Summary
The multi-subunit Sin3 co-repressor complex regulates gene transcription through deacetylation of nucleosomes. However, the full range of Sin3 activities and targets is not well understood. Here, we have investigated genome-wide binding of mouse Sin3 and RBP2 as well as histone modifications and nucleosome positioning as a function of myogenic differentiation. Remarkably, we find that Sin3 complexes spread immediately downstream of the transcription start site on repressed and transcribed genes during differentiation. We show that RBP2 is part of a Sin3 complex, and on a subset of E2F4 target genes, the coordinated activity of Sin3 and RBP2 leads to deacetylation, demethylation, and repositioning of nucleosomes. Our work provides evidence for coordinated binding of Sin3, chromatin modifications, and chromatin remodeling within discrete regulatory regions, suggesting a model in which spreading of Sin3 binding is ultimately linked to permanent gene silencing on a subset of E2F4 target genes.
Introduction
Chromatin structure and compaction can be altered by a combination of post-translational histone modifications and the specific positioning of nucleosomes. Histone acetyl-transferases (HATs) and deacetylases (HDACs), recruited by sequence-specific activators and repressors, respectively, antagonistically regulate the acetylation of lysines on amino-terminal histone tails and directly link histone modifications with gene expression (Grunstein, 1997). Mammalian HDAC1 is a deacetylase that is highly homologous to yeast Rpd3 (Ekwall, 2005). Studies in yeast and mammalian cells showed that HDAC1/Rpd3 is an enzymatic component of multi-protein complexes containing the Sin3 co-repressor protein. In mammalian cells, the Sin3 core complex consists of at least eight subunits (Alland et al., 2002; Hassig et al., 1997; Laherty et al., 1997; Zhang et al., 1997). However, there is considerable disagreement regarding a ‘holo-Sin3’ complex, most likely due to transient associations with, and heterogeneity of, sub-stoichiometric regulatory proteins, including Swi/Snf remodeling proteins, Retinoblastoma (RB) binding protein 2 (RBP2), and other proteins (Hayakawa et al., 2007; Nagl et al., 2007; Sif et al., 2001). Interestingly, RBP2 was recently shown to be a demethylase specific for di- and tri-methylated lysine 4 of histone H3 (Christensen et al., 2007; Klose et al., 2007). Thus, the Sin3 complex provides a versatile platform for chromatin modifying and remodeling activities.
Sin3/Rpd3 co-repressor complexes are recruited to promoter regions via sequence-specific repressors such as Ume6 or Mad in yeast and mammalian cells, respectively, resulting in localized deacetylation of histones within promoter regions and transcriptional silencing (Ayer et al., 1995; Kadosh and Struhl, 1997; Schreiber-Agus et al., 1995). Interestingly, in addition to its well-established role in promoter binding and gene repression, distinct Sin3/Rpd3 complexes have recently been identified in both budding and fission yeast and shown to deacetylate nucleosomes within the coding regions of active and repressed genes, preventing spurious forward and antisense transcription (Carrozza et al., 2005; Keogh et al., 2005; Li et al., 2007; Nicolas et al., 2007). However, the relationship between yeast and mammalian Sin3 complexes has not been completely resolved, and whether mammalian complex(es) exhibit regulatory activities (besides histone deacetylation) analogous to those in yeast is not known.
Expression of cell cycle genes is regulated by the reversible recruitment of the E2F/DP family of transcription factors and associated chromatin remodeling enzymes during discrete stages of the mammalian cell cycle (Blais and Dynlacht, 2007; Frolov and Dyson, 2004). In quiescent or early G1 cells, E2F4/DP and associated retinoblastoma tumor suppressor protein (pRb) family members p107 and p130 binds to the promoters of cell cycle regulated genes, in some instances recruiting the Sin3/HDAC complex and repressing transcription (Rayman et al., 2002). As cells progress into S phase, E2F4 and Sin3 dissociate from genes, leading to increased histone H3 and H4 acetylation and gene expression (Balciunaite et al., 2005; Rayman et al., 2002; Takahashi et al., 2000). This strongly suggests a model in which E2F4-pocket protein complexes periodically and reversibly recruit Sin3 to cell cycle regulated genes during cell cycle progression.
We have also postulated a role for E2F4 and HDACs in terminal differentiation of skeletal muscle cells, wherein cell cycle genes are de-acetylated during cell cycle arrest and are subsequently permanently silenced in differentiated myotubes through pRb-mediated methylation of histone H3 lysine 27 (Blais et al., 2007). However, the molecular mechanisms by which genes are permanently repressed in a step-wise fashion during differentiation are not understood, and a general description of changes at the level of chromatin remodeling is completely lacking.
In order to gain insight into the molecular mechanisms of permanent gene repression in differentiated cells, we performed extensive genome-wide transcription factor binding analyses in which we coupled chromatin immunoprecipitation (ChIP) with promoter microarrays (ChIP-on-chip). Here, we performed large-scale binding analyses for Sin3A, Sin3B, RBP2 and E2F4, and in parallel, we analyzed histone H3 acetylation and methylation levels, nucleosome positioning, and gene expression in growing and differentiated skeletal muscle cells. Our data suggest that Sin3 and RBP2 play a concerted role in repressing expression of a subset of E2F4 target genes through the modification and repositioning of nucleosomes in differentiated cells.
Results
Recruitment of Sin3 complexes to diverse sets of target genes
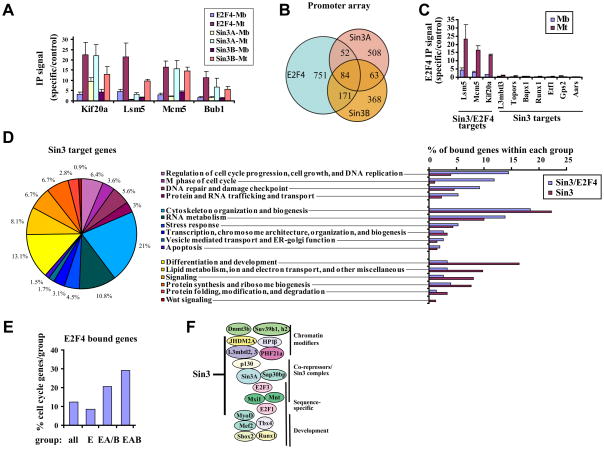
Our previous studies indicated that repression of E2F target genes in differentiated myotubes proceeds through a succession of steps in which promoters of cell cycle genes are de-acetylated and subsequently methylated on histone H3 (Blais et al., 2007). We asked how the first step, namely, histone de-acetylation, was achieved in growing C2C12 myoblasts and cells differentiated to produce myotubes through mitogen depletion. Using quantitative chromatin immunoprecipitation (qChIP), we showed that the Sin3 co-repressor is recruited to E2F4 target genes in terminally differentiated myotubes (Fig. 1A).
