Introduction
The greatest opportunity afforded by discovering the genetic basis of human heart disease is accurate prediction and prevention of illness. Hypertrophic cardiomyopathy (HCM) provides a paradigm to fulfill this opportunity. Human genetics research has identified many gene mutations that result in cardiac hypertrophy, of which HCM is the most common and well-characterized. Sarcomere gene mutations in HCM result in left ventricular hypertrophy (LVH), myocardial fibrosis and disarray, diastolic dysfunction, and increased risk for arrhythmias, sudden death and heart failure. Making the clinical diagnosis of HCM currently hinges on identifying unexplained hypertrophy, but LVH is a sign of established disease only. This finding cannot identify at-risk mutation carriers and cannot discriminate HCM from other forms of cardiac hypertrophy-either genetic or acquired.
In contrast, gene-based diagnosis is not constrained in this manner. Defining the mutation that causes LVH in an individual provides an accurate diagnosis for that patient and therefore, considerable information about their prognosis. Moreover, unlike clinical diagnosis which only identifies overt disease, genetic diagnosis can also identify patients at risk for developing disease. There are certainly many challenges to realizing the full potential of genetics, but such information provides unprecedented promise. Continued efforts to refine and clinically implement genetic testing in HCM will bring important payoffs in the future, and serve as a model for other genetic cardiovascular disease. By identifying at-risk individuals prior to clinical diagnosis, characterizing disease pathogenesis, and fostering development of novel therapies to delay or prevent phenotypic expression, genetic discoveries will improve the lives of our patients with HCM.
Genetics of HCM
The familial, autosomal dominant nature of HCM has long been recognized but the precise genetic etiology was discovered through genome-wide linkage studies in the 1980’s. This seminal work identified pathogenic mutations in genes encoding contractile proteins and established the paradigm that HCM is a disease of the sarcomere.1, 2 Over the past 20 years, more than 900 individual mutations have been identified, the majority (~75–80%) involving cardiac β-myosin heavy chain (MYH7) and cardiac myosin binding protein C (MYBPC3). Other sarcomere genes, cardiac troponin T (TNNT2), cardiac troponin I (TNNI3), α-tropomyosin (TPM1), the myosin light chains (MYL2, MYL3) and actin (ACTC) have also been established to cause HCM.
Genetic testing for HCM was initially available only in research laboratories and performed primarily to further basic understanding of disease. Clinical genetic testing can now be obtained through a variety of commercial laboratories, making diagnostic use feasible. Current testing is based on a candidate-gene approach, typically analyzing the 8 sarcomere genes most commonly implicated in HCM, listed above, and a small number of genes associated with metabolic/storage cardiomyopathies that may mimic HCM, including PRKAG2, LAMP2, and GLA, as discussed below. Information regarding clinical genetic testing can be obtained at http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests. Sarcomere mutations are found in approximately 65% of adult and pediatric patients with familial HCM and approximately 40% of patients with unexplained LVH but no family history of disease.3–7
In addition to these well-established sarcomere genes, mutations in other sarcomere-associated genes have been reported in association with HCM, including cardiac troponin C (TNNC1), and z-disc components telethonin (TCAP) and muscle LIM protein (CRP3).8–10 Mutations in these genes are extremely rare—identified in only a small number of isolated probands. Due to the lack of rigorous genetic support, they have not definitively been demonstrated to be disease-causing. Clinical genetic testing is not available.
Genotype Provides Insights Into HCMDiagnosis, Prognosis, and Treatment
Cardiac hypertrophy is a sign of disease rather than a specific diagnosis. Disease processes other than HCM can lead to the common finding of LVH, but have different prognoses. The underlying pathologic process cannot be determined on the basis of LV morphology alone. In contrast, the identification of a pathogenic sarcomere gene mutation establishes a definitive diagnosis of HCM and the exact genetic etiology of disease. This information provides both key insight into the patient’s prognosis, and guidance in the management of their family. Furthermore, leveraging genetic insights will teach us about the fundamental biology of HCM and drive development of improved treatment strategies (Table 1).
Table 1.
Benefits and limitations of genetic testing in HCM
| Examples and Consequences | Future Implications | ||
|---|---|---|---|
| Benefits | Confirm diagnosis in ambiguous situations |
|
|
| Definitive identification of at-risk family members |
|
|
|
| Accurate identification of disease phenocopies |
|
||
| Definition of disease etiology |
|
|
|
| Limitations | Genotype-phenotype correlations are still emerging | Results currently may not change management | More comprehensive and longitudinal studies of mutation carriers will identify more precise phenotypes |
| Incomplete knowledge of all genes associated with LVH | Negative genetic testing results are not informative | Next generation sequencing will allow a larger number of genes to be analyzed simultaneously and will detect a greater variety of mutations, including copy number variants | |
| Pathogenicity of DNA variants can be ambiguous |
|
|
|
| Genetic testing is expensive | Genetic testing may not currently be feasible for all patients | Next generation sequencing will substantially reduce costs |
Current Applications of Genetic Testing
Genetic testing identifies individuals at risk for developing disease
In medicine, we are rarely able to identify individuals who will develop disease years before typical clinical manifestations appear. Genetic testing in HCM provides extraordinary opportunities in this regard. It is particularly fruitful in the context of familial disease, since a sarcomere mutation can be identified in ~60% of probands, and half of their first degree relatives are predicted to carry the mutation. Genotype determination provides substantial prognostic insight through its ability to definitively and precisely identify relatives at risk for developing disease at an early stage, independently of other clinical features. Simply put, if the family’s pathogenic mutation is not inherited, there is no risk of HCM, either in that relative or their offspring. If a mutation is present, HCM is very likely to develop. Longitudinal clinical follow up can then be focused on pathogenic mutation carriers, as only they are at risk (Figure 1). Family members who have not inherited a pathogenic mutation can be reassured that neither they nor their offspring are at risk for disease development and do not require serial clinical evaluation or lifestyle restrictions. Figure 2 presents a general framework for incorporating genetic testing in HCM patients and families. Predictive testing in HCM families with a known pathogenic sarcomere mutation can also be used for reproductive planning through preimplantation genetic diagnosis to attempt to achieve a pregnancy with an embryo that does not carry the mutation.
