDH487 | + | + | + | + | + | + | + | 4 |
| IDH514 | + | + | + | + | + | + | + | 128 |
| IDH547 | - | - | - | + | + | + | - | - |
| IDH626 | + | + | + | + | + | + | + | 16 |
BCET, B.cereus enterotoxin
Clonal analysis by PFGE:
Twenty isolates were selected for PFGE analysis based on the PCR results, which represents different combination of hbl and nhe genes. The fingerprints generated by macrorestriction with SmaI comprised about 20-25 bands of approximately 5-500 Kb (Fig. 2), whereas with NotI approximately 5-13 bands of approximately 50-700 Kb found (Fig. 3). In the present study, PFGE banding patterns in two isolates were identical (M20885 and M20890) with both the enzymes tested. These isolates also exhibited identical virulence gene profiles.
Fig. 2.
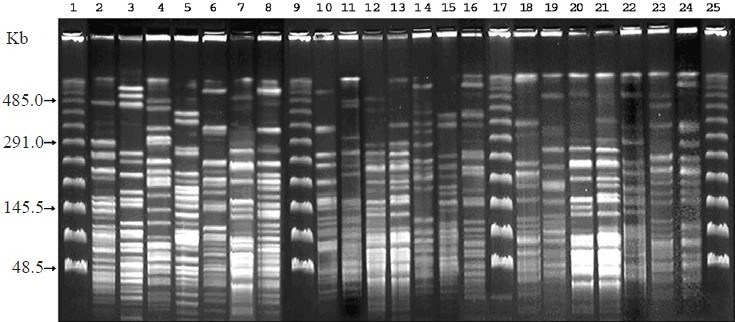
PFGE of SmaI digested genomic DNA of B. cereus isolates. Lanes 1, 9, 17, and 25 are bacteriophage lamda molecular size makers; 2, ATCC 9139; 3, L20166; 4, L21519; 5, M3139; 6, M8263; 7, M20890; 8, VTE2742; 10, M7985; 11, VTE2665; 12, M21325; 13, M21558; 14, IDH80; 15, IDH232; 16, L20190; 18, M78; 19, M103; 20, M20885; 21, M21518; 22, IDH106; 23, IDH195; 24, VTE2620.
Fig. 3.
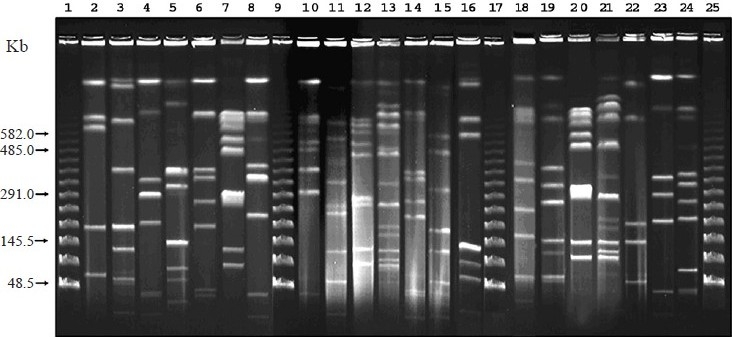
PFGE of Not I digested genomic DNA of B. cereus isolates. Lanes 1, 9, 17, and 25 are bacteriophage lamda molecular size makers; 2, ATCC 9139; 3, L20166; 4, L21519; 5, M3139; 6, M8263; 7, M20890; 8, VTE2742; 10, M7985; 11, VTE2665; 12, M21325; 13, M21558; 14, IDH80; 15, IDH232; 16, L20190; 18, M78; 19, M103; 20, M20885; 21, M21518; 22, IDH106; 23, IDH195; 24, VTE2620.
Haemolysin assay:
Majority (76%) of the B. cereus isolates exhibited haemolysis (Table IV). Except for isolates VTE2591 and L23188 expression of haemolysin was associated with presence of any of the hbl genes. Haemolytic activity was not detected in 13 isolates, which did not harbour any hbl genes (Table IV). Discontinuous haemolysis was noticed with B. cereus isolates harbouring hblCDA operon, whereas continuous haemolysis was recorded in isolates that lack any one of these genes.