Figure 1. Identification and characterization of Sin3 targets in differentiated C2C12 cells.
(A) Analysis of Sin3 and E2F4 binding to selected E2F4 target genes in myoblasts (Mb) and differentiated myotubes (Mt) by ChIP and quantitative real time PCR (qChIP). IP signal was defined as the ratio of IP/Input for specific versus a control amplicon (Gapdh). The average of three independent experiments is shown. Error bars represent standard deviation. (B) Venn diagrams showing overlap between E2F4 and Sin3 target genes identified in differentiated myotubes using genome-wide ChIP-on-chip. Target genes were deduced from at least two independent experiments (Fig. S3). (C) Verification of E2F4 binding to selected Sin3/E2F4 and Sin3 target genes by qChIP in growing myoblasts (Mb) and differentiated myotubes (Mt) as in panel A. (D) Distribution of GO annotations for Sin3 targets. In some instances, a gene is assigned to more than one category. The percentage refers to the number of bound genes within a particular category in relation to the total number of bound genes that have a GO annotation. The histogram depicts the distribution of GO categories among genes bound by Sin3 and E2F4 versus genes bound by Sin3 only. Percentages for the histogram were calculated as described above. (E) The percentage of cell cycle related genes that fall into the indicated groups bound by E2F4 is shown. E: genes bound by E2F4; EA/B: genes bound by E2F4 and Sin3A or Sin3B; EAB: genes bound by E2F4, Sin3A and Sin3B. (F) Sin3 binds to a cadre of genes involved in transcriptional regulation.
To ask whether Sin3 plays a global role in transcriptional regulation during differentiation or if it is restricted to E2F4 target genes in myotubes, we performed genome-wide factor location analysis with antibodies specific for mouse Sin3A and Sin3B. (Fig. S1A). We performed genome-wide ChIP-on-chip experiments with promoter arrays that contained approximately 17,000 known mouse genes centered on the region from -5.5 kb to +2.5 kb relative to the TSS at 250 bp resolution. Analysis of at least two independent experiments for each factor (Fig. S3) identified 707 target genes for Sin3A, 686 target genes for Sin3B, and 1058 target genes for E2F4 (Supplemental Table 1). A comparison of Sin3 and E2F4 target genes indicated limited overlap (Fig. 1B). The vast majority of Sin3 target genes (80% for Sin3A and 62% for Sin3B) were not bound by E2F4. Using qChIP analysis we confirmed that this population of Sin3 target genes did not bind E2F4 in myotubes (Fig. 1C). In addition, we verified Sin3A and Sin3B binding to a select group of Sin3 targets by qChIP and found that many genes were bound by both Sin3A and Sin3B (Fig. S1C). Although we and others have uncovered functional distinctions between the two isoforms (David et al., 2008) (and data not shown), our goal here was to capture as many Sin3 binding events as possible, without respect to isoform differences, and therefore, we will collectively refer to genes bound by Sin3A, Sin3B, or both, as Sin3 targets for simplicity, unless otherwise noted.
To functionally annotate our set of Sin3 target genes and to test whether genes bound by E2F4 and Sin3, or Sin3 only, play different physiological roles, we clustered them using the Gene Ontology (GO) program DAVID (Dennis et al., 2003) and extensive manual curation (Fig. 1D-F, and Supplemental Table 1). Generally, Sin3 target genes were enriched in categories related to cell cycle regulation, transcription, mitochondrial and cellular protein synthesis, lipid metabolism, stress response, and RNA metabolism, a result reminiscent of depletion and knockout experiments conducted in yeast, Drosophila, and mammalian cells (Dannenberg et al., 2005; Kurdistani et al., 2002; Pile et al., 2003; Robert et al., 2004). When compared to genes bound by Sin3 alone, Sin3/E2F4 targets were more strongly enriched in several functional categories, particularly those related to cell cycle regulation, DNA repair and DNA damage checkpoint (Fig. 1D and 1E). This strongly suggests that Sin3/E2F4 complexes represent a major co-repressor of cell cycle genes during differentiation. Genes bound exclusively by Sin3 were more highly enriched in processes related to mitochondrial and cellular protein synthesis, differentiation and development, and lipid metabolism. Interestingly, we also found a sub-cluster of Sin3-only targets involved in Wnt signaling. Thus, genes bound by Sin3 and E2F4 function within distinct cellular pathways as compared to genes that are bound by Sin3 only.
Sin3 binds activated and repressed genes at distinct positions
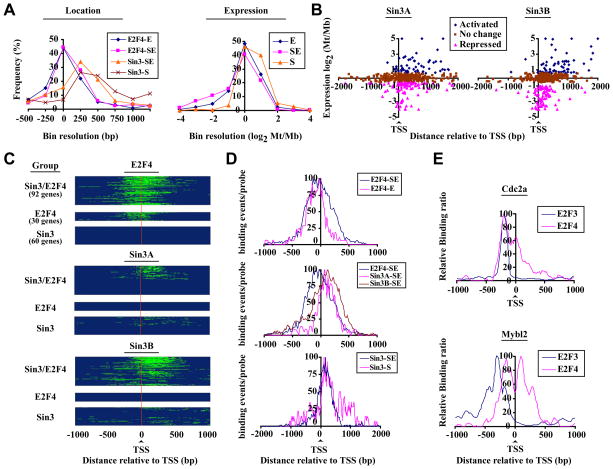
In mammalian cells, Sin3 is recruited to promoter regions by sequence-specific repressors to silence gene transcription (Silverstein and Ekwall, 2005). The identification of genes that differentially recruit Sin3 complexes (in the presence or absence of E2F4) prompted us to test whether these complexes differ with respect to binding site preferences (Fig. 2A, panel 1). Surprisingly, we found that Sin3 was bound primarily to regions downstream of the TSS rather than promoters. Interestingly, we also observed that E2F4 binding in the presence of Sin3 is centered about the TSS, whereas E2F4 binding in the absence of Sin3 is slightly biased toward the upstream region.