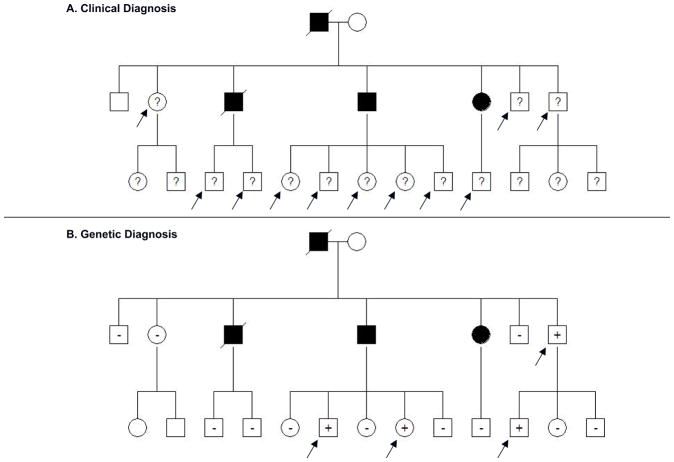
Figure 1. Genetic testing in families can identify individuals who are definitively at risk for developing disease and focus longitudinal follow up.
A. Pedigree of a family with HCM: Clinical screening is recommended for first degree relatives of patients with HCM to identify people with left ventricular hypertrophy and previously unrecognized disease. In this example, at least 11 family members (arrows) without LVH and of unknown genotype (?) are at risk for developing HCM and require longitudinal clinical evaluation.
B. With genetic testing, serial clinical screening can be appropriately focused on mutation carriers, since relatives who have not inherited the pathogenic mutation are not at risk for disease development. In this family, only the 4 mutation carriers without clinical HCM (arrows) would require longitudinal follow up to assess for disease onset.
Circles, females; squares males; solid symbols, clinical diagnosis of HCM; slash, deceased; ?, genotype unknown; +, mutation present; −, mutation absent
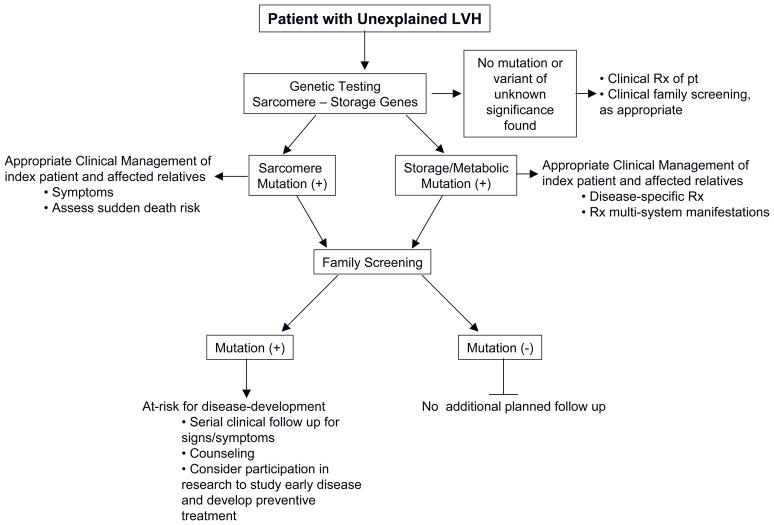
Figure 2.
A general framework for incorporating genetic testing in the approach to patients and families with unexplained LVH.
Such gene-based diagnostic strategies in families have economic as well as medical benefits. Because HCM follows autosomal dominant inheritance and the penetrance of LVH is age-dependent, serial clinical screening of first degree relatives is recommended, including physical examination, 12-lead EKG, and echocardiography.11 Cardiac magnetic resonance imaging, Holter monitoring, and exercise testing may also be pursued. This translates into roughly $1000 for each family member for each visit. Over time, this will cost ~$6000 to follow the child of an HCM patient through puberty when the frequency of screening is highest, and ~$20,000 over their lifetime. Compared with these longitudinal expenses, genetic testing is a relative bargain. It currently costs ~$3000 to identify a mutation in a family through genetic testing of a single affected individual. Predictive genetic testing for the rest of the family to determine whether or not the mutation has been inherited costs ~$400 per relative. Since only mutation carriers are at risk for developing HCM, genetic testing results will reduce the number of family members who require serial follow up, as illustrated in Figure 1. This will lead to substantial healthcare cost-savings, especially as the price of genetic testing falls in the future. An important caveat to this strategy is that there must be a high degree of confidence that the family’s mutation is pathogenic. If there is uncertainty as to whether an identified DNA sequence variant can cause disease, it should not be used to drive management.
Genetic testing clarifies ambiguous diagnoses
Clinicians may be confronted with patients who have mild LVH and confounding features such as mild to moderate hypertension or intensive athletic training. In these situations, there may be ambiguity as to whether the degree of hypertrophic remodeling can be explained by co-existing pressure load or physical conditioning, or whether a primary cardiomyopathy is present. Since HCM is a genetic condition associated with increased risk for arrhythmias and sudden death, clarifying the underlying etiology is of major importance to the patient and their family. Distinguishing HCM from hypertensive heart disease or athlete’s heart may be straightforward, for example, by documenting regression of LVH after controlling blood pressure or stopping athletic training. However, the diagnosis may remain unclear despite comprehensive clinical evaluation. Identifying a pathogenic sarcomere mutation in this setting would confirm the diagnosis of HCM and trigger appropriate management of the patient and their family, including cessation of competitive athletics, assessment of sudden death risk, and screening relatives.