Enterotoxin assay:
Qualitative tests on B. cereus enterotoxin production using BCET-RPLA kit of 54 isolates showed that 36 (67%) isolates were able to produce BCET on BHIG in a varied concentration ranging from 8 to >256 ng/ml (Table IV). Of these enterotoxigenic isolates, 3 per cent produced 4 ng/ml, 6 per cent produced 8 ng/ml, 19 per cent produced 16 ng/ml, 14 per cent produced 32 ng/ml, 16.5 per cent produced 64 ng/ml, 24 per cent produced 128 ng/ml and 16.5 per cent produced ≥256 ng/ml. There appears to be a association between presence of hbl genes and expression of BCET. Of the 18 isolates that were negative in the BCET, 15 did not harbour any hbl genes and in 3 hblC was absent. There was no association between combination of hbl genes and an activity of expressed BCET.
Invasion assay:
Based on the PCR results, which represents different combination of hbl and nhe genes, 20 isolates (same as in PFGE) were also included for invasion assay using HEp-2 cell line. Our result showed that none of the tested isolates invaded into the HEp-2 cells.
Susceptibility to antimicrobials:
The susceptibility of 54 B. cereus isolates was tested for 10 different antibiotics. All the isolates were susceptible for amikacin, ciprofloxain, gentamicin, and imipenem. Majority of the isolates were also susceptible for ofloxacin and azithromycin. a0 moxyclav and cephalosporins resistance was seen in most of the isolates (Table V).
Table V.
Antimicrobial resistance of B. cereus
| Antibiotics (µg/disc) | Zone of inhibition* (mm) R - S | % (n=54) |
||
|---|---|---|---|---|
| Sensitive (S) | Intermediate (I) | Resistant (R) | ||
| Amikacin (30) | ≤14-≥17 | 100 | 0 | 0 |
| Amoxyclav (30) | ≤19-≥20 | 0 | 0 | 100 |
| Azithromycin (15) | ≤13-≥18 | 80 | 20 | 0 |
| Cefexime (5) | ≤16-≥23 | 0 | 0 | 100 |
| Ceftriaxone (30) | ≤13-≥21 | 0 | 45 | 55 |
| Ceffotaxim (30) | ≤14-≥23 | 5 | 42 | 53 |
| Ciprofloxacin (5) | ≤15-≥21 | 100 | 0 | 0 |
| Gentamicin (10) | ≤12-≥15 | 100 | 0 | 0 |
| Imipenem (10) | ≤13-≥16 | 100 | 0 | 0 |
| Ofloxacin (5) | ≤12-≥16 | 95 | 5 | 0 |
Resistance/susceptible range (Hi-Media, Mumbai, India)
Discussion
B. cereus associated food poisoning is underreported as the types of illnesses are relatively mild and usually last for less than 24 h. Nevertheless, occasional reports of more severe form of diarrhoeal type of illnesses, ubiquitous presence and heat-stable endospore forming nature of the organism underscore the significance of the organism. The unique properties such as heat resistance, endospore forming ability, toxin production and psychrotrophic nature give ample scope for this organism to be a prime cause of public health hazard17.
Identification of B. cereus was almost constant in all age groups. Contaminated food and warm weather seems to support incidence of B. cereus18–21. In 78 per cent of the patients, we identified the B. cereus as a sole pathogen with all the maker virulence genes included in this study thereby indicating their role in causing the disease. However, in 22 per cent of the diarrhoeal patients, polymicrobial aetiology was detected; hence it is difficult to conclude the role of B. cereus in these cases. Since, complete data on viral aetiology are not available, we cannot rule out the possibility of mixed infection caused by enteric viruses. The importance of B. cereus mediated diarrhoea should be assessed through community based case and control studies.