Figure 2. Selective targeting of Sin3 to transcribed regions of differentially expressed groups of genes.
(A) Distribution of E2F4, Sin3A, and Sin3B binding (panel 1) and expression (panel 2) of distinct E2F4 and Sin3 target genes, deduced from promoter array data. Groups represent genes bound by E2F4 (E) or Sin3 (S) only or by both Sin3 and E2F4 (SE). Mb: myoblasts, Mt: myotubes. (B) Location of Sin3 binding (central probe) relative to the TSS was plotted as a function of relative expression during differentiation (Blais and Dynlacht, 2007). Activated: log2 Mt/Mb>0.58, repressed: Mt/Mb < −0.58. (C) Heatmaps of E2F4, Sin3A, and Sin3B binding deduced from tiling array data. Genes were grouped as described for panel A. Probes associated with factor binding are shown in green. A binding event (peak) was defined by a minimum of 3 probes with log2 (IP/Input) > 1, maximally spaced by 80 bp (Fig. S3). The results of three independent experiments are shown. (D) E2F4, Sin3A, and Sin3B binding profiles on genes grouped (as in panel A) according to factor occupancy. For each group, genes were aligned with respect to their TSS. Probes associated with a binding event (as determined by our peak finding algorithms) were set to 1, while all others were set to 0. The total number of binding events for all genes per group per probe was calculated, normalized for maximum binding, and plotted relative to the TSS. (E) Binding profiles for E2F3b and E2F4 on genes bound by E2F4 (Cdc2a) or by E2F4 and Sin3 (Mybl2).
Next, we analyzed whether genes bound by both E2F4 and Sin3 differ with respect to expression levels, when compared to genes bound by E2F4 or Sin3 alone (Fig. 2A, panel 2). Genes bound by E2F4 and Sin3 are more highly repressed (average log2 Mt/Mb of -0.90) compared to genes bound by E2F4 (average log2 Mt/Mb of −0.49). Genes bound by Sin3 alone, on average, did not show a change in expression, although Sin3 binds to a sub-cluster of highly expressed genes (Fig. 2A, panel 2 and Fig. 2B). Thus, Sin3 preferentially binds to the downstream regions of both repressed and activated genes. In budding yeast, a Sin3/Rpd3 complex was also shown to bind within coding regions of transcribed and infrequently transcribed genes (Carrozza et al., 2005; Keogh et al., 2005; Kurdistani et al., 2002; Li et al., 2007). To test whether the positional bias for mammalian Sin3 binding reflects differences in expression levels, we plotted the location of Sin3 binding (based on our promoter array data) as a function of gene expression during differentiation (Fig. 2B) (Blais et al., 2007). Sin3 (Sin3A and Sin3B) binding was restricted to regions immediately downstream of the TSS (median distance from TSS: +182 bp) on genes that were repressed (Fig. 2B). In contrast, Sin3 spreads further into downstream regions on genes that are not repressed, and this observed positional bias was even more pronounced on genes up-regulated in differentiated C2C12 myotubes (median distance from TSS: +568 bp). Surprisingly, our analysis also clearly showed that the majority of Sin3 target genes were not repressed: We found that only 20 percent of Sin3 target genes were repressed (log2<−0.58) in differentiated versus growing C2C12 cells (Fig. 2B), and E2F4 targets accounted for the majority (58%) of these repressed genes (data not shown). These observations fail to support the prevailing notion that mammalian Sin3 is generally recruited to proximal promoters via sequence-specific repressor proteins. Further, binding of Sin3 complexes is not restricted to genes that are repressed.
Sin3 complexes spread over regions downstream of the TSS
To define Sin3 and E2F4 binding events more precisely, we designed high-density tiling arrays (Supplemental Information) and performed ChIP-on-chip analysis on both factors (Supplemental Table 2). We observed multiple, overlapping E2F4 and Sin3 binding events downstream of the TSS per gene, strongly suggesting coordinated recruitment, or spreading, of Sin3/E2F4 co-repressor complexes (Fig. 2C-D and Fig. S4A). We also observed a small sub-population of E2F4 binding events specifically in the upstream promoter region that is apparently not associated with Sin3 binding. In agreement with the genome-wide promoter array analysis, our high density tiling array data confirmed that Sin3 binding is influenced by E2F4 binding: on E2F4 targets, Sin3 is restricted to positions immediately downstream of the TSS, while Sin3 exhibited more scattered binding within downstream regions in the absence of E2F4. In contrast, E2F4 binding in the absence of Sin3 is restricted to sites near the TSS and did not show a bias for downstream regions (Fig. 2C-D and Fig. S4B). Taken together, this suggests that E2F4 and Sin3 mutually influence each other’s binding position and behave like a complex on chromatin. To further verify our conclusions regarding spreading, we performed ChIP-on-chip with antibodies against another E2F family member, E2F3 as this factor is also present on a subset of E2F4 target genes in differentiated cells (Fig. 2E and data not shown). Here, we observed sharp distinctions in binding profiles. For example, we observed that E2F3 binds within a narrow window upstream of the TSS of the Cdc2a gene, reminiscent of E2F4 binding in the absence of Sin3. Furthermore, although both E2F3 and E2F4 are recruited to the Mybl2 gene, binding of E2F4 (in the presence Sin3) can be readily distinguished from that of E2F3. These data strengthen our findings regarding specific spreading of Sin3/E2F4 downstream of the TSS.
In subsequent studies, we focused our work primarily on Sin3/E2F4 target genes to gain insight into the role of these factors in silencing gene expression.
