Abstract
During their synthesis many water-soluble proteins and nearly all membrane proteins transit through a protein-conducting channel in the membrane, the Sec translocon, from where they are inserted into the lipid bilayer. Increasing evidence indicates that folding of the nascent protein begins already within the ribosomal exit tunnel in a sequence- and environment-dependent fashion. To examine the effects of the translocon on the nascent-chain folding, we have calculated the potential of mean force for α-helix formation of a 10-alanine oligopeptide as a function of its position within the translocon channel. We find that the predominant conformational states, α-helical and extended, reflect those found for the peptide in water. However, the translocon, via its surface properties and its variable diameter, shifts the equilibrium in favor of the α-helical state. Thus we suggest that the translocon facilitates not only the insertion of membrane proteins into the bilayer but also their folding.
Introduction
Proteins are synthesized by the ribosome initially as linear chains of amino acids, beginning at the peptidyl transferase center (PTC) deep within the ribosome and extending through its 100-Å-long exit tunnel to the surface1. This tunnel is largely cylindrical with a diameter ranging from 10 to 20 Å2 and, although originally presumed to be inert, many studies now suggest it plays an active role during protein synthesis3. For example, peptides that induce translational arrest, e.g., TnaC, proceed via specific interactions with the walls of the exit tunnel4,5. Furthermore, the activity of macrolides, a broad class of antibiotics that interfere with nascent-chain progression, can be modulated by altering their interaction with the exit tunnel6.
The ribosomal exit tunnel also plays a role in nascent protein folding. Numerous studies have demonstrated that the tunnel permits modest folding of the nascent chain into a secondary structure7–14 and possibly even some tertiary structure15,16. Recent cryo-electron microscopy maps display α-helical segments within a nascent chain inside the exit tunnel17,18. The exit tunnel can actively modulate folding as well; experiments using 5- and 10-residue poly-alanine (polyAla) indicate that specific zones inside the tunnel lower the free energy of the folded state by up to 1.1 kcal/mol10.
Upon exit of their first few residues, the signaling element, from the ribosome, nascent proteins destined for secretion or the membrane interact with the signal recognition particle19, which recruits a protein-conducting channel, the Sec translocon. The translocon, also known as the SecY and Sec61 complex in bacteria and eukaryotes, respectively, brings about the insertion of nascent membrane proteins into the lipid bilayer or the translocation of secreted proteins across it20–22. Transfer of the nascent chain to the channel is accomplished by docking of the ribosome to the translocon, which, despite inserting its two cytoplasmic loops into the exit tunnel, does not interfere with the extrusion of the nascent chain23–26. The translocon forms an hourglass-shaped channel with each half, cytoplasmic and periplasmic, tapering to a constrictive hydrophobic pore ring at its center (see Fig. 1)27,28. The ribosome-translocon system, thus, forms an almost continuous tunnel from the PTC to the opposite side of a membrane.
Figure 1.
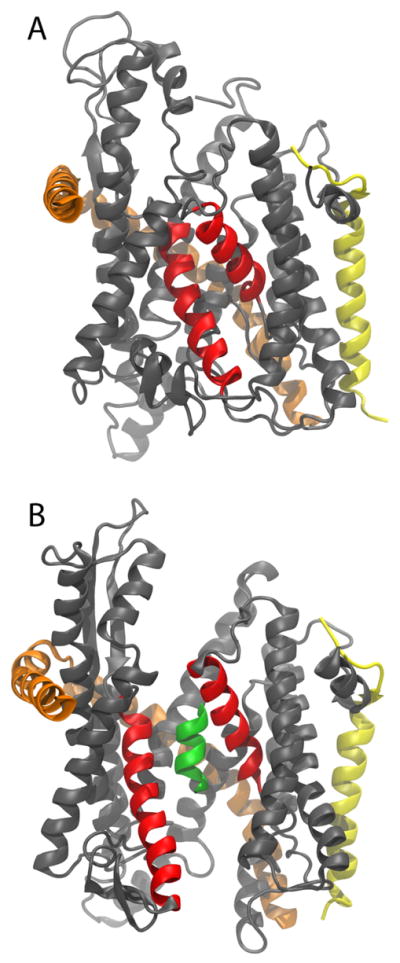
Structure of the SecY complex. The SecY complex is shown from within the membrane plane in a cartoon representation with its three subunits colored grey, orange, and yellow, respectively. The two helices lining the lateral gate are shown in red. (A) SecY complex with a closed lateral gate27.(B) SecY complex with an opened gate. Ala10 is shown inside the channel as a green helix.