Two different enterotoxic protein complexes have been characterized, nhe genes found in most of the strains in a higher proportion than hbl genes. Distribution of these genes in B. cereus were more in this study compared to other reports22. Our results confirmed previous speculation that in B. cereus two or more enterotoxins might be involved in causing diarrhoea in humans23. Hansen and Hendriksen11 reported that polymorphism among the genes is the likely explanation of the inability to detect all genes in some B. cereus isolates by PCR.
The NotI PFGE profile has low discriminatory power as it generates less number of DNA bands12. The same trend was observed with the clinical isolates of B. cereus. On the other hand, Zhong et al24 found only two to three fragments in the case of SmaI digestion, whereas we found 20-25 fragments. Overall, our study demonstrated that B. cereus isolated from diarrhoeal patients was not clonal in this region though the virulence gene profiles and expression of BCET was similar. M20885 and M20890 were isolated as a sole pathogen from two patients who were admitted to the IDH on the same day. Both the patients had identical clinical symptoms such as acute watery diarrhoea and dehydration. However, demographic information showed that they were not related. Since these two isolates were clonally identical as evidenced from the PFGE, these would have infected from a common source.
The determination of haemolytic activity on SBA led to interesting results. All the isolates that were positive for hbl genes in PCR exhibited haemolysis in SBA. Two isolates of B. cereus found weakly positive for haemolysis, but failed to show PCR amplification for hbl genes. This could be due to sequence variation in the binding site of the primers25. While studying the distribution of hblCDA and nheABC genes, Ngamwongsatit et al26 have shown that detection of these genes were increased when alternative primers were used, with high specificity of the assay. Similar to the finding of Thaenthanee et al27, we have detected higher number (51.8%) of B. cereus isolates that displayed discontinuous haemolysis.
To define the extent of toxin production by B. cereus, an analysis of BCET was made. The BCET-RPLA test does not specifically react with diarrhoeal toxin, but with the L2 component of HBL complex. Since, the non-enterotoxigenic isolates lack hbl gene specially the L2 component, the BCET-RPLA kit is useful in the identification of this virulent factor28. Expression of BCET was detected in more number of isolates in this study than from other reports23.
Our result showed that none of the tested isolates invaded into the HEp-2 cells. Rowan et al15 found 91 per cent Bacillus species, of which 100 per cent B. cereus isolates were unable to invade HEp-2 cells, whereas Rowan et al29 showed 49 per cent of different Bacillus species showed various levels of invasion on HEp-2 cells of which 75 per cent of B. cereus strains were non-invasive to HEp-2 cells.
Antimicrobial susceptibility testing guides for the empirical use of antibiotics and proper management of diarrhoea. In this study, majority of B. cereus isolates were resistant to amoxyclav and cephalosporins. This trend seems common in other regions as well30. B. cereus isolated from different sources was generally resistant to penicillin, ampicillin, cephalosporins, and trimehoprim and susceptible for clindamycin, erythromycin, chloramphenicol, vancomycin, imipenem, aminoglycosides and ciprofloxacin31.
In conclusion, our results showed the importance of B. cereus among hospitalized patients with acute diarrhoea in Kolkata. PCR amplification of toxin genes and PFGE analysis showed diverse virulence factors as well as of clonality of the isolates from acute diarrhoeal patients. Knowledge of spectrum of antibiotic susceptibility will possibly become a guide to empirical therapy to shorten the morbidity in acute stage, as microbial quality assay from stool specimens is not a routine practice in this part of the world. Community based studies are needed to know the diarrhoeal disease burden due to this pathogen in this region.
Acknowledgments
This work was supported by Department of Science and Technology, Government of India (No. SR/WOS-A/LS 169/2005).
