Abstract
Plasma levels of high density lipoprotein cholesterol (HDL-C) are inversely proportional to the incidence of cardiovascular disease. Recent applications of modern proteomic technologies have identified upward of 50 distinct proteins associated with HDL particles with many of these newly discovered proteins implicating HDL in nonlipid transport processes including complement activation, acute phase response and innate immunity. However, almost all MS-based proteomic studies on HDL to date have utilized density gradient ultracentrifugation techniques for HDL isolation prior to analysis. These involve high shear forces and salt concentrations that can disrupt HDL protein interactions and alter particle function. Here, we used high-resolution size exclusion chromatography to fractionate normal human plasma to 17 phospholipid-containing subfractions. Then, using a phospholipid binding resin, we identified proteins that associate with lipoproteins of various sizes by electrospray ionization mass spectrometry. We identified 14 new phospholipid-associated proteins that migrate with traditionally defined HDL, several of which further support roles for HDL in complement regulation and protease inhibition. The increased fractionation inherent to this method allowed us to visualize HDL protein distribution across particle size with unprecedented resolution. The observed heterogeneity across subfractions suggests the presence of HDL particle subpopulations each with distinct protein components that may prove to impart distinct physiological functions.
Keywords: high density lipoprotein, proteomics, lipoprotein, apolipoprotein, mass spectrometry
Introduction
Lipoproteins are dynamic particles composed of lipid and proteins called apolipoproteins.1 They are formed in the liver, small intestine and certain macrophages and are secreted into the bloodstream where they mediate transport and metabolism of lipids. Chylomicrometers (CM) and very low and low density lipoproteins (VLDL/LDL) act in the delivery of dietary or liver-derived triglycerides and cholesterol to peripheral tissues for use or storage. High density lipoproteins (HDL) are generally thought to mediate a process called reverse cholesterol transport (RCT),2 involving the efflux of cholesterol from peripheral cells and its transport to the liver for excretion or recycling. High plasma levels of LDL and VLDL have been correlated with increased risk for cardiovascular disease (CVD) but HDL levels are inversely correlated.3,4 While HDL’s role in RCT undoubtedly plays a major role in cardio-protection, recent studies indicate that HDL also possesses antioxidative5 and anti-inflammatory6,7 properties that likely contribute to its cardio-protective effects.
To identify a mechanistic basis for these observed functions, research quickly focused on HDL apolipoproteins.8 HDL are secreted from the liver as nascent phospholipid “discs” made stable by their association with the most abundant HDL protein apolipoprotein A-I (apoA-I). As these particles accumulate free cholesterol from peripheral tissues, HDL associated lecithin cholesterol acyl transferase (LCAT) esterifies fatty acids to free cholesterol to form cholesteryl esters which accumulate as a hydrophobic core in the particle, eventually resulting in a spherical morphology. Mature HDL can transfer cholesteryl esters to LDL in exchange for triglyceride via another HDL protein, cholesteryl ester transfer protein (CETP). Other proteins with roles outside of RCT have been identified on HDL. For example, paraoxonase-1 (Pon1) which may be responsible for HDL’s ability to prevent oxidation of LDL particles;9 oxidized LDL are a major contributing factor to atherosclerotic development.10,11
Recently, several groups have applied mass spectrometry (MS)-based proteomic technologies to identify nearly 100 protein components of HDL,12 although there is only substantial agreement among studies on about 35 of these.12 Many of these newly identified HDL associated proteins mediate functions that are surprisingly outside the realm of lipid transport and metabolism. For example, HDL was found to be a host for numerous protease inhibitors as well as mediators of the complement cascade,13 suggesting a possible role for HDL in innate immunity. This raises interesting new possibilities for functional roles of HDL and suggests that many more remain to be discovered.
Despite these advances, a full proteomic understanding of HDL remains incomplete. To date, nearly all mass spectrometry-based proteomics studies of HDL have utilized density gradient ultracentrifugation (UC) based methods for the isolation of HDL from human plasma. This method is optimal for MS-based proteomics as it has the advantage of quantitatively floating the relatively light lipid-containing proteins away from the dense nonlipoprotein associated proteins. However, the separation involves high shear forces and prolonged exposure to elevated salt concentrations that likely alter HDL functionality and composition. For example, Van’t Hooft et al. showed that about half of the apoE on HDL is dissociated during UC and, compared with gel filtration isolated HDL, UC isolated HDL interacted more avidly with the apoE receptor due to either changes in apoE conformation or depletion of other apolipoproteins.14 Thus, there is a significant need to analyze the HDL proteome using alternative separation techniques. An attractive alternative is gel filtration chromatography, which separates plasma components by size. The separation can be performed quickly under physiological salt and shear conditions and thus is less likely to alter the HDL proteome. However, the significant disadvantage of this technique (and most other non-UC techniques) is the overlap between HDL and many high abundance plasma proteins. For example, abundant immunoglobulins can have a MW in the range of 150–900 kDa (depending on class) which overlaps with the 150–360 kDa mass range of most HDL particles. In addition, the presence of extremely high abundance small proteins, such as human albumin (40–60 mg/mL), can significantly contaminate HDL fractions. These contaminants can reduce MS detection of the desired HDL proteins due to ion suppression effects, or by forcing the instrument to devote the majority of its duty cycle to the MS/MS analysis of peptide ions derived from abundant, contaminant proteins. These issues have been major roadblocks to the use of noncentrifugal methods for HDL proteomic analysis.
In this study, we approached this problem in two ways. First, we derived gel filtration conditions that separate HDL from the bulk of high abundance low MW proteins such as albumin. Then we developed an affinity technique to specifically analyze only those proteins that are associated with plasma phospholipids. As a result, we have identified several new HDL associated proteins, provided more evidence that ultracentrifugal separations of HDL may modify its proteome, and demonstrated distinct distribution patterns for a variety of proteins between the different size HDL particles.
Experimental Section
Plasma Collection
Venous blood was collected from fasted (≥12 h), apparently healthy normolipidemic (total cholesterol between 125–200 mg/dL; HDL-C ≥ 40 mg/dL; triglycerides <150 mg/dL) male donors (ages: 21, 22 and 34) by a trained phlebotomist using BD Vacutainer Plus Plastic Citrate Tubes containing buffered sodium citrate (0.105 M) as an anticoagulant. Cellular components were pelleted by centrifugation at ~ 1590× g for 15 min in a Horizon mini-E (Quest Diagnostics) at room temperature. Plasma was stored at 4 °C until gel filtration separation, always within 16 h. Samples were never frozen.