E2F4 and Sin3 are coordinately recruited as a co-repressor complex
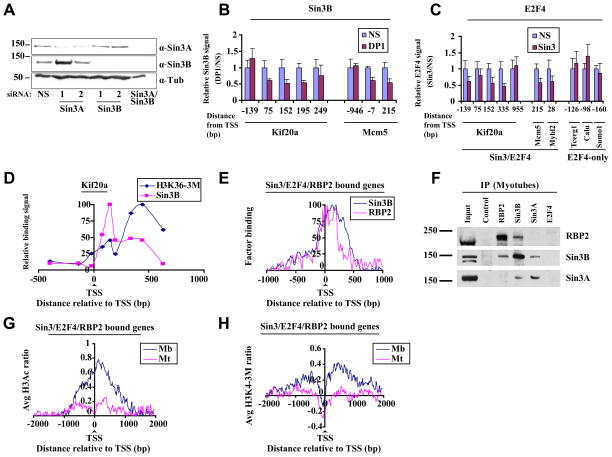
Using genome-wide and high density tiling arrays, we observed a striking overlap between E2F4 and Sin3 binding primarily downstream of the TSS, strongly suggesting that E2F4 and Sin3 are recruited as a complex. To further demonstrate a functional interaction, we transfected myotubes with siRNAs to deplete both isoforms of Sin3 (Fig. 3A) or DP1 (Fig. S1B) and assessed recruitment of E2F4 and Sin3B. All E2F transcription factors (with the exception of E2F7 and E2F8) bind to DNA as obligate heterodimers with the DP family of proteins (Blais and Dynlacht, 2004). Since DP1 is known to exclusively pair with E2Fs, we reasoned that depletion of this factor would be the most robust and effective way to eliminate both E2F4 activity and prevent promoter occupancy by compensatory repressor E2Fs, since it is well known that the remaining E2F family members can readily replace depleted factors at promoters. Indeed, depletion of DP1 resulted in a reduction of E2F4 binding (Fig. S1B). More importantly, Sin3B binding was preferentially and markedly reduced in downstream regions of several genes, including Kif20a and Mcm5, upon depletion of DP1 (Fig. 3B). As we have depleted all E2F activity present on our target genes, loss of other E2F family members, besides E2F4, might contribute to reduced Sin3B binding. Thus, to further strengthen our observation about a functional E2F4-Sin3 complex, we depleted Sin3A and Sin3B and tested for E2F4 binding. Interestingly, depletion of Sin3 also resulted in reduced binding of E2F4 to the Kif20a, Mcm5, and Mybl2 genes, again preferentially within the downstream region, but not on a set of genes bound by E2F4 only (Fig. 3C). Thus, in agreement with our location analysis, Sin3 and E2F4 appear to functionally interact on chromatin as a complex, and stable association of this complex with target genes in differentiated cells is mediated by both factors.
Figure 3. Sin3/E2F4-RBP2 function as a complex on target genes.
(A) Representative western blot indicates knock-down of Sin3A and Sin3B after transfection of myotubes with indicated siRNAs. Myotubes were transfected 48 h after induction of differentiation and were isolated 72h after transfection. (B) Histogram depicting Sin3B binding assessed by qChIP of selected genes in differentiated myotubes transfected with a non-specific control (NS) and DP1 siRNAs. Relative binding is expressed as a ratio of signals obtained in specific vs. NS siRNA transfected cells. The average of three independent experiments is shown. Error bars represent standard deviation. (C) As in B except that cells were transfected with control and Sin3A/Sin3B (Sin3) siRNAs and E2F4 binding was measured. (D) H3K36 tri-methylation levels and Sin3B binding were examined on the Kif20a gene using qChIP. Enrichment was calculated as percentage of input and normalized to maximum binding levels. The results of three independent experiments are shown. (E) Binding profiles for RBP2 and Sin3B binding to genes bound by Sin3, E2F4, and RBP2, as described in Fig. 2D. Only genes bound by all three factors were examined. (F) Endogenous Sin3B and RBP2 interact in vivo. Sin3 and RBP2 were immunoprecipitated using whole cell extracts of differentiated C2C12 cells (myotubes) with the respective antibodies and probed for either RBP2 or Sin3. (G) Genome-wide histone H3 acetylation (H3Ac) and (H) histone H3 lysine 4 tri-methylation (H3K4-3M) profiles of genes bound by Sin3, E2F4, and RBP2 in myoblasts and differentiated myotubes were obtained using tiling arrays. Genes were aligned relative to the TSS, and average histone H3Ac or H3K4-3M values per probe were calculated. The results of two independent experiments are shown.
Interestingly, we did not find a correlation between binding of Sin3/E2F4 complexes and H3K36 tri-methylation, a modification that mediates recruitment of Sin3 complexes to coding regions in budding yeast (Carrozza et al., 2005; Keogh et al., 2005; Nicolas et al., 2007), as maximal Sin3B binding and peak levels of H3K36 tri-methylation were offset on Kif20a (Fig. 3D). Although we cannot rule out a role for the H3K36 mark in recruitment of Sin3 complexes, these data suggest that it may not be obligatory for genes repressed by Sin3/E2F4 complexes.
Coordinated activity of Sin3 and RBP2 during differentiation
Recently, the Sin3 complex has also been linked to demethylase activity by virtue of its association with RBP2 (Hayakawa et al., 2007). Therefore, we tested whether RBP2 was recruited to Sin3 targets in differentiated myotubes using our high density tiling arrays. Our analysis showed that the majority of Sin3 target genes (58%) were bound by RBP2 (Supplemental Table 2). We confirmed RBP2 binding to selected Sin3/E2F4 target genes by qChIP (Fig. S1D). Interestingly, comparison of Sin3B and RBP2 binding profiles on genes bound by Sin3/E2F4 and RBP2 indicated a very striking positional overlap immediately downstream of the TSS (Fig. 3E), strongly suggesting coordinated binding of Sin3 and RBP2 within this region. To further verify this Sin3-RBP2 interaction, we asked whether Sin3 associates with RBP2 in myotube extracts. Indeed, co-immunoprecipitation experiments confirmed that RBP2 is part of a Sin3 complex in differentiated cells (Fig. 3F). Interestingly, when we immunoprecipitated Sin3B, but not Sin3A, RBP2 co-precipitated. In the reciprocal experiment, when we immunoprecipitated RBP2, Sin3B was detected. In agreement with previous reports we did not observe an association between either RBP2 or Sin3 and E2F4, suggesting either that this interaction occurs only on chromatin (Fig. 3B-C) or that the interaction is not stable under our immunoprecipitation conditions (Hayakawa et al., 2007; Silverstein and Ekwall, 2005).