References
- 1.Smith DP, Berrang ME, Feldner PW, Phillips RW, Meinersmann RJ. Detection of Bacillus cereus on selected retail chicken products. J Food Protect. 2004;67:1770–3. doi: 10.4315/0362-028x-67.8.1770. [DOI] [PubMed] [Google Scholar]
- 2.Kudaka J, Horikawa K, Uryu K, Matsuyuki S, Ogata K, Kawano K, et al. Symptoms of food-borne diseases and gastroenteritis in Kyushu, Japan. Kansenshogaku Zasshi. 2005;79:864–70. doi: 10.11150/kansenshogakuzasshi1970.79.864. [DOI] [PubMed] [Google Scholar]
- 3.Adams MR, Moss MO. Bacterial agents of food borne illness. Food microbiology. Cambridge, England: Royal Society of Chemistry; 1995. pp. 160–3. [Google Scholar]
- 4.Beecher DJ, Schoeni JL, Wong ACL. Enterotoxin activity of hemolysin Bl from Bacillus cereus. Infect Immun. 1995;63:4423–8. doi: 10.1128/iai.63.11.4423-4428.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan PA, Macmillan JD, Zilinskas BA. Molecular cloning and characterization of the genes encoding the L 1 and L 2 components of hemolysin BL from Bacillus cereus. J Bacteriol. 1997;179:2551–6. doi: 10.1128/jb.179.8.2551-2556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoeni JL, Wong ACL. Bacillus cereus food poisoning and its toxins. J Food Protect. 2005;68:636–48. doi: 10.4315/0362-028x-68.3.636. [DOI] [PubMed] [Google Scholar]
- 7.Granum PE, O’Sullivan K, Lund T. The sequence of the non-hemolytic enterotoxin operon from Bacillus cereus. FEMS Microbiol Lett. 1999;177:225–9. doi: 10.1111/j.1574-6968.1999.tb13736.x. [DOI] [PubMed] [Google Scholar]
- 8.Holbrook R, Anderson JM. An improved selective and diagnostic medium for the isolation and enumeration of Bacillus cereus in foods. Can J Microbiol. 1980;26:753–9. [Google Scholar]
- 9.Manual for laboratory identification of acute enteric infections. Geneva, Switzerland: 1987. World Health Organization (WHO) CDD/83-3/Rev. [Google Scholar]
- 10.Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43:755–60. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen BM, Hendriksen NB. Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl Environ Microbiol. 2001;67:185–9. doi: 10.1128/AEM.67.1.185-189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu PU, Ke S, Chen S. Use of pulsed field gel electrophoresis to investigate a pseudo-outbreak of Bacillus cereus in a pediatric unit. J Clin Microbiol. 1997;35:1533–5. doi: 10.1128/jcm.35.6.1533-1535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrell LJ, Andersen GL, Wilson KH. Genetic variability of Bacillus anthracis and related species. J Clin Microbiol. 1995;33:1847–50. doi: 10.1128/jcm.33.7.1847-1850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prüß BM, Dietrich R, Nibler B, Martlbauer E, Scherer S. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl Environ Microbiol. 1999;65:5436–42. doi: 10.1128/aem.65.12.5436-5442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowan NJ, Caldow G, Gemmell CG, Hunter IS. Production of diarrheal enterotoxins and other potential virulence factors by veterinary isolates of Bacillus species associated with nongastrointestinal infections. Appl Environ Microbiol. 2003;69:2372–6. doi: 10.1128/AEM.69.4.2372-2376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Performance standards for antimicrobial disk susceptibility tests. 9th ed. Wayne, PA: CLSI; 2006. Clinical Laboratory Standards Institute. Approved standard. CLSI document M2-A9. [Google Scholar]
- 17.Griffiths MW, Schraft H. Bacillus cereus food poisoning. In: Cliver DO, Riemann HP, editors. Foodborne diseases. London: Academic Press; 2002. pp. 261–70. [Google Scholar]