For the exit of individual transmembrane (TM) segments into the membrane, the translocon possesses a lateral gate at the interface of two halves of SecY, which, when opened, exposes the nascent chain to the lipid bilayer27,29. It is expected that TM segments will transit through the gate in a preformed helical state, due to the significant free-energy penalty of embedding an exposed peptide backbone into the hydrophobic core30. Biochemical experiments on the insertion of model peptides as well as Kv1.3, indeed, indicate that TM segments acquire secondary structure already within the ribosome and maintain that structure within the translocon7,9,13. However, not all TM segments form compact states early in their development; for example, both polyVal and glycophorin A (GpA) remain extended even within the translocon, at least initially7.
Based on the observed compaction of polyAla and polyLeu inside the ribosomal exit tunnel and translocon and the lack of compaction for polyVal, polyPro, and GpA, Mingarro et al.7 suggest that the folding propensity of a nascent chain in those locations generally reflects that for amino acids in an aqueous environment, rather than their membrane-insertion propensity. Yet at least one exception has been observed: the TM segment of VSV-G has a compact structure inside the ribosome, but unfolds upon emergence into the cytoplasm9. In addition, computational studies have demonstrated that confinement, e.g., by the ribosome or translocon, can stabilize entropically α-helices31–34. Such confinement may, however, be insuffcient to alter significantly the folding pathways of those polypeptides tested in Mingarro et al., hence, leaving open the question of what is the precise effect of the ribosome-translocon system on the folding free-energy landscape of polypeptides.
To resolve how confinement in a native environment alters the free energy of folding quantitatively, we determined a two-dimensional potential of mean force (PMF) for helix formation of deca-alanine (Ala10) inside the translocon. The two coordinates used are translocation of the peptide chain along the channel axis z and the average i, i + 4 hydrogen-bonding distance ξ along the backbone, i.e., the helicity of Ala10 Translocation was restricted over a 30-Å range, from z = 20 Å to z = −10 Å, where. z = 0 corresponds roughly to the center of SecY and z is positive in the cytoplasmic half of the channel. Distances between successive hydrogen-bonding pairs were restrained in the second coordinate such that hydrogen bonds form or break concomitantly. The 2D PMF was obtained using adaptive biasing forces (ABF) in NAMD35–37 and required about 300 ns of total simulation time (see Methods).
Methods
Simulations of the translocon began from the crystal structure of the closed state (PDB 1RHZ)27. The channel was embedded in a POPC lipid bilayer, solvated, and ionized with Na+ and Cl− ions to a strength of 100 mM, using the visualization and analysis program VMD38. The lateral gate was then opened to a width of 8 A, in accordance with experimental measurements 39, by applying forces that drove in a near-equilibrium fashion the conformation of SecY toward that of a different structure of SecY with a partially open gate40,41. Finally, Ala10 was inserted in a helical conformation into the center of the channel, which was then equilibrated for 2 ns while the gate and Ala10 were restrained.
MD simulations were carried out using NAMD 2.736 with the CHARMM force field42,43. The temperature was held at 310 K using Langevin dynamics; the membrane area was held fixed while the pressure normal to the membrane was held at 1 atm using the Langevin piston method44. The equations of motion were integrated employing the RESPA multiple time-step algorithm45 with a time step of 2 fs being used for all bonded interactions, 2 fs for short-range non-bonded interactions, and 4 fs for long-range electrostatic interactions. Long-range electrostatic forces were calculated using the particle-mesh Ewald method. Bonds involving hydrogen atoms were constrained to their equilibrium length, employing the Rattle algorithm46.
All PMFs were calculated using adaptive biasing forces (ABF), implemented with the collective variables module of NAMD 2.735–37. As noted above, two reaction coordinates were utilized, namely the position of the center of mass of Ala10 relative to that of the channel and the average hydrogen-bonding distance along the backbone. To improve sampling uniformity, the full reaction-coordinate space was subdivided into multiple regions, which were simulated independently. The resulting data were then combined to produce the PMF.