Not all cardiac hypertrophy is HCM: Genetic testing identifies phenocopies
Cardiac hypertrophy is a relatively nonspecific phenotype that may reflect the final common pathway for a number of different disease processes. It does not indicate etiology or reveal underlying pathophysiology. In contrast, genetic testing affords a level of discrimination not attainable by cardiac imaging or even histological evaluation. Gene-based diagnosis can identify the precise disease process underlying a patient’s hypertrophy at the molecular level. As a result of broader application of genotyping, phenocopies of HCM have been identified where cardiac hypertrophy is caused by mutations in genes distinct from those which encode sarcomere proteins (Table 2). For example, a separate category of metabolic cardiomyopathies has been described in families and sporadic patients with unexplained LVH who commonly also have concomitant conduction abnormalities (progressive atrioventricular block, atrial fibrillation, ventricular pre-excitation). In these individuals, cardiac hypertrophy is caused by mutations in PRKAG2, encoding the γ2 regulatory subunit of adenosine monophosphate-activated protein kinase, and in LAMP2, encoding the X-linked lysosome associated membrane protein. Mutations in these genes are rare but may be present in roughly 2–12% of individuals with a clinical diagnosis of HCM but no sarcomere mutation.12–15 Although the presence of pre-excitation may be suggestive, these conditions cannot be reliably differentiated on the basis of cardiac imaging alone (Figure 3).
Table 2.
Mutations in genes associated with phenocopies of HCM, resulting in metabolic or storage cardiomyopathy.
| Protein | Gene | Chromosome | Associated Disease | Comments |
|---|---|---|---|---|
| γ-Subunit, AMP Kinase | PRKAG2 | 7q36 | Pre-excitation and conduction disease | |
| Lysosome assosiated membrane protein | LAMP2 | Xq24 | Danon Disease | Cardiomyopathy, skeletal myopathy, and neurologic involvement may be present; Pre-excitation on EKG; Rapid progression to end stage heart failure in adolescence, particularly males; High risk of sudden death |
| α-Galactosidase | GLA | Xq22 | Fabry Syndrome | Assess plasma or lymphocyte α–Gal activity (males); Consider enzyme replacement |
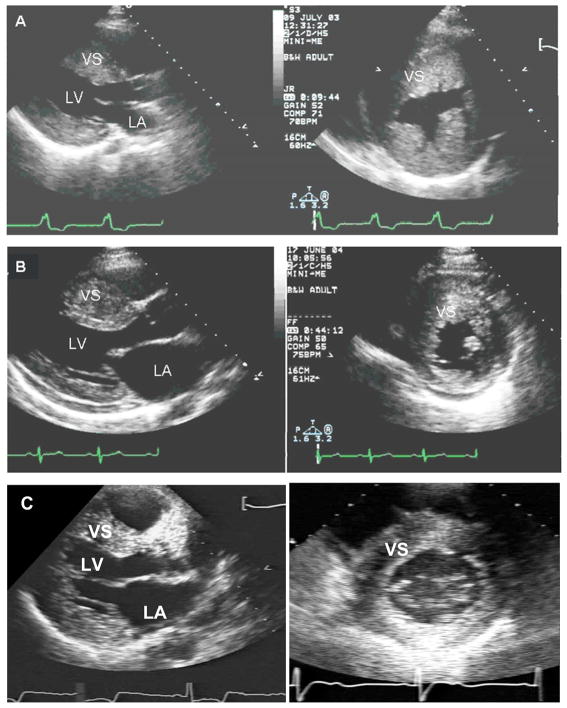
Figure 3. Phenocopies mimic the clinical appearance of HCM and cannot be reliably differentiated by echocardiography, although the underlying disease process, prognosis and approach to management are different.
A. Parasternal long and short axis echocardiographic images from an 18 year-old male with a LAMP2 mutation. There is marked, diffuse LV hypertrophy with a maximal LV septal wall thickness of 35 mm.
B. Parasternal images from a 20 year old female with a myosin heavy chain (MYH7) mutation, also showing marked, diffuse LVH, with a maximal wall thickness of 30 mm. By imaging alone, these patients cannot be discriminated although his disease course is predicted to be quite different, with a considerably worse prognosis in patient A.
C. Parasternal images from a 45 year-old male with a GLA mutation and cardiac-restricted Fabry disease. Maximal septal wall thickness is 22 mm.
LV= left ventricle; LA= left atrium; VS= ventricular septum
Distinguishing HCM from phenocopies is important because the molecular pathways triggered by mutations in PRKAG2 and LAMP2, or other non-myofilament genes, are almost certainly different from those triggered by sarcomere gene mutations. As such, applying HCM management tenets may not be appropriate. In the case of LAMP2 cardiomyopathy, genetic diagnosis provides important prognostic information owing to the typically severe and lethal natural history of this disorder, particularly in affected males.16 In the case of Fabry disease, an X-linked recessive disorder caused by mutations in the gene encoding the lysosomal hydrolase, α-galactosidase (GLA), clinical management may be specifically changed. GLA mutations cause enzyme deficiency and glycosphyngolipid accumulation in the heart, kidneys, nervous system and skin. Classic Fabry disease occurs at a prevalence of ~1/40,000 and commonly presents in childhood or adolescence. However a cardiac-predominant variant of Fabry disease has been described and may account for 2–3% of unexplained LVH in adult males.17, 18 Accurate diagnosis of Fabry disease is important as there is potentially effective α-galactosidase enzyme replacement therapy.19
Genotype influences phenotype and prognosis in HCM
HCM is a highly complex and heterogeneous disease regarding not only the number of associated mutations, but also the variable degree of LVH, symptom burden, and risk for sudden death or heart failure. The factors that drive this broad clinical spectrum have not been fully elucidated. Although genotype certainly influences phenotype, the relationship may not be obvious. For example, family members with the same mutation may have very different clinical manifestations. As such, the results of genetic testing alone may not identify exactly which individuals will benefit from an ICD or whether competitive sports can be safely pursued. However, this perspective reflects a somewhat restrictive view of its clinical utility. With comprehensive and longitudinal study of mutation carriers and collaborative bench-to-bedside investigation, more cohesive patterns and greater insights to disease manifestations and pathogenesis will emerge.