Plasma Separation by Gel Filtration Chromatography
Three-hundred seventy microliters of plasma from a single subject was applied directly to three Superdex 200 gel filtration columns (10/300 GL; GE Healthcare) arranged in series on an ÄKTA FPLC system (GE Healthcare). The sample processed at a flow rate of 0.3 mL/min in standard Tris buffer (STB) (10 mM Tris, 0.15 M NaCl, 1 mM EDTA, 0.2% NaN3). Eluate was collected as 47 1.5-mL fractions on a Frac 900 fraction collector (GE healthcare) maintained at 4 °C. Each fraction was assessed for protein, phospholipid and total cholesterol by colorimetric kits from Wako (Richmond, VA). For ether delipidation protein shift experiments, plasma (5 mL) was delipidated with butanol-di-isopropyl ether (40:60, 10 mL) according to a procedure described by Cham and Knowles.15 The volume of freshly delipidated plasma was then adjusted with STB to match the protein concentration of normal plasma and applied to triple Superdex 200 columns exactly as with normal plasma. Fractions collected from delipidated plasma were not subjected to CSH treatment (described below).
Purification of Phospholipid-Containing Particles Using Calcium Silicate Hydrate (CSH)
To isolate lipoprotein particles from coeluting proteins in the collected fractions, we used a commercially available synthetic calcium silicate hydrate called Lipid Removal Agent (Supelco). This compound, developed for the removal of lipids in biopharmaceutical production, tightly binds lipids and lipoproteins. In a centrifuge tube, 45 μg of CSH (from 100 mg/mL stock solution in 50 mM ammonium bicarbonate) per 1 μg of PL in 400 μL of fraction were mixed gently for 30 min at room temperature. The CSH was then pelleted by centrifugation (~2200× g for 2 min) in a minicentrifuge (Fisher) and the supernatant containing lipid-free plasma proteins was removed. The CSH was then washed with 50 mM ammonium bicarbonate (AB). All PL-containing fractions from each subject’s FPLC separation were carried through this process individually.
Western Blotting for ApoA-I
Purified human apoA-I, UC isolated HDL, supernatant from CSH procedure, and SDS elution were run on 4–15% PAGE, then transferred to a PVDF membrane. Membranes were probed with rabbit antihuman apoA-I antibody (Calbiochem, 178422).
Mass Spectrometry Analysis of Fractions
HDL particles were subjected to trypsin digestion while still bound to the CSH. One and a half micrograms of sequencing grade trypsin (Promega) in 25 μL of 50 mM AB was added to each CSH pellet and incubated at 37 °C overnight on a rotating plate. To collect the digested peptides, the CSH was washed with 125 μL of 50 mM AB. Peptides were first reduced and then carbamidomethylated with dithiothreitol (200 mM; 30 min at 37 °C) and iodoacetamide (800 mM; 30 min at room temperature), respectively. Peptide solutions were then lyophilized to dryness and stored at −20 °C until analyzed by mass spectrometry. Dried peptides were reconstituted in 15 μL of 0.1% formic acid in water. An Agilent 1100 series Autosampler/HPLC was used to draw 0.5 μL of sample and inject it onto a C18 reverse phase column (GRACE; 150 × 0.500 mm) where an acetonitrile concentration gradient (5–30% in water with 0.1% formic acid) was used to elute peptides for online ESI–MS/MS by a QStar XL mass spectrometer (Applied Biosystems). Column cleaning was performed automatically with 2 cycles of a 5–85% acetonitrile gradient lasting 15 min each between runs.
MS Data Analysis
To identify the protein composition of particles contained in the various gel filtration fractions, peak lists generated from analysis of each fraction were scanned against the UniProtKB/Swiss-Prot Protein Knowledgebase (release 57.0, 03/2009) using both the Mascot (version 2.1) and X!Tandem (version 2007.01.01.1) search engines. Search criteria included: human taxonomy, variable modifications of Met oxidation and carbamidomethylation, both peptide tolerance and MS/MS tolerance were set to ±0.15 Da, and up to 3 missed tryptic cleavage sites were allowed. Scaffold software (version Scaffold_2_04_00, Proteome Software) was used to validate MS/MS based peptide and protein identifications. Peptide identification required a value of 90% probability (using data from both Mascot and X!Tandem) using the Peptide Prophet algorithm.16 Positive protein identification also required a value of 90% probability by the Protein Prophet algorithm.17 Also, a minimum of 2 peptides were required unless the protein in question was found with single peptide hits in multiple consecutive fractions that were consistent across all subjects. Since equal volumes of sample were applied to the MS analysis, not equal protein contents, we reasoned that the relative amount of a given protein present in a given fraction should be proportional to the number of spectral counts (i.e., the number of MS/MS spectra assigned to a particular protein) in each fraction. In no case were conclusions made about the relative abundance of two different proteins on the basis of peptide counting. We have previously demonstrated that this approach provides a semiquantitative abundance pattern across the fractions that matches well with patterns derived from immunological analyses.18
Results
Optimization of Gel Filtration Resolution for Human Plasma HDL
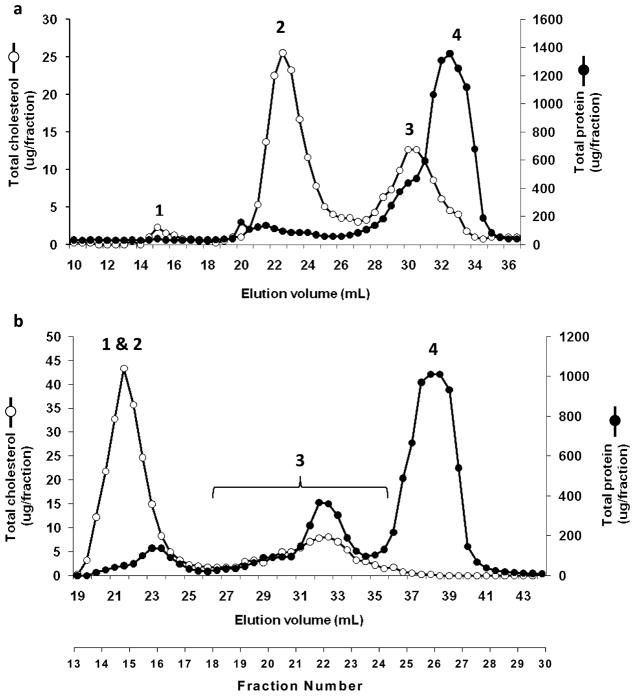
A widely used method for gel filtration-based separations of plasma lipoproteins involves the application of plasma to two Superose 6 columns (GE Healthcare) connected in series. This method has proven useful in analysis of the lipid content of plasma lipoproteins.19 We began by evaluating this method in terms of its effectiveness in separating HDL proteins from the high abundance, nonlipidated proteins such as albumin. Figure 1a shows the protein and total cholesterol distribution of human plasma separated by the tandem Superose 6 protocol. The cholesterol peaks 1, 2, and 3 represent VLDL, LDL and HDL respectively. While the lipid peaks were well distinguished, it is clear that the HDL cholesterol peak underwent substantial overlap with the majority of plasma proteins (peak 4). To optimize separation of HDL from the abundant protein peak, we developed a method that utilized three Superdex 200 columns arranged in series (Figure 1b). In addition to the extra resolving power contributed by a third column, the Superdex 200 matrix pore size distribution allowed for greater resolution within the HDL size range. In this case, VLDL and LDL cholesterol ran together (peak 1 and 2) while HDL cholesterol distributed in a broad peak 3. It is clear that this protocol separates the majority of HDL cholesterol from the free plasma proteins evident in peak 4. Figure 2 shows an SDS-PAGE analysis that compared total human HDL isolated by ultracentrifugation vs a pooled sample of gel filtered HDL, volumes 30–35 mL in Figure 1b. The major HDL proteins apoA-I and apoA-II are visible in both samples. However, the GF sample still contained human albumin. We also observed higher molecular weight proteins in the GF sample that may or may not be truly associated with the HDL particles. Since further optimizations of the gel filtration protocol failed to significantly improve HDL separation from albumin, we elected to pursue methods that would allow affinity isolation of those proteins that were specifically associated with lipid.