The collective variables module was also employed to enforce a number of restraints to the system. Harmonic potentials were applied between pairs of potential hydrogen-bond partners such that all respective distances were nearly identical; these restraints caused the symmetric formation or deformation of the α-helical state. Additionally, distance restraints were applied to eight pairs of residues across the lateral gate to ensure that its 8-Å opening was maintained. In the periplasmic region of the translocon, i.e., for z < 0, an additional restraint was required to keep Ala10 from rotating too far off the translocation axis. Forces were applied only when the polar angle of Ala10 was greater than 30°.
Calculation of the channel diameter was performed using HOLE47. The diameter plotted is 2 Å larger than that calculated by HOLE to account for the algorithm’s tendency to underestimate the size of irregular volumes28. The interaction area was calculated over all simulations: for each trajectory frame, the translocation coordinate and the fraction of Ala10 in contact with lipids, or with lipids and hydrophobic residues were determined. The data were then sorted into 0.2-Å bins in z and averaged over all fractional area values observed for each bin.
Results
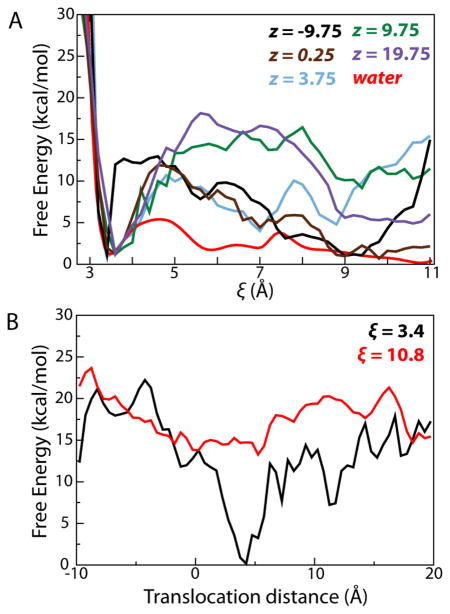
As a basis for comparison with the PMF obtained for helix formation in the translocon, we first determined the 1D PMF for folding along the i, i + 4 hydrogen-bond length ξ of Ala10 in a purely aqueous environment, shown in Fig. 2A. Two folding states are markedly favored, namely a helical state around ξ=3.4 and a family of extended states with ξ> 9. A stable intermediate state of slightly higher free energy is also found between the helical and the extended states with two barriers around ξ= 4.5 and ξ= 7.5 separating the local minima. That both helical and extended states are well represented in the PMF is expected for Ala10. It is known from both theory48–50 and experiment51,52 that polyAla forms helices in water. Additionally, since Ala10 used here is zwitterionic due to its charged termini, which mimic the properties of an N-terminal signal sequence, the large dipole moment of the extended state contributes to its relatively low free energy.
Figure 2.
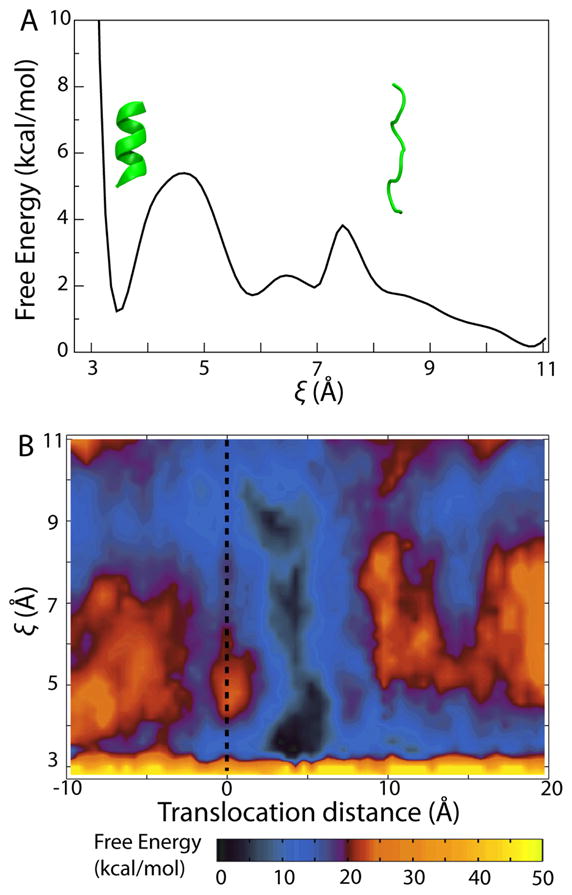
Potential of mean force (PMF) for folding of Ala10. (A) One-dimensional PMF for Ala10 in water as a function of the i, i + 4 hydrogen-bond ξ. The two inset proteins (green) represent example configurations for two values of ξ, respectively. (B) Two-dimensional PMF for Ala10 in the translocon as a function of the concerted-helix-formation coordinate ξ and translocation distance. The dotted line indicates the position of the translocon’s constrictive pore ring.