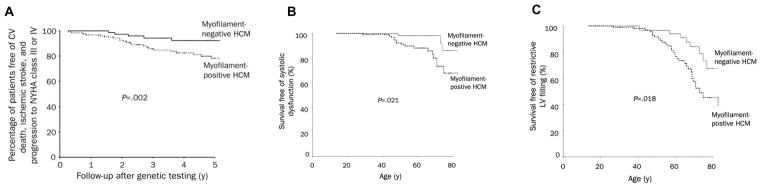
Prognostic insight can be gained comparing individuals with and without sarcomere mutations. In addition to the HCM phenocopies discussed above, sarcomere mutations are currently not identified in ~30–60% of individuals with a clinical diagnosis of HCM. The molecular basis of unexplained LVH in the absence of a sarcomere mutation is unknown and may represent complex rather than Mendelian genetics. Sarcomere-negative subjects cannot be reliably differentiated on the basis of cardiac imaging or clinical features alone. However, making the distinction may be important from a diagnostic and prognostic standpoint, as the outcome of patients with sarcomere mutations appears to be worse than those without. In one of the few longitudinal studies of a genotyped HCM cohort, investigators demonstrated an increased risk of cardiovascular death, nonfatal stroke, or progression to NYHA III/IV functional class in HCM patients with sarcomere mutations as compared to those with negative genetic testing.20 As shown in Figure 4, mutation-positive HCM patients were significantly more likely to achieve the composite endpoint, and develop systolic and severe diastolic dysfunction as compared to mutation-negative patients. Multivariate analysis showed that the presence of a sarcomere mutation was the strongest independent predictor of an adverse outcome with a hazard ratio of 4.27 (95% confidence interval 1.43, 12.48; p=0.008). Genotype outperformed clinical variables such as age, the presence of LV outflow tract obstruction, and atrial fibrillation.
Figure 4. Clinical outcomes are worse for HCM patients with sarcomere mutations (myofilament-positive), compared to those without mutations (myofilament-negative).20.
A. Myofilament-positive patients have a higher probability of cardiovascular (CV) death, nonfatal ischemic stroke, or progression to severe heart failure symptoms (New York Health Association (NYHA) functional classes III or IV).
B. Myofilament-positive patients have a higher probability of developing systolic dysfunction, defined as LV ejection fraction <50%.
C. Myofilament-positive patients have a higher probability of developing restrictive LV filling pattern, defined as transmitral early deceleration time <120 ms or a ratio of peak mitral early to late diastolic filling velocity (E/A ratio) of ≥ 2 in conjunction with a deceleration time of ≥ 150 ms.
Adapted with permission from Olivotto I. et al. Mayo Clinic proceedings 2008;83(6):630–8.
Gene dosage likely also influences prognosis in HCM. Approximately 3–5% of probands with HCM have more than one sarcomere mutation (compound or double heterozygosity).21, 22 Individuals with more genetic “hits” tend to have more severe disease expression,20, 22, 23 even if the variant results in relatively mild disease in isolation.24, 25 Disease can be particularly severe in rare cases of triple mutations26 and homozygosity.24
At the level of the individual gene, prognostic insights have been derived by comparing MYH7 and MYBPC3 mutations, as well as by assessing mutation type and the functional domain affected. MYBPC3 mutations have been associated with delayed, even elderly-onset disease,7 and may have a higher rate of incomplete penetrance than MYH7 mutations.27 In children, MYBPC3 missense mutations (resulting in an amino acid substitution) were more common, whereas truncation mutations predominate in adults.4 In MYH7 mutations, amino acid substitutions resulting in a change of charge in the ATP hydrolysis active site or in the head–rod junction may be associated with a higher risk of heart failure and reduced survival.28
At the level of the individual nucleotide, a small number of specific point mutations have been associated with more predictable severe phenotypes in unrelated HCM families.29–33 For example, the clinical course of MYH7 missense mutations in which arginine is replaced by glutamine at amino acid residue 403 (Arg403Gln) and Arg719Trp have shown a markedly increased risk of sudden death or near-universal development of end-stage heart failure requiring cardiac transplantation, respectively. Certain TNNT2 mutations (Arg92Trp, Arg92Gln, Ile79Asn) have also been associated with an increased risk of sudden death in certain families. However, these mutations are rare, accounting for ~0.2–0.6% of probands referred for genetic testing (personal communication, Dr. H. Rehm, Director, Laboratory for Molecular Medicine) so a strategy of screening only for previously characterized mutations will not have appreciable clinical yield. Furthermore caution is needed in generalizing these observations to individual patients, as exceptions have been well documented and mutations are not consistently malignant or benign.34 Nonetheless, identifying both potential trends and exceptions help to further understanding of myocardial biology and disease pathogenesis.
Future Applications of Genetics: Changing Clinical Destiny
Our current approach to diagnosing and managing HCM is unsatisfying as it is largely reactive and palliative. Patients are diagnosed when unexplained LVH is identified and treated to alleviate symptoms. To truly transform medicine, we need more precise information regarding pathogenesis and therapies that prevent disease. This can be achieved by integrating basic science discovery with patient management in HCM. A combined genetic and clinical approach allows identification of individuals at risk for developing disease independently of clinical findings, better characterization of the full spectrum of phenotypes caused by sarcomere mutations, and development of rational new strategies to change the natural history of disease, based on mechanistic insights.
Gene-based diagnosis: Redefining phenotype
Genetic testing can identify individuals who carry pathogenic sarcomere mutations, and are therefore at high risk for developing disease, before a clinical diagnosis of HCM can be made. As such, this population provides a remarkable window into the future, casting new light on the earliest cellular responses to sarcomere gene mutations and teaching us about the biology of disease. Moreover, preclinical mutation carriers are a key target of future trials designed to prevent development or progression of HCM.