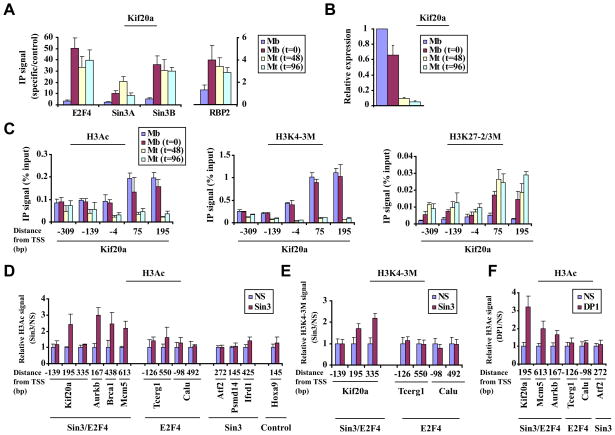
Next we asked whether Sin3/RBP2 binding overlaps with HDAC and demethylase activity within this downstream region on a subset of E2F4 target genes using our tiling arrays (see Fig. S3C). We observed a profound decrease in H3Ac and H3K4-3M levels immediately downstream of the TSS in differentiated myotubes as compared to proliferating myoblasts (Fig. 3G and 3H). Further, we asked whether the recruitment and activities of Sin3 and RBP2 are also temporally coordinated during differentiation. In order to address this question, we isolated chromatin from cells during a differentiation time course and analyzed E2F4, Sin3, and RBP2 binding to an E2F4 target gene, Kif20a. Each factor exhibited substantial or maximal binding to the Kif20a gene at the earliest stages of differentiation, when cells are transiently arrested by contact inhibition (represented by t=0) (Fig. 4A), although the gene was not fully repressed at this stage (Fig. 4B). This phenomenon was similarly observed for several other E2F4 targets genes (data not shown). Interestingly, H3Ac and H3K4 methylation levels remain high at this stage, but 48 hours after induction of differentiation, both marks showed a precipitous decrease specifically within the region immediately downstream of the TSS (Fig. 4C). Previously, we had shown that the acquisition of the H3 lysine 27 di-/tri-methylation mark (H3K27-2/3M) is associated with repression of E2F4 genes in differentiated cells (Blais et al., 2007). Therefore, we tested H3K27-2/3M levels during differentiation. Interestingly, H3K27-2/3M levels on the Kif20a gene peaked 48–96 hours after induction of differentiation, most prominently within the region immediately downstream of the TSS.
Figure 4. Coordinated removal of “active” chromatin marks by Sin3 and RBP2 leads to repression.
(A) Analysis of Sin3, E2F4, and RBP2 binding to a selected E2F4 target gene (Kif20a) during a differentiation time course as in Fig. 1A. Cycling myoblasts (Mb), arrested myoblasts (Mb, t=0), and myotubes after 48 or 96 hours after induction of differentiation (Mt, t=48 or t=96, fully differentiated myotubes) were examined. (B) Gene expression analysis using quantitative real time RT-PCR for a differentiation time course, as described in panel 4A. (C) Histone H3 acetylation (H3Ac, panel 1) Histone H3 lysine 4 tri-methylation (H3K4-3M, panel 2), and Histone H3 lysine 27 di-/tri-methylation (H3K27-2/3M, panel 3) levels were measured by qChIP during a differentiation time course as in panel 4A. Enrichment by IP was measured as a function of input signal. (D and E) Histograms depicting H3Ac (D) and H3K4 tri-methylation (E) levels, assessed by qChIP on selected genes in differentiated myotubes transfected with a non-specific control (NS) or Sin3A/Sin3B (Sin3) siRNA. See Fig. 3B for details. (F) H3Ac levels were measured after knock-down of DP1 as described in panels D and E. All panels, averages of at least 3 independent experiments are shown and error bars represent standard deviation.
To directly assess a functional role for Sin3 complexes in localized histone de-acetylation and de-methylation, we tested the impact of ablating both isoforms by transfecting myotubes with specific and non-specific (scrambled) siRNAs (Fig. 3A). Since RBP2 (Fig. 3F) and HDAC1/2 are part of a Sin3 complex (Hayakawa et al., 2007; Silverstein and Ekwall, 2005), we reasoned that depletion of Sin3 activity should result in changes in H3 acetylation and H3K4 tri-methylation on genes that recruit this Sin3 complex. Depletion of Sin3 in differentiated C2C12 myotubes resulted in robust localized increases in H3Ac and H3K4 tri-methylation levels on gene(s) bound by E2F4 and Sin3 (Fig. 4D-E). In striking contrast, we did not observe changes in H3Ac on genes bound by Sin3 or E2F4 only or on a control gene not bound by either factor (Hoxa9), nor did we observe changes in H3K4 methylation on genes bound by E2F4 only. Although we cannot formally rule out the possibility that elevated acetylation levels are a consequence of enhanced transcription after depletion of Sin3 (see Fig. 7A), we note that increased acetylation levels were most obvious near Sin3 binding sites immediately downstream of the TSS on multiple genes, and we did not observe re-acetylation of transcribed regions further downstream on the same genes (Fig. 4D, and data not shown).
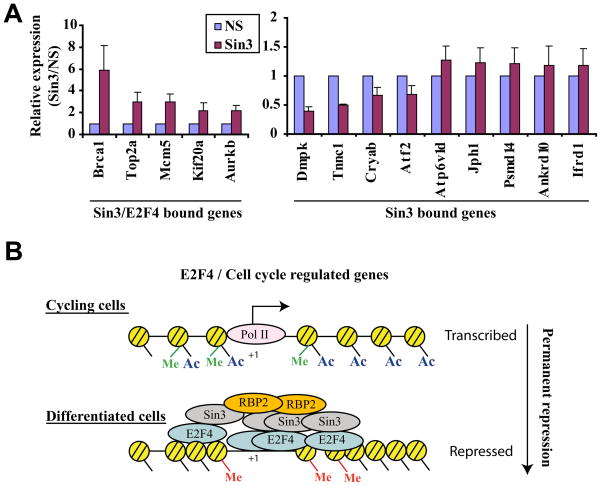
Figure 7. Expression of Sin3 target genes after knock-down of Sin3.
(A) Gene expression analysis using quantitative real time RT-PCR of selected genes after ablation of Sin3. Myotubes were transfected 48 h after induction of differentiation and were isolated 96 after transfection. Expression levels of selected genes in Sin3 siRNA-transfected cells were compared to non-specific control (NS) transfected cells. The average of three independent experiments is shown. Error bars represent standard deviation. (B) Model detailing Sin3 function in differentiated muscle cells on a subset of E2F4 target genes. Me: methylation of H3K4 (green) and H3K27 (red); Ac: acetylation. See text for details.