Many of the features observed in the 1D PMF of folding of Ala10 in water are reproduced in the PMF of folding at different points inside the translocon, shown in Fig. 2B. Minima for both α-helical (ξ= 3.4) and extended (ξ> 9) states are displayed almost uniformly throughout the channel. In contrast, the barriers separating these states varies with position. Moreover, the free energy of a given Ala10 folding state varies along the channel axis, reaching a global minimum near the pore ring. In particular, the PMF for folding near the center of the translocon, specifically around z = 5 Å, appears quite similar to that of Ala10 in a neat aqueous environment. This similarity supports the suggestion of Mingarro et al. that the free-energy landscape of a peptide in the ribosome-translocon system reflects its conformational equilibrium in water7.
Projecting the 2D PMF along ξ for different values of z, as shown in Fig. 3A, permits a comparison with the 1D PMF of folding of Ala10 in water. At each position within the translocon, clear minima are observed at ξ= 3.4, the α-helical state, and at ξ≥ 9, the extended states. At nearly all positions, the free-energy difference between the helical state and the minimum of the extended states amounts to between 0 and 3.5 kcal/mol, with the helical state being noticeably more favored on the cytoplasmic side of the channel. Similarly, all PMFs display barriers at intermediate values of ξ; the heights of these barriers are significantly larger than those observed for folding of Ala10 in water. This increase in the barrier height, ranging from 8 to 15 kcal/mol, suggests that although Ala10 still finds free-energy minima in both helical and extended states, transitions between them are dramatically slowed upon entering the channel, in agreement with coarse-grained simulations of protein folding under confinement33. Ala10, thus, most likely retains the folded state it possessed prior to its entrance despite the significantly different environment presented by the translocon.
Figure 3.
Projections of the 2D PMF in Fig. 2B along individual coordinates. (A) Projections of the 2D PMF along different values of z, colored as indicated in the legend, as a function of ξ. The 1D PMF for Ala10 in water is included for comparison. The free energy of the α helical state was chosen as a reference point for all PMFs. (B) Projections of the 2D PMF for the α-helical (ξ= 3.4) and extended (ξ= 10.8) states as a function of position in the channel.
The effect of the tapering of the channel to the constrictive pore ring, a hydrophobic gasket-like seal at its center53, is made most apparent by projecting the 2D PMF along the helical (ξ= 3.4) and extended (ξ= 10.8) states (see Fig. 3B). The free energy of the α-helical state roughly mirrors the diameter of the channel, shown in Fig. 4A, reaching a minimum just above the pore ring, whereas the maximally extended state shows little deviation in free energy throughout the channel. Confinement is expected to stabilize the folded state, as observed for Ala10, but also to destabilize the unfolded state, due to a loss of configurational entropy32,33. Yet, on account of the restraints imposed on Ala10 to keep successive hydrogen bonds equidistant, sampling of bent conformations is limited (the propriety of these restraints is discussed below). The entropic effect on folding is, nonetheless, clearly manifested, both in the change in free energy of the helical state across the channel and in the increasing free-energy difference between the extended and the helical states (see Fig. 3A).
Figure 4.