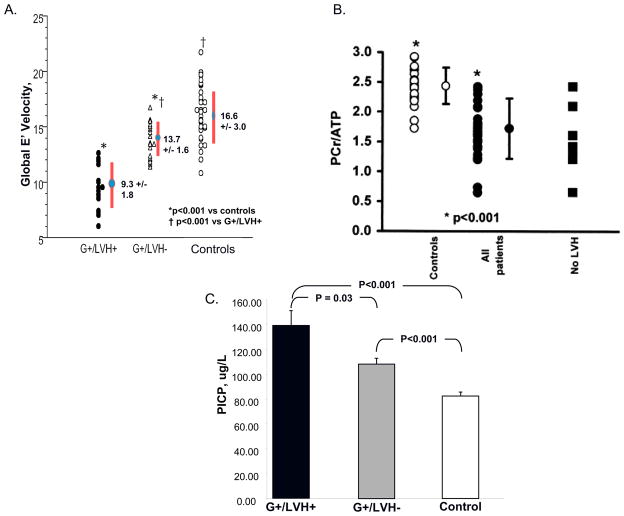
Using this approach, novel early phenotypes have been identified in prehypertrophic sarcomere mutation carriers, providing key information about the development of myocardial function, energetics, biochemistry, and fibrogenesis in disease pathogenesis (Figure 5). At the functional level, mutation carriers have been found to have diastolic dysfunction before LVH develops, as reflected by reduced early myocardial relaxation velocities (E′ velocity) on tissue Doppler interrogation (Figure 5A).35–38 At the biophysical level, impaired myocardial energetics has been proposed as a unifying mechanism by which sarcomere mutations give rise to cardiac hypertrophy and heart failure.39–41 31P magnetic resonance spectroscopy (MRS) studies have demonstrated a significantly decreased ratio of phosphocreatine to ATP (PCr/ATP) both in the early, prehypertrophic stage, and in patients with clinically overt HCM (Figure 5B).40 These data indicate a compromised energetic state and lend further support to the primary role energy deficiency in the pathogenesis of HCM.40
Figure 5. Early phenotypes of sarcomere mutations have been identified by studying mutation carriers prior to the development of clinical manifestations of HCM.
A. Impaired relaxation is detectable as reduced early myocardial relaxation velocities (E′) on tissue Doppler interrogation in mutation carriers who have not yet developed LVH (G+/LVH−) compared to normal controls. E′ velocities fall further with the development of overt HCM (G+/LVH+).36 Adapted with permission from Ho CY, et al. Circulation 2002;105(25):2992–7.
B. Mutation carriers have impaired myocardial energetics as manifest by decreased Phosphocreatine/adenosine triphosphate (PCr/ATP) ratio. This is present all patients, including mutation carriers without LVH. Filled circles, HCM patients; open circles, controls; filled squares, mutation carriers without LVH. 40 Adapted with permission from Crilley JG, et al. Journal of the American College of Cardiology 2003;41(10):1776–82.
C. Mutation carriers show evidence of increased collagen synthesis, as manifest by increased serum levels of C-terminal propeptide of type I procollagen (PICP), before LVH develops (G+/LVH+). Compared to controls, PICP levels are significantly increased in all sarcomere mutations carriers, with (G+/LVH+) and without (G+/LVH−) hypertrophy. The threshold for significance is P <0.017 and adjusted for age and family relations.
Reproduced with permission from Ho, C.Y., et al, N Engl J Med 2010;363(6):552–563.
At the histological level, myocardial fibrosis is a hallmark of HCM and postulated to be the substrate underlying sudden cardiac death, ventricular tachyarrhythmias, LV dysfunction, and heart failure.42–46 The trigger for increased myocardial fibrosis in HCM is unknown. Cardiac gene expression studies performed on young, prehypertrophic HCM mice showed early activation of pathways involved in fibrosis and collagen deposition even when cardiac histology is normal.47 Recent studies in humans also show evidence of increased collagen synthesis in sarcomere mutation carriers before the development of LVH. Increased levels of C-terminal propeptide of type I procollagen indicate that a pro-fibrotic milieu is present early in human HCM (Figure 5C) in the absence of cardiac hypertrophy or visible fibrosis on cardiac magnetic resonance imaging.48
These studies indicate that sarcomere mutations have considerable adverse impact on the heart before the development of clinically recognized disease. Diastolic dysfunction, impaired myocardial energetics, and increased collagen synthesis appear to be early phenotypes that are intrinsic manifestations of the underlying sarcomere mutation, rather than a secondary reaction to the abnormalities in myocardial structure and function that accompany clinically overt disease. The presence of such early phenotypes may identify individuals at risk for arrhythmias, sudden death, or heart failure. Furthermore, identification of new phenotypes may highlight previously unrecognized pathways that contribute to disease development that may be targeted therapeutically. Finally, quantitative traits such as E′ velocity, PCr/ATP ratio, and serum biomarkers may serve as surrogate endpoints to monitor treatment effect, thereby fostering development of novel treatment strategies to change the natural history of HCM.
Translating Genetic Discoveries: Preventing Disease
Greater understanding of the molecular pathogenesis of HCM provides the necessary rationale to develop new therapies designed to slow or prevent disease development and improve outcomes in HCM. Disease-modifying studies are in active development in animal models of HCM. These genetically modified animals carry sarcomere mutations which cause human disease and replicate the HCM phenotype.49, 50 For example, in mice carrying the Arg403Gln missense mutation in MYH7, abnormalities in intracellular Ca2+ homeostasis (present at 4 weeks of age) are one of the earliest detectable manifestations of sarcomere mutations, preceding the development of diastolic abnormalities (~age 6 weeks), and visible LVH, fibrosis and disarray (~age 20 weeks).51, 52 Early treatment with the L-type calcium channel blocker, diltiazem, appeared to mitigate development of hypertrophy and fibrosis if started prior to development of LVH.52 More recently, early treatment with losartan in mice with an Arg719Trp MYH7 mutation has shown promising results in mitigating the development of fibrosis, potentially by inhibiting TGF-β-mediated pathways.53 A pilot human randomized control trial is ongoing, comparing diltiazem to placebo in sarcomere mutation carriers who have not yet developed LVH (http://clinicaltrials.gov/ct2/show/NCT00319982). A collaborative, NIH-sponsored multicenter effort has recently been established to facilitate clinical translational research in early HCM and support future trials of disease prevention.