Furthermore, we reasoned that ablation of E2F activity should promote alterations similar to those observed upon depletion of Sin3 if there were a physical (and functional) link between E2F4 and a Sin3-RBP2 complex. Indeed, depletion of DP1 resulted in increased H3Ac on genes bound by Sin3 and E2F4 but not on control genes bound by E2F4 or Sin3 only (Fig. 4F).
These data strongly suggest that Sin3 complexes, comprised of Sin3, HDAC, and RBP2 are recruited by E2F4, resulting in highly specific modification of nucleosomal targets immediately downstream of the TSS .
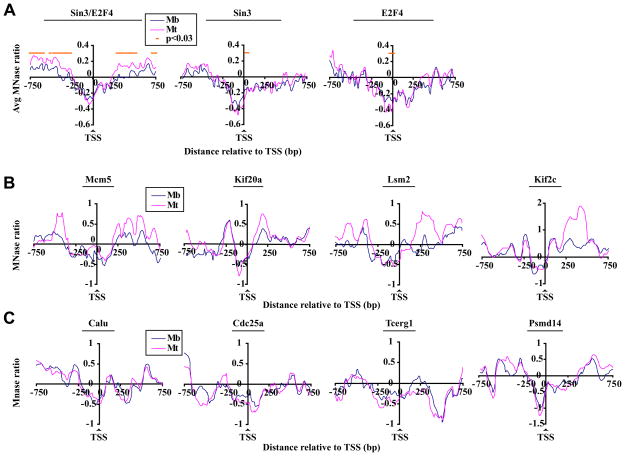
Sin3 impacts local chromatin structure
Next, we asked whether genes bound by E2F4 and Sin3 exhibit differences in nucleosome occupancy (see Fig. S3C), since acetylation of nucleosomes is tightly linked to chromatin remodeling in certain settings (Gregory et al., 2001; Neely and Workman, 2002). Remarkably, genes bound by both Sin3 and E2F4 showed a significant (p<0.03) enhancement of nucleosomes immediately downstream of the TSS in differentiated myotubes as compared to cycling myoblasts (Fig. 5A-B and Fig. S5A, S6C). In sharp contrast, we did not observe any significant differences in nucleosome density over identical regions of genes bound only by Sin3 (Fig. 5A, panel 2, Fig. 5C, panel 4 and Fig. S5B) or E2F4 (Fig. 5A, panel 3 and Fig. 5C, panels 1–3).
Figure 5. High resolution nucleosome density profiles of Sin3 target genes.
(A) Average nucleosome density profiles for genes bound by Sin3 and E2F4 (panel 1), Sin3 only (panel 2), and E2F4 only (panel 3) in myoblasts and differentiated myotubes deduced from our tiling arrays (see Fig. 2C). Genes were aligned relative to the TSS, and average MNase values per probe were calculated for each group. The results of at least two (myotubes) or three (myoblasts) experiments are shown. Mb, myoblasts; Mt, myotubes. (B) Nucleosome density profiles for individual genes bound by Sin3 and E2F4 in myoblasts and myotubes. MNase ratio is given by the moving average (window size 140 basepairs, step size 5 probes). (C) Genes bound by E2F4 only (panel 1–3) or a gene bound by Sin3 only (Psmd14, panel 4) were analyzed as described in panel B.
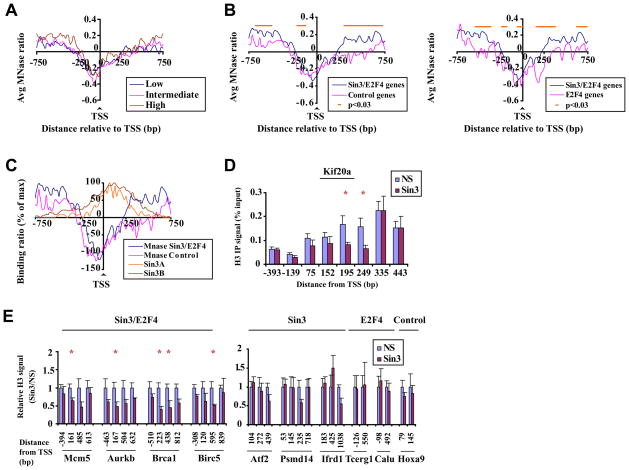
We considered the possibility that differences in nucleosome occupancy within this downstream region simply reflected markedly different gene expression levels in each group of Sin3 targets. To test this possibility, we first compared nucleosome density profiles of genes with low, intermediate, and high expression levels in differentiated myotubes, irrespective of Sin3 and E2F4 binding (Fig. 6A). For this comparison, average expression levels were clearly different, yet we did not observe distinct, associated nucleosome density profiles. Second, we compared genes bound by E2F4 and Sin3 with a set of control genes (i.e., those not bound by E2F4 or Sin3; Supplemental Table S2) that exhibit a similar expression profile (wherein the distribution of absolute expression values is not different between Sin3/E2F4 and control groups; Fig. S5C) in differentiated myotubes and found that genes bound by E2F4 and Sin3 displayed significantly enhanced nucleosome density at positions immediately downstream of the TSS (Fig. 6B panel 1). Thus, we conclude that in differentiated myotubes, nucleosome density patterns are not simply a reflection of transcription rates; rather, they are shaped in part by the presence of E2F4 and Sin3. To further dissect the influence of E2F4 and Sin3 on chromatin structure, we compared nucleosome binding profiles of genes bound by E2F4 and Sin3 versus genes bound exclusively by E2F4. Importantly, this analysis indicated that E2F4 binding alone was not sufficient to generate a nucleosome density profile similar to genes bound by both E2F4 and Sin3 (Fig. 6B, panel 2). Moreover, ChIP-on-chip analysis of Sin3 binding and nucleosome densities indicated a partial overlap of Sin3 binding and enhanced nucleosome densities (Fig. 6C), providing an explanation for the underlying alterations of nucleosome positioning in the immediate vicinity of Sin3 binding.
Figure 6. Sin3 complex directly impacts acetylation levels and nucleosome structure downstream of the TSS.