Figure 1.
Elution profiles from Superose 6 (2×) and Superdex 200 (3×) size exclusion chromatography configurations. Three-hundred seventy microliters of fresh human plasma from a normal male donor was analyzed by a tandem Superose 6 setup (a) or a triple Superdex setup (b) as described in the Experimental section. Total protein (●, determined by Lowry assay) and total cholesterol (○, enzymatic assay) profiles across the fractions are shown. Peak designations refer to 1, VLDL; 2, LDL; 3, HDL; and 4, lipid-free plasma proteins.
Figure 2.
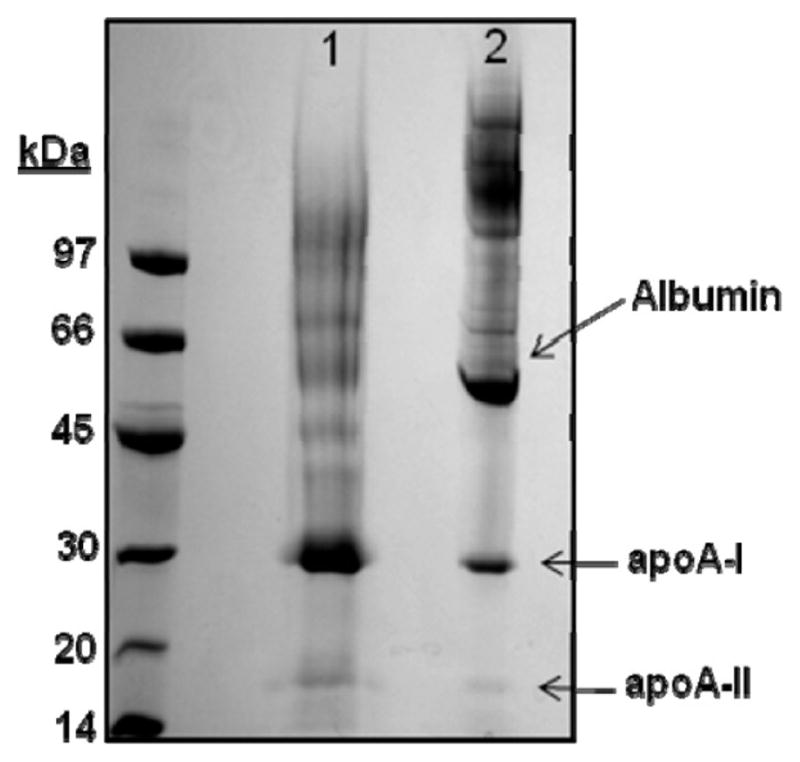
SDS PAGE comparison of total HDL preparations derived from ultracentrifugation (UC) and gel filtration (GF) chromatography. A 4–15% SDS-PAGE analysis of total HDL isolated by UC (lane 1) or pooled HDL fractions from the triple Superdex 200 gel filtration separation (lane 2) is shown. The gel was stained with coomassie blue.
Selection of Lipid Bound Proteins Using Calcium Silicate Hydrate
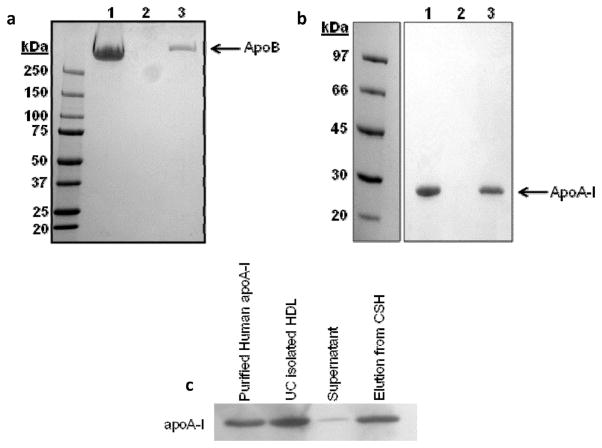
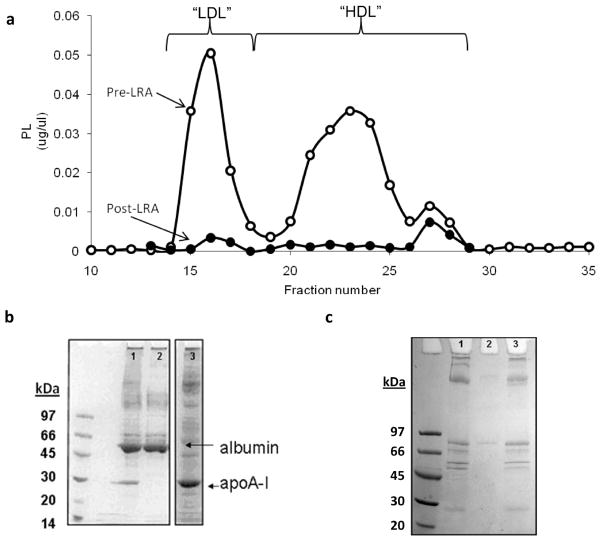
After exploring a number of potential strategies for isolating lipid-bound proteins, we developed a method that utilizes a commercially available hydrated calcium silicate resin (CSH) with a high binding affinity for phospholipid.20 In optimization experiments, we determined that 150 μg of CSH could bind about 1 μg of plasma phospholipid in STB at pH 8.0. We first tested the ability of CSH to sequester HDL and LDL that had been previously purified by ultracentrifugation. For both HDL and LDL, exposure to CSH resulted in the loss of 99.9% of both phospholipid and cholesterol in the flow-through, indicating the quantitative binding of both lipoproteins to the resin (Table 1). Additionally, the major protein components of both LDL (apoB, Figure 3a) and HDL (apoA-I, Figure 3b), were removed from solution after CSH (lanes 1 and 2) and were recovered from the resin with an SDS wash (lane 3). Figure 3c shows the HDL experiment as analyzed by Western blot using an anti-apoA-I antibody, confirming that apoA-I was almost completely removed from the supernatant (Figure 3c). We next assessed the capacity of CSH to remove phospholipid-containing particles from fractions produced by gel filtration chromatography. Figure 4a compares the phospholipid profile of plasma separated by the triple Superdex protocol before and after the fractions were treated with CSH. CSH was capable of quantitatively removing nearly all PL associated with the VLDL/LDL peak as well as the major HDL peak. Interestingly, it failed to bind a small amount of PL that comigrates with plasma proteins. This may represent sequestered phospholipids or lyso-PC’s that are tightly associated with small proteins. SDS-PAGE revealed that the CSH specifically removed apoA-I from the supernatant while allowing albumin and several other proteins to wash through (Figure 4b, lanes 1 and 2). When the resin was boiled in sample buffer and analyzed by SDS-PAGE, it is clear that apoA-I and not albumin was retained on the resin (Figure 4b, lane 3). Similarly, the same analysis performed with fraction 16 in the VLDL/LDL range showed that apoB (Figure 4c, band at the top of the gel) was also selectively retained on the resin.