Other strategies have been trialed in animal models to reverse the effects of established disease by targeting myocardial fibrosis. Administration of angiotensin II receptor blockers (losartan),54 HMG-CoA reductase inhibitors (simvastatin),55 aldosterone antagonists (spironolactone),56 and the antioxidant, N-acetylcysteine57 have shown encouraging results in decreasing myocardial fibrosis and collagen content. Notably, all of these trials were performed on animal models with specific genetic substrate and may only be applicable to patients with sarcomere mutations, not other forms of cardiac hypertrophy. As such, genotype may profoundly influence disease management and prognosis in the future.
Limitations of Genetic Testing
When the genetic underpinnings of HCM were first described, there was great optimism and perhaps unrealistic expectations that this knowledge would quickly revolutionize the prognosis and management of disease.29, 30, 58 However, as is typically the case with complex biological systems, practical application of these scientific breakthroughs has not been immediate or straightforward. Significant obstacles are created by the marked heterogeneity of HCM and by difficulties in determining whether novel DNA sequence variants are pathogenic and capable of causing disease. Therefore, the utility of incorporating genotype data into clinical management has been appropriately criticized. As indicated in Table 1, it is important to recognize the advantages, limitations, implications, and applications of genetic testing both now and in the future.
The daunting clinical and genetic variability of HCM limit robust genotype-phenotype correlations. In considering just sarcomere genes, mutations can occur in tens of thousands of base pairs of DNA and only ~1000 have been identified so far, often only in a single proband. Given the great variability in the genetic cause of HCM, it is not surprising that clinical outcome cannot be accurately predicted from identifying single mutations. Moreover, although mutations trigger development of disease, they are not the sole determinant of the final phenotype as genetic modifiers, environmental influences, and comorbid illnesses exert important influences which are currently not well understood. Mutations initially characterized as “benign” or “malignant” in the context of studying large families29–31 do not always result in a consistent phenotype in unrelated probands.34, 59 These unavoidable complexities diminish the application of genotyping for rigorous prediction of risk. Identifying the exact sequence variant responsible for causing HCM typically will not directly impact prognosis or management decisions for the individual patient, such as the need for an implantable cardiovertor-defibrillator (ICD), indications for surgery, exercise recommendations, or initiation of medical therapy.
Also, sarcomere mutations are not universally identified in all patients with a clinical diagnosis of HCM. Depending on other characteristics such as family history, over 50% of patients may have negative, and therefore non-informative, genetic testing from analysis of sarcomere genes. This statistic again highlights that LVH is a crude and non-specific tool for diagnosing disease. However, the imperfect yield of HCM genetic testing also reflects incomplete knowledge of all genes that cause LVH. Ongoing gene discovery efforts will identify new genes associated with HCM, but this process takes many years.
Other technical issues that limit the yield of genetic testing will be solved more quickly. Next-generation sequencing platforms, and ultimately whole exome strategies, will soon be able to interrogate millions of basepairs of DNA faster and cheaper than current strategies.60 In addition to improving the availability of genetic testing because of lowered costs, this technology will allow two major advances. First, many more genes can be interrogated simultaneously; an improvement from the existing strategy of examining a fixed subset of genes (sarcomere) for specific diseases (HCM). More comprehensive sequencing will identify potential interactions between different genetic pathways, beyond the sarcomere. For example, variants in genes implicated in other cardiomyopathies, inherited arrhythmias, calcium regulation and myocardial metabolism may interact to shape disease phenotype. Second, new sequencing strategies will also be able to determine if variation in the number of copies of sarcomere gene sequences cause HCM. Copy number variants have been implicated in congenital heart disease, neurologic disease, and malignancy,61 and may indeed be relevant in HCM, but undetected by traditional sequencing strategies.
Conclusions
Genotype analysis offers unique information regarding prognosis in HCM that is relevant both now and in the future. Genetic testing provides definitive diagnosis of disease, preclinical recognition of at-risk individuals, and a powerful means to dissect disease pathogenesis. Identifying the genetic basis of disease is a critical first step in the long journey that will allow us to advance and transform medicine, based on mechanistic insight, early diagnosis, and disease prevention.
Acknowledgments
Funding Sources
The author is supported by funding from the National Institutes of Health.