(A) Average nucleosome density profiles for genes that were classified according to absolute expression levels (based on Affymetrix gene expression arrays) in differentiated myotubes (Blais et al., 2007): low (average: 100, range 1–200), intermediate (average: 485, range 201–1000), and high (average: 4024, range 1000+) abundance transcripts. (B) Average nucleosome density profiles for genes bound by Sin3 and E2F4, a control group of genes (65 genes not bound by Sin3/E2F4 but similarly expressed; see Supplemental Table 2), or genes bound by E2F4 only in differentiated myotubes, with median log2 (Mt/Mb) expression values for each group being −1.4, −1.6 and −0.54 respectively. (C) Binding profiles of Sin3A and Sin3B relative to nucleosome density profiles of the same genes. Binding and MNase ratios were normalized for maximum levels. (D) Histogram depicting H3 levels on distinct regions of an individual Sin3/E2F4 target gene in myotubes transfected with a non-specific control (NS) or Sin3 silencing (Sin3) siRNA. The asterisk indicates a histone H3 signal significantly lower than the NS control (p < 0.01 by t test). The average of at least 3 independent experiments is shown. Error bars represent standard deviation. (E) Analyses were performed as panel D except that histone H3 signal is expressed relative to the signal obtained in cells transfected with NS control.
Sin3 is essential for the permanent repression of genes by maintaining chromatin structure
To examine a direct role for Sin3 on localized nucleosome accumulation, chromatin derived from myotubes transfected with either control or specific Sin3 siRNAs was immunoprecipitated with antibodies against histone H3, and enriched DNA was analyzed using qPCR. Histone H3 levels in control qPCR samples mirrored nucleosome density profiles obtained on the Kif20a gene using our tiling arrays (Fig. 6D and Fig. 5B, panel 2). Remarkably, cells depleted of Sin3A and Sin3B showed specific and highly significant (p<0.01) decreases in histone H3 levels downstream of the TSS. Importantly, we did not observe any significant decreases in histone H3 levels either upstream or far downstream of the TSS (Fig. 6D). Analysis of additional Sin3/E2F4 target genes showed consistent diminution of histone H3 levels immediately downstream of the TSS upon Sin3 ablation (Fig. 6E). In contrast, genes bound exclusively by Sin3 or E2F4 and an unrelated control gene not bound by either Sin3 or E2F4 (Hoxa9) did not exhibit significant reductions in histone H3 levels at corresponding locations.
Next, we sought to determine whether Sin3 binding and its effect on downstream chromatin structure are critical for gene expression. To address this question, differentiated myotubes were transfected with control or specific siRNAs targeting Sin3A and Sin3B (Fig. 3A), and differences in RNA abundance were analyzed using quantitative RT-PCR. We found that expression of genes targeted by both E2F4 and Sin3 was markedly enhanced upon depletion of Sin3, indicative of de-repression (Fig. 7A). Similar results were obtained using a different combination of specific siRNAs targeting Sin3 (data not shown). In striking contrast, we observed diminished or unaltered expression of several genes that bind Sin3 only. These data highlight the functional distinctions, including differences in nucleosome organization and gene expression, between Sin3-only and Sin3/E2F4 target genes.
Discussion
Our studies provide evidence that Sin3 may play an essential role in heterochromatin formation and permanent gene repression by means of promoter binding and spreading, localized de-acetylation and de-methylation of one or more proximal nucleosomes, and subsequent repositioning of nucleosomes on a subset of E2F4 regulated genes. Thus, the Sin3/E2F4 complex provides a paradigm for the coordinated modification of chromatin ultimately leading to the formation of heterochromatin and permanent gene silencing in differentiated cells.
A model for stable gene repression in terminally differentiated cells
A mammalian Sin3B complex is reversibly recruited to cell cycle genes in quiescent and early G1 cells (Balciunaite et al., 2005; Rayman et al., 2002), and the activity of Sin3B appears essential for transient cell cycle withdrawal (David et al., 2008). Our detailed analysis suggests a model in which Sin3 is also a critical component for the permanent repression of cell cycle genes during differentiation (Fig. 7B). Several observations lend credence to this model: (1) In differentiated cells, distinct Sin3/E2F4 complexes (and on a subset of Sin3/E2F4 target genes, RBP2) bind to downstream regions and locally alter both histone modifications and chromatin structure. Moreover, our genome-wide location, factor ablation, and bioinformatics analyses show a clear functional relationship between Sin3 and E2F4 at target sites. In addition, although we cannot formally rule out the possibility that Sin3 associates with other repressors within the same region, extensive computational analysis with search algorithms designed to identify enriched motifs failed to identify enrichment of known Sin3-associated repressor proteins (Supplemental information and Supplemental Table 3). (2) The activities of Sin3 and RBP2 result in the coordinated removal of acetylation and methylation marks, and this coincides with the timely acquisition of the repressive H3K27 methylation mark and altered nucleosome architecture. (3) Ablation of Sin3 activity leads to loss of nucleosome density and this co-incides with re-acetylation, reflecting hyper-acetylation and subsequent re-expression of cell cycle regulated genes in differentiated cells. (4) Consistent with a role in permanent gene silencing, our recent conditional knock-out studies in mice suggest both overlapping and redundant roles for Sin3 isoforms in maintenance of the differentiated state (C.V.O, G. David, and B.D.D., unpublished).
How Sin3 stably associates with this downstream region is not known. Recruitment of the Sin3 complex creates localized regions of histone H3 hypo-acetylation and methylation in a coordinated manner (Fig. 4). Interestingly, in vitro experiments suggest that the Sin3 complex shows a preference for the hypo-acetylated amino-terminal tail of histone H3, resulting in strong, repressor-independent anchoring of the Sin3 complex (Vermeulen et al., 2006). Here, we have observed a mutual dependency on E2F4 and Sin3 for the stable association of Sin3/E2F4 complexes. Thus, we propose that in differentiated myotubes, Sin3 is stably associated with downstream promoter regions by E2F4 and interaction with hypo-acetylated histone tails. Other chromatin modifications, in particular H3K36 methylation, could possibly also play a role in directing mammalian Sin3 recruitment to target genes
A role for Sin3 complex in chromatin remodeling
Our nucleosome density experiments and gene expression studies suggest that binding of Sin3 to cell cycle regulated genes has a profound effect on chromatin structure and gene expression (Figs. 5–7A). How do Sin3/E2F4 complexes alter chromatin structure? It is known that Sin3 and E2F/pocket protein complexes interact with a variety of factors involved in chromatin function. For example, the Sin3 complex was shown to interact with Brg1, the catalytic subunit of the Swi/Snf chromatin remodeling enzyme (Nagl et al., 2007; Sif et al., 2001). Given this connection, it is reasonable to speculate that, in the proper setting, the recruitment and activity of Sin3-HDAC and Swi/Snf (or functionally related remodeling enzymes) are tightly coordinated to promote nucleosome re-positioning.