Table 1.
Quantitative Binding of Ultracentrifugally Isolated LDL and HDL Lipids by CSH
| initial [lipid] (μg/mL) | post CSH [lipid] (μg/mL) | % bound by CSH | ||
|---|---|---|---|---|
| LDL | Phospholipid | 359 | 2 | 99.9 |
| Cholesterol | 251 | <1 | 99.9 | |
| HDL | Phospholipid | 260 | 4 | 99.9 |
| Cholesterol | 72 | <1 | 99.9 |
Figure 3.
Ability of calcium silicate hydrate (CSH) to bind ultracentrifugally isolated human plasma lipoproteins. UC isolated LDL (a) or HDL (b) were analyzed by SDS-PAGE prior to incubation with CSH (lane 1 of each panel). The resulting flow through is shown in lane 2 and the proteins retained on the resin after boiling with SDS sample buffer is shown in lane 3 of each panel. The gels were stained with coomassie blue. (c) Same experiment as in (b), except that apoA-I was detected by Western blot using an antihuman apoA-I antibody.
Figure 4.
Ability of CSH to bind phospholipid-containing particles from fractions collected by gel filtration chromatography. (a) Two identical samples of human plasma were fractionated on the triple Superdex gel filtration set up. One set of fractions was incubated with CSH under the conditions described in the Experimental section (●) while the other fraction was left untreated (○). The traces show the phospholipid content of each fraction as determined by enzymatic assay. (b) Triple Superdex gel filtration fraction 23 (lane 1) was incubated with CSH for 30 min and then the supernatant containing unbound components was removed (lane 2). The CSH was washed with buffer and bound proteins were recovered from CSH by boiling in SDS-sample buffer (lane 3). SDS-PAGE was carried out on a 4–15% gel and stained with coomassie brilliant blue. (c) ApoB containing lipoproteins (fraction 16) were analyzed in the same manner as described for (b).
Given that PL binds extremely tightly to the resin by an unknown mechanism, we have not identified a practical way to recover CSH-bound lipoproteins intact, at least not in a manner consistent with subsequent mass spectrometry analysis. However, we found that we could trypsinize the particles while still in contact with the resin. The resulting peptides were then eluted from the resin and used for MS analysis. Since the lipid remains associated with the resin, it was not necessary to perform subsequent delipidation steps prior to the MS analysis. The obvious disadvantage of this approach is the possibility that certain hydrophobic peptides may remain associated with the lipid after trypsinization. However, we found that overall peptide detection and sequence coverage of most of the lower abundance HDL proteins analyzed by the CSH method was comparable to those isolated by UC without the CSH step (Supplemental Table 1, Supporting Information).
ESI–MS/MS Analysis of Lipid-Associated Proteins
Phospholipid-associated proteins in each PL-containing fraction were identified by LC–ESI–MS/MS and subsequent database searching using criteria described in the experimental section. Our analysis identified 81, 98, and 103 proteins across all fractions for the three subjects studied, of these, 79 were common across all subjects (detailed peptide identification information can be found in Supplement 4, Supporting Information). Many of these proteins had been shown to associate with UC-isolated HDL in previous studies, however many were potentially new phospholipid-associated proteins.
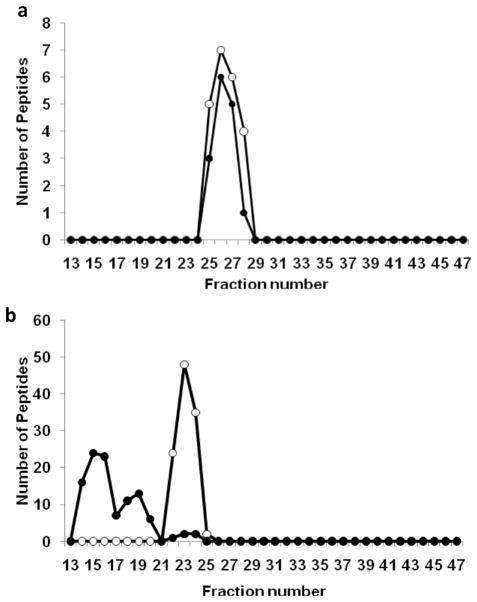
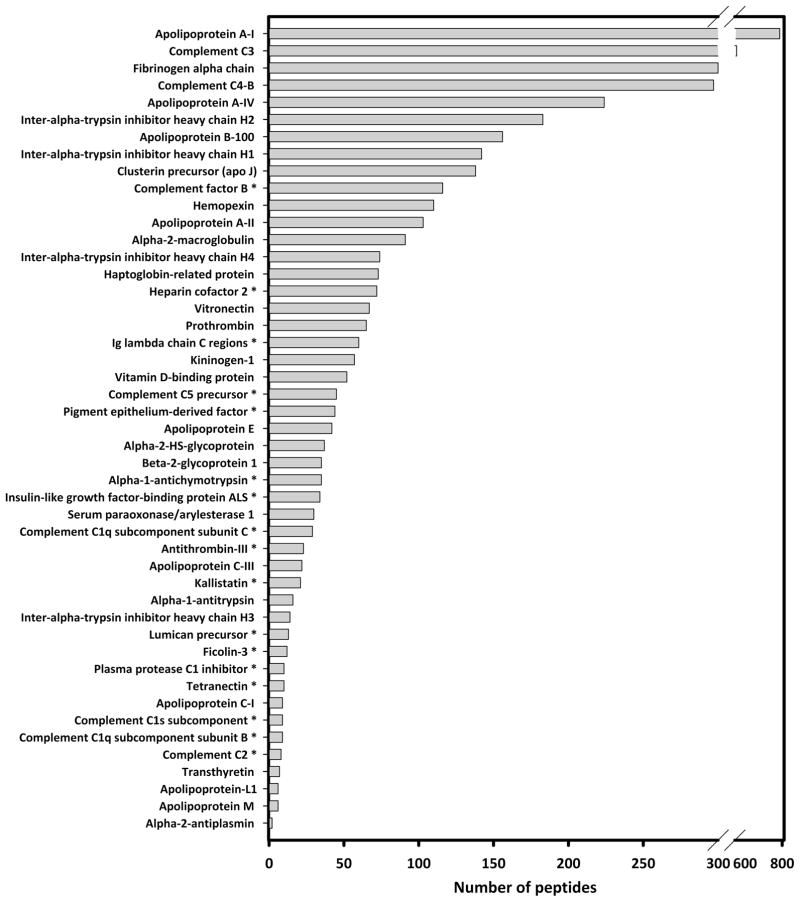
To evaluate the potential for phospholipid independent (i.e., nonspecific) binding to the resin, we performed a set of experiments where human plasma was first subjected to an ether based delipidation procedure shown to cause minimal protein denaturation,15 prior to separation on the columns. The rationale was that lipid-associated proteins will migrate with a different apparent size after the lipid is removed. However, those that are not lipid-associated, but bind to CSH nonspecifically, should elute at the same volume. By monitoring shifts in protein elution patterns in delipidated vs control samples, we distinguished proteins that were most likely associated with lipid. An example of each case is shown in Figure 5. Plasminogen is a common plasma zymogen, the precursor for plasmin, an enzyme responsible for dissolving blood clots. This protein is not known to be associated with lipid and its elution patterns failed to shift when