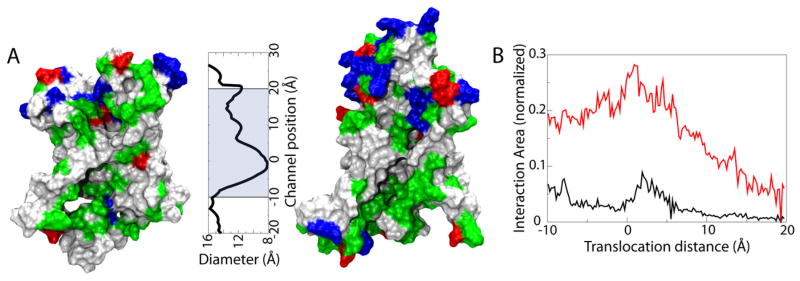
Interaction between Ala10 and the channel. (A) Surface properties of the translocon interior. The N-terminal (left) and C-terminal (right) halves of the translocon interior are shown as molecular surfaces colored according to residue type (non-polar is white, polar green, basic blue and acidic red). The channel diameter as a function of channel position is plotted to scale with the channel surfaces. The shaded area indicates the range over which the PMF was calculated. (B) Fraction of the area of Ala10 in contact with lipids (black) and all hydrophobic regions (red) as a function of channel position.
Beyond purely geometric considerations, the nature of the translocon interior can also affect the free-energy landscape of Ala10. The translocon features a predominantly polar interior on its cytoplasmic side, a hydrophobic constriction in the central pore ring, and a mixed hydrophobic/polar periplasmic side, illustrated in Fig. 4A. In addition to interacting with the interior of the translocon, the nascent chain is known to interact with lipids across the lateral gate54–57; in the partially opened translocon model used here (see Methods), the gate opening is ~8 Å wide, large enough to allow lipids to interact with Ala10 inside. To characterize the interaction of lipids and Ala10, we measured the fraction of surface area of Ala10 in contact with lipids at different positions within the channel. Shown in Fig. 4B, the interaction peaks at 10% near the global free-energy minimum; when the translocon interior is accounted for as well, the total fraction of Ala10 interacting with hydrophobic residues or lipids reaches over 25%.
The 1D PMF projections in Fig. 3, when correlated with the surface properties of the translocon interior, suggest that predominantly polar surfaces stabilize the helical state of Ala10 relative to the unfolded state, while more hydrophobic surfaces have little effect. The narrow pore ring is an exception, as the helical state is found to have here a significantly lower free energy than the extended states. Given the large contact area of Ala10 with hydrophobic residues and lipids (up to 30% at its peak), direct interactions between the surface and Ala10 play a significant role in determining the free energy of folding. These observed surface-dependent effects on folding of Ala10 are in good agreement with coarse-grained simulations of polyAla inside a nanotube of varying hydrophobicity58. In those simulations, small hydrophobic patches embedded in an otherwise hydrophilic nanotube maximized helical stability, with the effect being most pronounced at large (≥20 Å) nanotube diameters 58. The confinement of water is also predicted to have an effect on the free-energy landscape of Ala10 folding59,60, for example by promoting helix formation for polyAla in a predominantly hydrophilic environment, such as the cytoplasmic half of the translocon, as demonstrated in Fig. 3B.
Conclusions
In order to interpret our simulation results for Ala10 in the translocon, it is necessary to consider assumptions inherent in the setup of the simulations. For example, SecY was modeled in a laterally open state, partially exposing Ala10 to the lipid bilayer. Although direct interactions of the nascent chain and the lipids have been observed experimentally54–57, the degree of lateral gate opening should be considered a flexible property, with an opening of at least 8 Å required to support translocation 39, 11 Å for the intercalation of the signal sequence 61, and at least 13–14 Å for lateral exit 26. An opening of 8 Å was modeled here, leading to a pore-ring diameter of 8 Å. This diameter hardly accommodates the Ala10 α-helix, which itself is 8-Å wide. The tight conditions faced by Ala10 at the pore ring explains why the free-energy minimum for the α-helix is found almost 4 Å above the ring. Should the gate, and concomitantly the pore ring, be allowed to expand further, the free-energy profile should shift downward along the channel axis, being otherwise unaltered.
While a nascent chain under physiological conditions extends beyond the translocon on both sides, a short 10-residue oligopeptide, Ala10, was used here. This peptide has greater conformational freedom than a typical nascent protein, being able, in principle, to rotate in all directions and to fold back upon itself. To limit such freedom and ensure that Ala10 remains linear, hydrogen bonds along its back-bone were enforced to form or break concomitantly, effectively causing α-helix formation to be highly cooperative. Although cooperativity is expected to be only moderate for short oligopeptides, it is proportional to peptide length, making the effect of the imposed restraints akin to embedding Ala10 into a longer nascent chain49,62,63. The exclusion of specific intermediate states, e.g., those with partial helicity, from the PMF calculation does not affect the relative free energies of the end states but may serve to raise the barrier between them. Without such exclusions, it may be expected that transitions between the free energy minima would be enhanced, although the final disposition of Ala10 would remain unaffected.