Nucleosome positioning within promoter regions plays a critical role in regulating gene expression by limiting the access of transcription factors. The presence of Sin3/E2F4 complexes could therefore physically block access of the pre-initiation complex and recruitment of factors to promoters at least in part by increasing the density of nucleosomes locally. Such regions could represent localized, facultative heterochromatin in differentiated skeletal muscle cells, as we have previously observed the presence of the repressive H3K27 methylation mark on a subset of E2F4/pRb target genes (Blais et al., 2007), many of which have been shown to bind Sin3 in this study. Our work also highlights an especially important role for nucleosomes immediately downstream of the TSS. Further work will be required to elucidate the importance of TSS-proximal nucleosomes implicated in this study, to define the chromatin marks associated with this region, to determine whether higher order chromatin compaction can occur, and to examine whether PolII binding is blocked.
Materials and Methods
Cell culture
The C2C12 murine myoblast cell line (obtained from ATCC) was cultured as described (Blais et al., 2007). Briefly, cells were grown in DMEM supplemented with 10% fetal bovine serum. Differentiated cells were obtained by culturing confluent C2C12 cells in DMEM supplemented with 2% horse serum. Differentiated cells were separated from undifferentiated cells using diluted trypsin.
Antibodies
Antibodies against E2F3 (sc-878), E2F4 (sc-1082), Sin3A (sc-767, sc-994), and Sin3B (sc-768) were obtained from Santa Cruz. Antibody against DP1 was obtained from BD Pharmingen (556462). Antibodies against RBP2 (1416 and 2470) were described previously (Benevolenskaya et al., 2005; Klose et al., 2007). Antibodies against histone H3 (ab1791), histone H3K4-3M (ab8580), and anti-H3Ac (K9Ac + K14Ac, 06-599) were obtained from Abcam and Millipore. Anti-H3K27-2/3M (clone 7B11; gift of D. Reinberg) has been characterized previously (Sarma et al., 2004).
ChIP, ChIP-on-chip, and data analysis
Cells were cross-linked, harvested, and lysed as described previously (Takahashi et al., 2000). For conventional ChIP, nuclei were collected by centrifugation, resuspended in sonication buffer EDTA, (1 mM 0.5 mM EGTA, 10 mM Tris at pH 8, 0.5 % N-lauroyl-sarcosine and protease inhibitors), and sonicated on ice to an average length of 350 bp. To enrich for histone H3 or acetylated H3 after knock-down of Sin3, 1 μg and 5μg of chromatin was used per immunoprecipitation, respectively. Mononucleosome preparation, ChIP-on-chip experiments, and data analysis are detailed in Supplemental Methods.
RNAi
Cells were grown to confluence, induced to differentiate for 48 hrs, and transfected in triplicate with specific siRNA duplexes (Dharmacon) or a scrambled non-specific control using siMPORTER (Millipore), according to the manufacturer's instructions. Differentiated myotubes were harvested at 72 hrs (ChIP) or 96 hrs (RT-PCR) after transfection using diluted trypsin. To isolate RNA and protein, cells were resuspended in Trizol and processed according to the manufacturer’s instructions. siRNA sequences appear in the Supplemental Methods section.
RT-PCR
RT-PCR was used to analyze gene expression after knock-down of Sin3. RT-PCR was performed using 250 ng of total RNA per reverse transcriptase reaction (Invitrogen). cDNA was diluted and quantified twice by real-time PCR using the SYBR green method, and the results expressed relative to the nonspecific control siRNA.
Co-immunoprecipitation
Cells were lysed in lysis buffer E (50 mM Tris·HCl (pH 7.9), 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.5% NP-40) supplemented with complete protease inhibitor cocktail (Roche Molecular Biochemicals). The cell lysates were diluted with equal volume of NETN buffer (20 mM Tris·HCl (pH 7.9), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with protease inhibitors, and immunoprecipitated with the specified antibodies.
Supplementary Material
Acknowledgments
We are grateful to members of the Dynlacht lab, and G. David for comments on the manuscript and helpful suggestions during the course of this study. We thank A. Bhattacharjee and G. Das for assistance with experiments. This work was supported by NIH grant CA077245 to B.D.D., CA076120 to W.G.K, Susan Komen postdoctoral fellowship to C.V.O., and Department of Defense Breast Cancer Research Program Concept Award W81XWH-07-1-0581 to Q. Y. W.G.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alland L, David G, Shen-Li H, Potes J, Muhle R, Lee HC, Hou H, Jr, Chen K, DePinho RA. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol Cell Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Balciunaite E, Spektor A, Lents NH, Cam H, Te Riele H, Scime A, Rudnicki MA, Young R, Dynlacht BD. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25:8166–8178. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci USA. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005;21:608–615. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- Gregory PD, Wagner K, Horz W. Histone acetylation and chromatin remodeling. Exp Cell Res. 2001;265:195–202. doi: 10.1006/excr.2001.5187. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. Embo J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Workman JL. Histone acetylation and chromatin remodeling: which comes first? Mol Genet Metab. 2002;76:1–5. doi: 10.1016/s1096-7192(02)00014-8. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- Pile LA, Spellman PT, Katzenberger RJ, Wassarman DA. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: implications for the regulation of energy metabolism. J Biol Chem. 2003;278:37840–37848. doi: 10.1074/jbc.M305996200. [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Nishioka K, Reinberg D. Tips in analyzing antibodies directed against specific histone tail modifications. Methods Enzymol. 2004;376:255–269. doi: 10.1016/S0076-6879(03)76017-0. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Current Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Walter W, Le Guezennec X, Kim J, Edayathumangalam RS, Lasonder E, Luger K, Roeder RG, Logie C, Berger SL, Stunnenberg HG. A feed-forward repression mechanism anchors the Sin3/histone deacetylase and N-CoR/SMRT corepressors on chromatin. Mol Cell Biol. 2006;26:5226–5236. doi: 10.1128/MCB.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.