plasma was delipidated (Figure 5a). However, a protein with an established association to HDL particles, complement component C3, showed a dramatic change in its elution pattern when plasma was delipidated prior to separation (Figure 5b). All previously known HDL associated proteins identified in this study were found to undergo some degree of elution profile shift in response to delipidation. Moreover, apolipoprotein B, the primary protein component of LDL, showed similar behavior. Of the 79 proteins that passed our identification criteria for all three subjects studied, 43 proteins exhibited significant elution volume shifts in two independent delipidation experiments. Four proteins whose association with HDL has been previously established were detected but at levels too low to determine if a definitive shift had occurred. These were included in the analysis giving a total of 47 lipid associated proteins. Those proteins that failed to undergo a profile shift (i.e., those that bind the resin via mechanisms independent from PL) are shown in supplement Table 2. The lipid-associated proteins are shown in Figure 6 along with the sum of their peptides across all PL-containing fractions collected from individual triple Superdex runs on all 3 subjects. Of the 47 proteins identified as lipid associated, 17 are newly identified as being associated with lipidated particles in plasma; these proteins are indicated with asterisks in Figure 6. Of these 17 proteins, 14 were found to elute within fractions 19–29, which is where the majority of the apoA-I elutes. These fractions likely correspond to “HDL” as traditionally defined by gradient ultracentrifugation (see Discussion). The other 3 proteins, complement C1q subcomponent subunits B and C and ficolin-3, comigrate with LDL/VLDL sized particles in fractions 13–18.
Figure 5.
Examples of elution profile shifts for proteins upon ether delipidation of fresh human plasma. Protein distribution profiles of selected proteins from untreated (○) and ether delipidated plasma (●) after separation by the triple Superdex setup. The distribution of each protein is represented as spectral count per fraction measured by ESI–MS/MS. (a) Plasminogen, which fails to exhibit a molecular size shift in response to ether delipidation and therefore is not associated with lipid, and (b) complement C3 which does shift, indicating an association with lipid. Representative data shown from two independent experiments are shown.
Figure 6.
Lipid-associated proteins identified in the plasmas of 3 normolipidemic donors. The proteins included in this list met all identification criteria laid out in the Experimental section and showed a shift in elution pattern after ether delipidation, indicating lipid association. The proteins are arranged according to the sum of identified peptides across all gel filtration fractions for all 3 subjects. Proteins indicated with an asterisk have not been previously described as lipid associated proteins, to our knowledge.
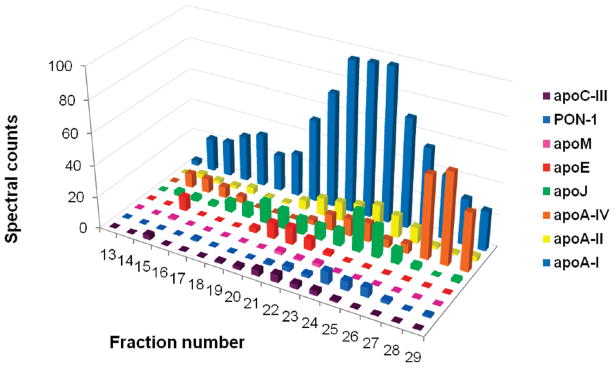
Using HDL subfractions defined by density ultracentrifugation, we previously showed that HDL associated proteins can be grouped into different classifications depending on their distribution patterns between dense and light fractions of HDL.18 Figure 7 displays the relative distribution of selected common HDL proteins across the gel filtration fractions while the elution patterns of all detected lipid-associated proteins are shown as a heat map in Figure 8. Larger, apoB containing lipoproteins eluted in earlier fractions. ApoB protein abundance peaked at fraction 16 while apoA-I protein abundance peaked later in fraction 24. These protein distribution patterns correlated with the PL peaks indicated as “LDL” or “HDL” in Figure 4. The major HDL proteins, apoA-I and apoA-II, were found in nearly all PL-containing fractions. Interestingly, the other lipid associated proteins distributed across fractions in distinct patterns. For example, several of the complement proteins identified seemed to be grouped primarily in fractions 22–24 while apoA-IV was concentrated to the smallest particles eluting in fractions 27 and 28.
Figure 7.
Distribution patterns of common HDL associated proteins across gel filtration fractions. For each protein, the number of spectral counts in a given fraction is represented by bar height. The values represent the sum of counts from 3 subjects.
Figure 8.