Footnotes
Disclosures
None
References
- 1.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 2.Watkins H, MacRae C, Thierfelder L, Chou YH, Frenneaux M, McKenna W, Seidman JG, Seidman CE. A disease locus for familial hypertrophic cardiomyopathy maps to chromosome 1q3. Nat Genet. 1993;3:333–337. doi: 10.1038/ng0493-333. [DOI] [PubMed] [Google Scholar]
- 3.Konno T, Chang S, Seidman JG, Seidman CE. Genetics of hypertrophic cardiomyopathy. Current opinion in cardiology. 2010 doi: 10.1097/HCO.0b013e3283375698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita H, Rehm HL, Menesses A, McDonough B, Roberts AE, Kucherlapati R, Towbin JA, Seidman JG, Seidman CE. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morita H, Larson MG, Barr SC, Vasan RS, O’Donnell CJ, Hirschhorn JN, Levy D, Corey D, Seidman CE, Seidman JG, Benjamin EJ. Single-gene mutations and increased left ventricular wall thickness in the community: the Framingham Heart Study. Circulation. 2006;113:2697–2705. doi: 10.1161/CIRCULATIONAHA.105.593558. [DOI] [PubMed] [Google Scholar]
- 6.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, Kristinsson A, Roberts R, Sole M, Maron BJ, Seidman JG, Seidman CE. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy [see comments] N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 7.Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 8.Bos JM, Poley RN, Ny M, Tester DJ, Xu X, Vatta M, Towbin JA, Gersh BJ, Ommen SR, Ackerman MJ. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Molecular genetics and metabolism. 2006;88:78–85. doi: 10.1016/j.ymgme.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, Hoffmann K, Pilz B, Martiniak Y, Gehmlich K, van der Ven PF, Furst DO, Vornwald A, von Hodenberg E, Nurnberg P, Scheffold T, Dietz R, Osterziel KJ. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–1395. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 10.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, Roberts R, Willerson JT, Marian AJ. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circulation research. 2007;100:766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maron BJ, Seidman JG, Seidman CE. Proposal for contemporary screening strategies in families with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:2125–2132. doi: 10.1016/j.jacc.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 12.Blair E, Redwood C, Ashrafian H, Oliveira M, Broxholme J, Kerr B, Salmon A, Ostman-Smith I, Watkins H. Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10:1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 13.Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, Ahmad F, Lozado R, Shah G, Fananapazir L, Bachinski LL, Roberts R, Hassan AS. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 14.Arad M, Benson DW, Perez-Atayde AR, McKenna WJ, Sparks EA, Kanter RJ, McGarry K, Seidman JG, Seidman CE. Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J Clin Invest. 2002;109:357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arad M, Maron BJ, Gorham JM, Johnson WH, Jr, Saul JP, Perez-Atayde AR, Spirito P, Wright GB, Kanter RJ, Seidman CE, Seidman JG. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 16.Maron BJ, Roberts WC, Arad M, Haas TS, Spirito P, Wright GB, Almquist AK, Baffa JM, Saul JP, Ho CY, Seidman J, Seidman CE. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. Jama. 2009;301:1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakao S, Takenaka T, Maeda M, Kodama C, Tanaka A, Tahara M, Yoshida A, Kuriyama M, Hayashibe H, Sakuraba H. An atypical variant of Fabry’s disease in men with left ventricular hypertrophy. N Engl J Med. 1995;333:288–293. doi: 10.1056/NEJM199508033330504. [DOI] [PubMed] [Google Scholar]
- 18.Sachdev B, Takenaka T, Teraguchi H, Tei C, Lee P, McKenna WJ, Elliott PM. Prevalence of Anderson-Fabry disease in male patients with late onset hypertrophic cardiomyopathy. Circulation. 2002;105:1407–1411. doi: 10.1161/01.cir.0000012626.81324.38. [DOI] [PubMed] [Google Scholar]
- 19.Banikazemi M, Bultas J, Waldek S, Wilcox WR, Whitley CB, McDonald M, Finkel R, Packman S, Bichet DG, Warnock DG, Desnick RJ. Agalsidase-beta therapy for advanced Fabry disease: a randomized trial. Ann Intern Med. 2007;146:77–86. doi: 10.7326/0003-4819-146-2-200701160-00148. [DOI] [PubMed] [Google Scholar]
- 20.Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, Ommen SR, Theis JL, Vaubel RA, Re F, Armentano C, Poggesi C, Torricelli F, Cecchi F. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clinic proceedings. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 21.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 22.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, Ackerman MJ. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 23.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. Journal of medical genetics. 2005;42(10):e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho CY, Lever HM, DeSanctis R, Farver CF, Seidman JG, Seidman CE. Homozygous mutation in cardiac troponin T: implications for hypertrophic cardiomyopathy. Circulation. 2000;102:1950–1955. doi: 10.1161/01.cir.102.16.1950. [DOI] [PubMed] [Google Scholar]
- 25.Saltzman AJ, Mancini-DiNardo D, Li C, Chung WK, Ho CY, Hurst S, Wynn J, Care M, Hamilton RM, Seidman GW, Gorham J, McDonough B, Sparks E, Seidman JG, Seidman CE, Rehm HL. Short communication: the cardiac myosin binding protein C Arg502Trp mutation: a common cause of hypertrophic cardiomyopathy. Circulation research. 106:1549–1552. doi: 10.1161/CIRCRESAHA.109.216291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girolami F, Ho CY, Semsarian C, Baldi M, Will ML, Baldini K, Torricelli F, Yeates L, Cecchi F, Ackerman MJ, Olivotto I. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. Journal of the American College of Cardiology. 2010;55:1444–1453. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 27.Christiaans I, Birnie E, van Langen IM, van Spaendonck-Zwarts KY, van Tintelen JP, van den Berg MP, Atsma DE, Helderman-van den Enden AT, Pinto YM, Hermans-van Ast JF, Bonsel GJ, Wilde AA. The yield of risk stratification for sudden cardiac death in hypertrophic cardiomyopathy myosin-binding protein C gene mutation carriers: focus on predictive screening. European heart journal. 2009;31:842–848. doi: 10.1093/eurheartj/ehp539. [DOI] [PubMed] [Google Scholar]