- 18.Thaikruea L, Pataraarechachai J, Savanpunyalert P, Naluponjiragul U. An unusual outbreak of food poisoning. Southeast Asian J Trop Med Public Health. 1995;26:78–85. [PubMed] [Google Scholar]
- 19.Luby S, Jones J, Dowda H, Kramer J, Horan J. A large outbreak of gastroenteritis caused by diarrheal toxin-producing Bacillus cereus. J Infect Dis. 1993;167:1452–5. doi: 10.1093/infdis/167.6.1452. [DOI] [PubMed] [Google Scholar]
- 20.Miwatani T, Honda T, Higashitsutsumi M, Tanaka R, Sakaue Y, Nakabayashi T, et al. Bacterial aetiology of infantile diarrhoea in Papua New Guinea. J Trop Pediatr. 1990;36:101–3. doi: 10.1093/tropej/36.3.101. [DOI] [PubMed] [Google Scholar]
- 21.Cho SH, Shin HH, Choi YH, Park MS, Lee BK. Enteric bacteria isolated from acute diarrheal patients in the Republic of Korea between the year 2004 and 2006. J Microbiol. 2008;46:325–30. doi: 10.1007/s12275-008-0015-4. [DOI] [PubMed] [Google Scholar]
- 22.Al-Khatib MS, Khyami-Horani H, Badran E, Shehabi A. Incidence and characterization of diarrheal enterotoxins of fecal Bacillus cereus isolates associated with diarrhea. Diagn Microbiol Infect Dis. 2007;59:383–7. doi: 10.1016/j.diagmicrobio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Ombui JN, Schmieger H, Kagiko MM, Arimi SM. Bacillus cereus may produce two or more diarrheal enterotoxins. FEMS Microbiol Lett. 1997;149:245–8. doi: 10.1111/j.1574-6968.1997.tb10336.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhong W, Shou Y, Yoshida TM, Marrone BL. Differentiation of Bacillus anthracis, B. cereus, and B. thuringiensis by using pulsed-field gel electrophoresis. Appl Environ Microbiol. 2007;73:3446–9. doi: 10.1128/AEM.02478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantynen V, LindstrÖm K. A rapid PCR based DNA test for enterotoxic Bacillus cereus. Appl Environt Microbiol. 1998;64:1634–9. doi: 10.1128/aem.64.5.1634-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ngamwongsatit P, Buasri W, Pianariyanon P, Pulsrikaran C, Ohba M, Assavanig A, et al. Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int J Food Microbiol. 2008;121:352–6. doi: 10.1016/j.ijfoodmicro.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Thaenthanee S, Wong ACL, Panbangred W. Phenotypic and genotypic comparisons reveal a broad distribution and heterogeneity of hemolysin BL genes among Bacillus cereus isolates. Int J Food Microbiol. 2005;105:203–12. doi: 10.1016/j.ijfoodmicro.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Ghelardi E, Celandroni F, Salvetti S, Barsotti C, Baggiani A, Senesi S. Identification and characterization of toxigenic Bacillus cereus isolates responsible for two food poisoning outbreaks. FEMS Microbiol Lett. 2002;208:129–34. doi: 10.1111/j.1574-6968.2002.tb11072.x. [DOI] [PubMed] [Google Scholar]
- 29.Rowan NJ, Deans K, Anderson G, Gemmell CG, Hunter IS, Chaithong T. Putative virulence factor expression by clinical and food isolates of Bacillus spp. After growth in reconstituted infant milk formulae. Appl Environ Microbiol. 2001;67:3873–81. doi: 10.1128/AEM.67.9.3873-3881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whong CMZ, Kwaga JKP. Antibiograms of Bacillus cereus isolates from some Nigerian foods. Niger Food J. 2007;25:178–83. [Google Scholar]
- 31.Turnbull PCB, Sirianni NM, LeBron CI, Samaan MN, Sutton FN, Reyes AE, et al. MCIs of selected antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringensis, and Bacillus mycoides from a range of clinical and environmental sources as determined by the Etest. J Clin Microbiol. 2004;42:3626–34. doi: 10.1128/JCM.42.8.3626-3634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