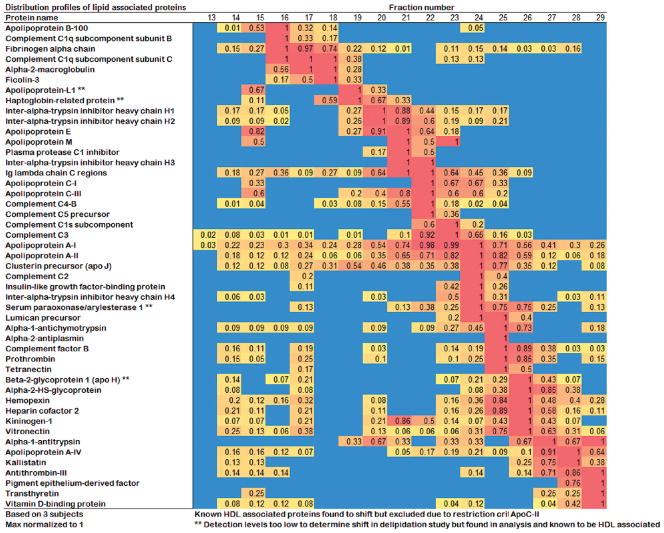
Triple Superdex distribution profiles for identified lipid-associated proteins. For each fraction, the relative abundance (determined by peptide count) is shown. A value of 1.0 was assigned to the fraction containing the highest peptide count for that particular protein and all other fractions were scaled from there. The relative abundance of each can also be assessed by the color of the square with blue representing 0 detected peptides and red representing the highest number.
Discussion
HDL is, by definition, distinguished in terms of particle density as originally exploited for separation by gradient ultracentrifugation.21 Thus, one immediately encounters a nomenclature issue when attempting to separate these particles by methods that do not rely on density. In such a case, it is tempting to define HDL on the basis of its major protein apoA-I. However, it is clear from Figures 7, 8, and previous studies18 that apoB containing lipoproteins such as LDL also contain significant amounts of apoA-I. In this study, we have separated fresh plasma across a broad size range that includes the traditional VLDL, LDL, and HDL particle sizes. By treating all fractions with a phospholipid binding agent, we have technically measured proteins that are associated with plasma phospholipid, rather than any specific lipoprotein class. Nevertheless, to relate these gel filtration results to traditional definitions of HDL, we elected to use the presence of apoB, the core constituent of LDL as the key distinguisher. We therefore defined fractions 14–18 as the VLDL/LDL fraction due to the presence of apoB. The remaining fractions 19–29 were considered “HDL”, though it is recognized that GF and UC isolate overlapping, but possibly distinct sets of particles. We suggest these particles might be better referred to as PL-rich lipoproteins.
A recent study reported a proteomic analysis of fractions of human plasma collected by gel filtration on a single Superdex 200 column.22 This study identified the majority of known HDL associated proteins but made no attempt to distinguish HDL-associated proteins from the multitude of abundant plasma proteins which coelute. In this work, we have overcome a major barrier standing in the way of using non-UC based methods to separate human plasma HDL for proteomic analysis. The use of the calcium silicate hydrate combined with optimized gel filtration conditions resulted in the identification of some 14 new proteins that associate with phospholipid in the HDL fractions.
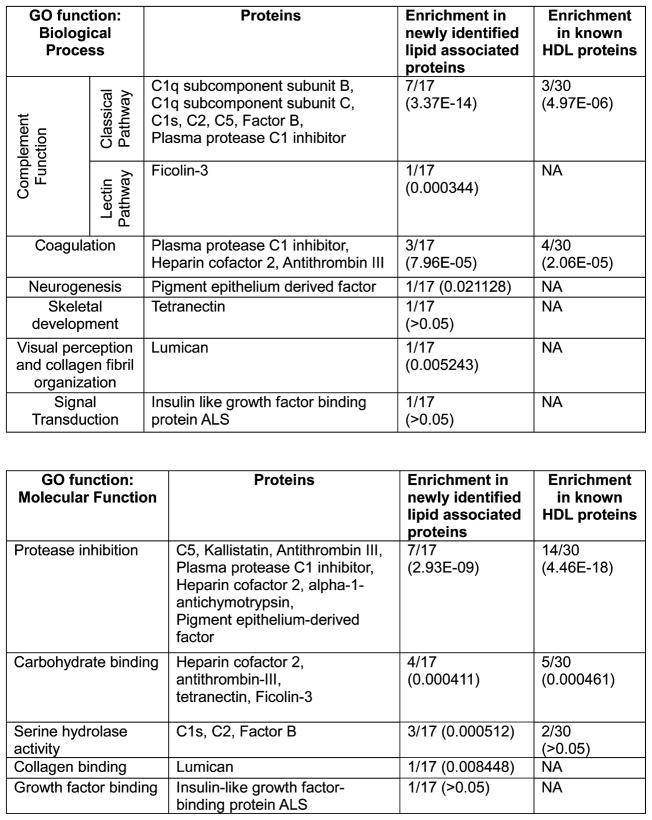
We examined the Biological Process and Molecular Function Gene Ontology (GO) annotations for the 17 newly lipid associated proteins as well as the 30 previously identified HDL associated proteins found in this study. Our results were consistent with previous HDL proteome studies but also pointed out some potentially new functions (Figure 9). For each annotated function, we calculated its enrichment among either previously known (# of proteins with given function/30) or the newly identified (# of proteins with given function/17) lipid associated proteins found in this study. The statistical significance is given by p-values in parentheses. In addition to those identified by Vaisar et al.,13 we have identified 8 phospholipid associated proteins with known functions in the complement cascade. Three of these were found to comigrate with apoB containing lipoproteins—complement C1q subcomponent subunits B and C which function in activation of the classical pathway23 and ficolin-3, which is involved in complement activation via the lectin pathway.24 The remainder, complement C1s, C2, C5, factor B and plasma protease C1 inhibitor, were distributed across the HDL containing fractions. The addition of these proteins to HDL’s repertoire further implicates the lipoprotein class in the complement pathway and innate immunity. This study also confirmed 6 other complement proteins previously described in Vaisar’s study, bringing the total count of HDL associated proteins with roles in complement function to about 14. Interestingly, plasma protease C1 inhibitor also plays a role in blood coagulation along with heparin cofactor 2 and antithrombin III. Isolated HDL have been found to possess anticoagulant properties,25 however the physical basis for this is not well understood. The presence of these proteins on HDL may begin to explain this observation.
Figure 9.
Gene Ontology functional associations of newly identified lipoprotein associated proteins. Identified proteins are grouped by functional category (left column) and enrichment of a particular function for either newly identified or previously established proteins is presented as the number of proteins possessing function divided by total number of proteins in group. P values are given in parentheses.
Others have identified several HDL proteins belonging to the serine protease inhibitor (SERPIN) superfamily.13,18,26,27 In this study we have identified 2 previously unreported SERPIN proteins. The first, alpha-1-antichymotrypsin (AACT) is an acute phase protein secreted by the liver under inflammatory conditions and has inhibitory activity against several proteases.28,29 The second, pigment epithelium derived factor (PEDF), is a member of the serine protease superfamily but has no known protease inhibitor activity.30 GO analysis annotated this protein as a positive regulator of neurogenesis, promoting the development and maintenance of motor neurons.31 Recently, a causal role for PEDF in obesity induced insulin resistance has been identified.32
The remaining newly identified proteins were linked to functions which have not yet been attributed to HDL. Known functions include skeletal development (tetranectin),33 collagen fibril organization and visual perception (lumican),34,35 and signal transduction (insulin like growth factor binding protein acid labile subunit; ALS).36 The latter is a liver secreted protein whose function is to bind to, and increase the half-life of, insulin like growth factor (IGF) in the plasma. Humans deficient in ALS exhibit decreased levels of plasma IGF-I, IGF-II and IGF binding protein 3 due to increased clearance.37 The reason for HDL localization of these proteins is not yet apparent and invites future study.