The PMFs in Figs. 2 and 3 demonstrate that the dominant conformational states of Ala10, both helical and extended, are identical to those found in water, in agreement with experimental studies7,14. The translocon does, however, exert, through its surface properties and through its hourglass-like shape, a biasing effect of a few kcal/mol in favor of the helical state. This helical bias may be necessary for the insertion of TM segments into the membrane, not all of which form stable helices in water alone9. Transition from an extended state to a helical one inside the translocon will, however, be significantly slower than in water as the barrier between the states is raised, by up to 15 kcal/mol in the case of Ala10 (not accounting for the possible effect of excluding partially helical states). A slow, yet inevitable transition to a helical state explains why two populations were observed experimentally for polyVal inside the translocon7. One population was extended, as polyVal would be in water, but the other was more compact, as required for its insertion into the membrane7. Further translocon-based assays focusing on sequences that only weakly favor the extended state over the helical one in water could confirm the modest helical bias predicted here.
Put together, the results presented here indicate that the translocon, specifically the monomeric SecY, is well suited to facilitate the insertion of TM segments. The mostly polar cytoplasmic half-channel functions essentially as an extension of the ribosomal exit tunnel, allowing whatever secondary structure preformed in the ribosome to be retained in the translocon. Next, for those TM segments that are not strong helix formers outside the membrane, the constrictive and hydrophobic pore ring at the center of the translocon tilts the free-energy landscape to stimulate folding, albeit slowly. The timescale of translation by the ribosome (on the order of 1 s/residue) should, however, provide ample time for TM segments to fold once fully ensconced within the channel. The multi-faceted pore ring also helps to define the energetic threshold separating membrane integration versus excretion into the periplasm41,64.
The application of the methods developed here to more realistic polypeptides, including native TM segments that fold in water, as well as those that do not, should further help elucidate the active role played by the translocon in membrane-protein development.
Acknowledgments
This work was supported by the National Institutes of Health (R01-GM067887 and P41-RR005969) and the National Science Foundation (PHY0822613). The authors acknowledge computer time provided by the Texas Advanced Computing Center, the National Center for Supercomputing Applications through the National Resources Allocation Committee (MCA93S028), the Grand Équipement National de Calcul Intensif and the Centre Informatique National de l’Enseignement Supérieur. J. G. was supported during completion of this report by an Argonne National Laboratory Director’s Postdoctoral Fellowship.
References
- 1.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 2.Voss NR, Gerstein M, Steitz TA, Moore PB. J Mol Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 3.Kramer G, Boehringer D, Ban N, Bukau B. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 4.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trabuco LG, Harrison CB, Schreiner E, Schulten K. Structure. 2010;18:627–637. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starosta AL, Karpenko VV, Shishkina AV, Mikolajka A, Sumbatyan NV, Schluenzen F, Korshunova GA, Bogdanov AA, Wilson DN. Chem Biol. 2010;17:504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Mingarro I, Nilsson I, Whitley P, von Heijne G. BMC Cell Biol. 2000;1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosolapov A, Tu L, Wang J, Deutsch C. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Woolhead CA, McCormick PJ, Johnson AE. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Deutsch C. Nat Struct Mol Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Deutsch C. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Deutsch C. J Mol Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu LW, Deutsch C. J Mol Biol. 2010;396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson JH, Woolhead CA, Bernstein HD. Mol Microbiol. 2010;78:203–217. doi: 10.1111/j.1365-2958.2010.07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosolapov A, Deutsch C. Nat Struct Mol Biol. 2009;16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien EP, Hsu SD, Christodoulou J, Vendruscolo M, Dobson CM. J Am Chem Soc. 2010;132:16928–16937. doi: 10.1021/ja106530y. [DOI] [PubMed] [Google Scholar]