In addition to discovering new HDL-associated proteins, the increased fractionation potential of GF has allowed us to visualize HDL protein distribution across particle size with unprecedented resolution. Figure 8 shows that most of the identified proteins were distributed in distinct patterns across the different sized particles. In a previous study, we separated human HDL into only 5 density subfractions by UC and also found that individual HDL proteins can be grouped with respect to their distributions among the subfractions.18 In that study, we found that many of the lower abundance proteins tended to cluster in the densest subfractions which are generally suggestive of a smaller particle size. The results of the current study confirmed many of these classifications. For example, apoA-IV, alpha-1-antitrypsin and transthyretin were found exclusively in the densest HDL3c fractions by UC and also in the smallest sized fractions by GF analysis. As in the UC study, common HDL proteins like apoA-I and apoA-II were distributed across the entire HDL range. However, there were several examples of proteins that exhibited different GF elution patterns than expected from the UC data. For example, apoL-I was exclusively found in the most dense particles by UC, but appeared in quite large HDL particles by GF. ApoE could be found distributed through all density subfractions by UC, but was focused in a rather tight pattern of larger HDL particles by GF. Although the correlation between particle density and size is not absolute, these observations lend support to the idea that high salt or sheer conditions encountered during UC separation of plasma may alter the distribution of certain proteins across HDL subpopulations. Indeed, the fact that 14 new proteins were identified by GF indicates that UC may even completely remove some of the more weakly associated HDL proteins. This highlights the importance of developing alternative separation and analysis strategies for characterizing the HDL proteome.
We have previously proposed that protein clusters detected in our UC study may be indicative of distinct subsets of HDL particles which might display unique biological functions. The current study revealed several interesting observations that support this idea. First, we noticed the tight comigration of apoL-I and haptoglobin related protein (HGRP). These are the active protein components of the trypanosome lytic factor (TLF) which is an HDL particle shown to have specific lytic activity against Trypansoma bruceii, a protozoan parasite from the class of organisms responsible for African Sleeping Sickness.38 These proteins coelute in larger sized HDL fractions and also seem to comigrate to a lesser extent in LDL sized particles. This pair comprises one of the few biochemically characterized HDL subspecies with a defined function. Second, we noted several additional comigrating pairs including apoE and apoM, apoC-III and complement C4–B, complement C2 and insulin-like growth factor binding protein, and complement factor B and prothrombin. Although it is possible that these proteins may have comigrated by pure coincidence, it is reasonable to propose that they may participate in potential structural interactions that sequester them to the same set of HDL particles, perhaps to perform an as yet undefined function like the apoL-I and HGRP pair. In addition to sharing the similar GO functions, many of these newly identified proteins interact with known HDL proteins through physical interactions. When we examined the 47 proteins in the human protein–protein interaction network from the Human Protein Reference Database (HPRD),39 we found 8 out of the 14 new HDL proteins have a direct interaction with the known HDL proteins, and an additional 3 can be connected to known HDL proteins by one intermediate protein (Supplemental Figure 3, Supporting Information). This supports the possibility of potential roles for these new proteins in HDL function. Further work, using additional orthogonal separation techniques as well as immunoprecipitation experiments, will be required to test this hypothesis.
Conclusions
We have developed new separation and analysis technologies that remove practical barriers to evaluating the HDL proteome using noncentrifugal separation techniques. This method identified new HDL associated proteins and added more weight to the growing body of evidence that distinct HDL subparticles exist that contain distinct sets of interacting proteins. Currently, therapeutic studies on cardiovascular disease are directed at raising total plasma HDL cholesterol (i.e., niacin and CETP inhibitors) without regard for the specific subspecies that may be altered. Further study of HDL subspecies may result in the identification of particles with superior cardioprotective properties or altogether new HDL functions. Knowledge of such subspecies may help to focus development of HDL modification strategies.
Supplementary Material
Footnotes
Supporting Information Available: Supplemental figures and tables. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Scott M. Gordon, Email: Gordonst@mail.uc.edu.
Jingyuan Deng, Email: dengjn@email.uc.edu.
L. Jason Lu, Email: long.lu@cchmc.org.
References
- 1.Scanu AM, Wisdom C. Serum lipoproteins structure and function. Annu Rev Biochem. 1972;41:703–730. doi: 10.1146/annurev.bi.41.070172.003415. [DOI] [PubMed] [Google Scholar]
- 2.Franceschini G, Maderna P, Sirtori CR. Reverse cholesterol transport: physiology and pharmacology. Atherosclerosis. 1991;88(2–3):99–107. doi: 10.1016/0021-9150(91)90073-c. [DOI] [PubMed] [Google Scholar]
- 3.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. Am J Med. 1977;62(5):707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 4.Gordon DJ, Probstfield JL, Garrison RJ, Neaton JD, Castelli WP, Knoke JD, Jacobs DR, Jr, Bangdiwala S, Tyroler HA. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 1989;79(1):8–15. doi: 10.1161/01.cir.79.1.8. [DOI] [PubMed] [Google Scholar]
- 5.Mackness MI, Arrol S, Durrington PN. Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett. 1991;286(1–2):152–154. doi: 10.1016/0014-5793(91)80962-3. [DOI] [PubMed] [Google Scholar]
- 6.Cockerill GW, Rye KA, Gamble JR, Vadas MA, Barter PJ. High-density lipoproteins inhibit cytokine-induced expression of endothelial cell adhesion molecules. Arterioscler, Thromb Vasc Biol. 1995;15(11):1987–1994. doi: 10.1161/01.atv.15.11.1987. [DOI] [PubMed] [Google Scholar]