- 17.Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. Nat Struct Mol Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 18.Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Mol Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Halic M, Beckmann R. Curr Opin Struct Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Rapoport TA. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 21.Driessen AJM, Nouwen N. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 22.Mandon EC, Trueman SF, Gilmore R. Curr Opin Cell Biol. 2009;21:501–507. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ménétret JF, Schaletzky J, Clemons WM, Jr, Osborne AR, Skånland SS, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, Rapoport TA, Akey CW. Mol Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 24.Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2026. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 28.Gumbart J, Schulten K. J Gen Physiol. 2008;132:709–719. doi: 10.1085/jgp.200810062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumbart J, Schulten K. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 30.White SH, von Heijne G. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 31.Zhou HX, Dill KA. Biochemistry. 2001;40:11289–11293. doi: 10.1021/bi0155504. [DOI] [PubMed] [Google Scholar]
- 32.Ziv G, Haran G, Thirumalai D. Proc Natl Acad Sci USA. 2005;102:18956–18961. doi: 10.1073/pnas.0508234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittal J, Best RB. Proc Natl Acad Sci USA. 2008;105:20233–20238. doi: 10.1073/pnas.0807742105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Cao D. Eur Phys J E. 2010;32:307–318. doi: 10.1140/epje/i2010-10634-y. [DOI] [PubMed] [Google Scholar]
- 35.Darve E, Pohorille A. J Chem Phys. 2001;115:9169–9183. [Google Scholar]
- 36.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hénin J, Forin G, Chipot C, Klein ML. J Chem Theor Comp. 2010;6:35–47. doi: 10.1021/ct9004432. [DOI] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.du Plessis DJF, Berrelkamp G, Nouwen N, Driessen AJM. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmer J, Nam Y, Rapoport TA. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gumbart J, Chipot C, Schulten K. Proc Natl Acad Sci USA. 2011;108:3596–3601. doi: 10.1073/pnas.1012758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacKerell AD, Jr, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 43.MacKerell AD, Jr, Feig M, Brooks CL., III J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 44.Feller SE, Zhang YH, Pastor RW, Brooks BR. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 45.Tuckerman M, Berne BJ, Martyna GJ. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 46.Andersen HC. J Chem Phys. 1983;52:24–34. [Google Scholar]
- 47.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP. J Mol Graphics. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 48.Tirado-Rives J, Maxwell DS, Jorgensen WL. J Am Chem Soc. 1993;115:11590–11593. [Google Scholar]
- 49.Young WS, Brooks CL. J Mol Biol. 1996;259:560–572. doi: 10.1006/jmbi.1996.0339. [DOI] [PubMed] [Google Scholar]
- 50.Topol IA, Burt SK, Deretey E, Tang TH, Perczel A, Rashin A, Csizmadia IG. J Am Chem Soc. 2001;123:6054–6060. doi: 10.1021/ja0038934. [DOI] [PubMed] [Google Scholar]
- 51.O’Neil KT, DeGrado WF. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 52.Chakrabartty A, Kortemme T, Baldwin RL. Prot Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gumbart J, Schulten K. Biophys J. 2006;90:2356–2367. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martoglio B, Hofmann MW, Brunner J, Dobberstein B. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 55.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 56.Urbanus ML, Scotti PA, Fröderberg L, Sääf A, de Gier JWL, Brunner J, Samuelson JC, Dalbey RE, Oudega B, Luirink J. EMBO Rep. 2001;2:524–529. doi: 10.1093/embo-reports/kve108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCormick PJ, Miao Y, Shao Y, Lin J, Johnson AE. Mol Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- 58.O’Brien EP, Stan G, Thirumalai D, Brooks BR. Nano Lett. 2008;8:3702–3708. doi: 10.1021/nl8019328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucent D, Vishal V, Pande VS. Proc Natl Acad Sci USA. 2007;104:10430–10434. doi: 10.1073/pnas.0608256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lucent D, Snow CD, Aitken CE, Pande VS. PLoS Comput Biol. 2010;6:e1000963. doi: 10.1371/journal.pcbi.1000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egea PF, Stroud RM. Proc Natl Acad Sci USA. 2010;40:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams S, Cosgrove TP, Gilmanshin R, Fang KS, Callendar RH, Woodruff WH, Dyer RB. Biochemistry. 1996;36:691–698. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 63.Klimov DK, Betancourt MR, Thirumalai D. Folding Des. 1998;3:481–496. doi: 10.1016/s1359-0278(98)00065-0. [DOI] [PubMed] [Google Scholar]
- 64.Junne T, Kocik L, Spiess M. Mol Biol Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]