- 7.Tolle M, Pawlak A, Schuchardt M, Kawamura A, Tietge UJ, Lorkowski S, Keul P, Assmann G, Chun J, Levkau B, van der Giet M, Nofer JR. HDL-associated lysosphingolipids inhibit NAD(P)H oxidase-dependent monocyte chemoattractant protein-1 production. Arterioscler, Thromb Vasc Biol. 2008;28(8):1542–1548. doi: 10.1161/ATVBAHA.107.161042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidsson P, Hulthe J, Fagerberg B, Camejo G. Proteomics of apolipoproteins and associated proteins from plasma high-density lipoproteins. Arterioscler, Thromb Vasc Biol. 2010;30(2):156–163. doi: 10.1161/ATVBAHA.108.179317. [DOI] [PubMed] [Google Scholar]
- 9.Shih DM, Gu L, Xia YR, Navab M, Li WF, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, Lusis AJ. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394(6690):284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- 10.Hoff HF, O’Neil J, Chisolm GM, III, Cole TB, Quehenberger O, Esterbauer H, Jurgens G. Modification of low density lipoprotein with 4-hydroxynonenal induces uptake by macrophages. Arteriosclerosis. 1989;9(4):538–549. doi: 10.1161/01.atv.9.4.538. [DOI] [PubMed] [Google Scholar]
- 11.Shao B, Oda MN, Vaisar T, Oram JF, Heinecke JW. Pathways for oxidation of high-density lipoprotein in human cardiovascular disease. Curr Opin Mol Ther. 2006;8(3):198–205. [PubMed] [Google Scholar]
- 12.Gordon S, Durairaj A, Lu J, Davidson WS. High-Density Lipoprotein Proteomics: Identifying New Drug Targets and Biomarkers by Understanding Functionality. Current Cardiovascular Risk Reports 4. 2010;4(1):1–5. doi: 10.1007/s12170-009-0069-9. Current Medicine Group LLC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117(3):746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van’t HF, Havel RJ. Metabolism of apolipoprotein E in plasma high density lipoproteins from normal and cholesterol-fed rats. J Biol Chem. 1982;257(18):10996–11001. [PubMed] [Google Scholar]
- 15.Cham BE, Knowles BR. A solvent system for delipidation of plasma or serum without protein precipitation. J Lipid Res. 1976;17(2):176–181. [PubMed] [Google Scholar]
- 16.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 18.Davidson WS, Silva RA, Chantepie S, Lagor WR, Chapman MJ, Kontush A. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler, Thromb Vasc Biol. 2009;29(6):870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Usui S, Hara Y, Hosaki S, Okazaki M. A new on-line dual enzymatic method for simultaneous quantification of cholesterol and triglycerides in lipoproteins by HPLC. J Lipid Res. 2002;43(5):805–814. [PubMed] [Google Scholar]
- 20.An Trinh. LRA (Lipid Removal Agent): Synthetic calcium silicate hydrate for the selective removal of lipids, endotoxins and other bio-organic molecules. Sigma-Aldrich Catalogue; 2010. [Google Scholar]
- 21.Havel RJ, EDER HA, BRAGDON JH. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J Clin Invest. 1955;34(9):1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins LA, Mirza SP, Kissebah AH, Olivier M. An integrated approach for the comprehensive characterization of lipoproteins from human plasma using FPLC and nano-HPLC-tandem mass spectrometry. Physiol Genomics. 2010;40(3):208–215. doi: 10.1152/physiolgenomics.00136.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenner AJ. Functional aspects of the C1q receptors. Behring Inst Mitt. 1993;(93):241–253. [PubMed] [Google Scholar]
- 24.Matsushita M, Endo Y, Hamasaki N, Fujita T. Activation of the lectin complement pathway by ficolins. Int Immunopharmacol. 2001;1(3):359–363. doi: 10.1016/s1567-5769(00)00045-x. [DOI] [PubMed] [Google Scholar]
- 25.Cuchel M, Rader DJ. The role of high density lipoproteins in thrombosis. Sci World J. 2002;2:89–95. doi: 10.1100/tsw.2002.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heller M, Stalder D, Schlappritzi E, Hayn G, Matter U, Haeberli A. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 2005;5(10):2619–2630. doi: 10.1002/pmic.200401233. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5(5):1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 28.Han YP, Yan C, Garner WL. Proteolytic activation of matrix metalloproteinase-9 in skin wound healing is inhibited by alpha-1-antichymotrypsin. J Invest Dermatol. 2008;128(9):2334–2342. doi: 10.1038/jid.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plotnick MI, Rubin H, Schechter NM. The effects of reactive site location on the inhibitory properties of the serpin alpha(1)-antichymotrypsin. J Biol Chem. 2002;277(33):29927–29935. doi: 10.1074/jbc.M202374200. [DOI] [PubMed] [Google Scholar]
- 30.Becerra SP, Sagasti A, Spinella P, Notario V. Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J Biol Chem. 1995;270(43):25992–25999. doi: 10.1074/jbc.270.43.25992. [DOI] [PubMed] [Google Scholar]
- 31.Houenou LJ, D’Costa AP, Li L, Turgeon VL, Enyadike C, Alberdi E, Becerra SP. Pigment epithelium-derived factor promotes the survival and differentiation of developing spinal motor neurons. J Comp Neurol. 1999;412(3):506–514. [PubMed] [Google Scholar]
- 32.Crowe S, Wu LE, Economou C, Turpin SM, Matzaris M, Hoehn KL, Hevener AL, James DE, Duh EJ, Watt MJ. Pigment epithelium-derived factor contributes to insulin resistance in obesity. Cell Metab. 2009;10(1):40–47. doi: 10.1016/j.cmet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Wewer UM, Ibaraki K, Schjorring P, Durkin ME, Young MF, Albrechtsen R. A potential role for tetranectin in mineralization during osteogenesis. J Cell Biol. 1994;127(6 Pt 1):1767–1775. doi: 10.1083/jcb.127.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quantock AJ, Meek KM, Chakravarti S. An x-ray diffraction investigation of corneal structure in lumican-deficient mice. Invest Ophthalmol Vis Sci. 2001;42(8):1750–1756. [PubMed] [Google Scholar]
- 35.Ezura Y, Chakravarti S, Oldberg A, Chervoneva I, Birk DE. Differential expression of lumican and fibromodulin regulate collagen fibrillogenesis in developing mouse tendons. J Cell Biol. 2000;151(4):779–788. doi: 10.1083/jcb.151.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Twigg SM, Kiefer MC, Zapf J, Baxter RC. Insulin-like growth factor-binding protein 5 complexes with the acid-labile subunit. Role of the carboxyl-terminal domain. J Biol Chem. 1998;273(44):28791–28798. doi: 10.1074/jbc.273.44.28791. [DOI] [PubMed] [Google Scholar]
- 37.Domene HM, Hwa V, Argente J, Wit JM, Camacho-Hubner C, Jasper HG, Pozo J, van Duyvenvoorde HA, Yakar S, Fofanova-Gambetti OV, Rosenfeld RG. Human acid-labile subunit deficiency: clinical, endocrine and metabolic consequences. Horm Res. 2009;72(3):129–141. doi: 10.1159/000232486. [DOI] [PubMed] [Google Scholar]
- 38.Thomson R, Samanovic M, Raper J. Activity of trypanosome lytic factor: a novel component of innate immunity. Future Microbiol. 2009;4:789–796. doi: 10.2217/FMB.09.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.