- 28.Woo A, Rakowski H, Liew JC, Zhao MS, Liew CC, Parker TG, Zeller M, Wigle ED, Sole MJ. Mutations of the beta myosin heavy chain gene in hypertrophic cardiomyopathy: critical functional sites determine prognosis. Heart. 2003;89:1179–1185. doi: 10.1136/heart.89.10.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watkins H, Rosenzweig A, Hwang DS, Levi T, McKenna W, Seidman CE, Seidman JG. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy [see comments] N Engl J Med. 1992;326:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 30.Moolman JC, Corfield VA, Posen B, Ngumbela K, Seidman C, Brink PA, Watkins H. Sudden death due to troponin T mutations. J Am Coll Cardiol. 1997;29:549–555. doi: 10.1016/s0735-1097(96)00530-x. [DOI] [PubMed] [Google Scholar]
- 31.Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O’Donoghue A, Spirito P, Matsumori A, Moravec CS, Seidman JG. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 32.Marian AJ, Roberts R. The molecular genetic basis for hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2001;33:655–670. doi: 10.1006/jmcc.2001.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tardiff JC. Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes. Heart failure reviews. 2005;10:237–248. doi: 10.1007/s10741-005-5253-5. [DOI] [PubMed] [Google Scholar]
- 34.Ackerman MJ, VanDriest SL, Ommen SR, Will ML, Nishimura RA, Tajik AJ, Gersh BJ. Prevalence and age-dependence of malignant mutations in the beta-myosin heavy chain and troponin T genes in hypertrophic cardiomyopathy: a comprehensive outpatient perspective. J Am Coll Cardiol. 2002;39:2042–2048. doi: 10.1016/s0735-1097(02)01900-9. [DOI] [PubMed] [Google Scholar]
- 35.Ho CY, Carlsen C, Thune JJ, Havndrup O, Bundgaard H, Farrohi F, Rivero J, Cirino AL, Andersen PS, Christiansen M, Maron BJ, Orav EJ, Kober L. Echocardiographic Strain Imaging to Assess Early and Late Consequences of Sarcomere Mutations in Hypertrophic Cardiomyopathy. Circulation: Cardiovascular Genetics. 2009;2:314–321. doi: 10.1161/CIRCGENETICS.109.862128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho CY, Sweitzer NK, McDonough B, Maron BJ, Casey SA, Seidman JG, Seidman CE, Solomon SD. Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation. 2002;105:2992–2997. doi: 10.1161/01.cir.0000019070.70491.6d. [DOI] [PubMed] [Google Scholar]
- 37.Nagueh SF, Bachinski LL, Meyer D, Hill R, Zoghbi WA, Tam JW, Quinones MA, Roberts R, Marian AJ. Tissue Doppler imaging consistently detects myocardial abnormalities in patients with hypertrophic cardiomyopathy and provides a novel means for an early diagnosis before and independently of hypertrophy. Circulation. 2001;104:128–130. doi: 10.1161/01.cir.104.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagueh SF, McFalls J, Meyer D, Hill R, Zoghbi WA, Tam JW, Quinones MA, Roberts R, Marian AJ. Tissue Doppler imaging predicts the development of hypertrophic cardiomyopathy in subjects with subclinical disease. Circulation. 2003;108:395–398. doi: 10.1161/01.CIR.0000084500.72232.8D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spindler M, Saupe KW, Christe ME, Sweeney HL, Seidman CE, Seidman JG, Ingwall JS. Diastolic dysfunction and altered energetics in the alphaMHC403/+ mouse model of familial hypertrophic cardiomyopathy. J Clin Invest. 1998;101:1775–1783. doi: 10.1172/JCI1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crilley JG, Boehm EA, Blair E, Rajagopalan B, Blamire AM, Styles P, McKenna WJ, Ostman-Smith I, Clarke K, Watkins H. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol. 2003;41:1776–1782. doi: 10.1016/s0735-1097(02)03009-7. [DOI] [PubMed] [Google Scholar]
- 41.Ashrafian H, Watkins H. Reviews of translational medicine and genomics in cardiovascular disease: new disease taxonomy and therapeutic implications cardiomyopathies: therapeutics based on molecular phenotype. J Am Coll Cardiol. 2007;49:1251–1264. doi: 10.1016/j.jacc.2006.10.073. [DOI] [PubMed] [Google Scholar]
- 42.Basso C, Thiene G, Corrado D, Buja G, Melacini P, Nava A. Hypertrophic cardiomyopathy and sudden death in the young: pathologic evidence of myocardial ischemia. Human pathology. 2000;31:988–998. doi: 10.1053/hupa.2000.16659. [DOI] [PubMed] [Google Scholar]
- 43.Shirani J, Pick R, Roberts WC, Maron BJ. Morphology and significance of the left ventricular collagen network in young patients with hypertrophic cardiomyopathy and sudden cardiac death. J Am Coll Cardiol. 2000;35:36–44. doi: 10.1016/s0735-1097(99)00492-1. [DOI] [PubMed] [Google Scholar]
- 44.Choudhury L, Mahrholdt H, Wagner A, Choi KM, Elliott MD, Klocke FJ, Bonow RO, Judd RM, Kim RJ. Myocardial scarring in asymptomatic or mildly symptomatic patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2002;40:2156–2164. doi: 10.1016/s0735-1097(02)02602-5. [DOI] [PubMed] [Google Scholar]
- 45.Varnava AM, Elliott PM, Mahon N, Davies MJ, McKenna WJ. Relation between myocyte disarray and outcome in hypertrophic cardiomyopathy. The American journal of cardiology. 2001;88:275–279. doi: 10.1016/s0002-9149(01)01640-x. [DOI] [PubMed] [Google Scholar]
- 46.Varnava AM, Elliott PM, Sharma S, McKenna WJ, Davies MJ. Hypertrophic cardiomyopathy: the interrelation of disarray, fibrosis, and small vessel disease. Heart. 2000;84:476–482. doi: 10.1136/heart.84.5.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 48.Ho CY, Lopez B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, Kwong R, Gonzalez A, Colan SD, Seidman JG, Diez J, Seidman CE. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med. 2010;363:552–563. doi: 10.1056/NEJMoa1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 50.Marian AJ, Wu Y, Lim DS, McCluggage M, Youker K, Yu QT, Brugada R, DeMayo F, Quinones M, Roberts R. A transgenic rabbit model for human hypertrophic cardiomyopathy. J Clin Invest. 1999;104:1683–1692. doi: 10.1172/JCI7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fatkin D, McConnell BK, Mudd JO, Semsarian C, Moskowitz IG, Schoen FJ, Giewat M, Seidman CE, Seidman JG. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest. 2000;106:1351–1359. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semsarian C, Ahmad I, Giewat M, Georgakopoulos D, Schmitt JP, McConnell BK, Reiken S, Mende U, Marks AR, Kass DA, Seidman CE, Seidman JG. The L-type calcium channel inhibitor diltiazem prevents cardiomyopathy in a mouse model. J Clin Invest. 2002;109:1013–1020. doi: 10.1172/JCI14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE, Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest. 2010 doi: 10.1172/JCI42028. Epub 2010 Sept 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim DS, Lutucuta S, Bachireddy P, Youker K, Evans A, Entman M, Roberts R, Marian AJ. Angiotensin II blockade reverses myocardial fibrosis in a transgenic mouse model of human hypertrophic cardiomyopathy. Circulation. 2001;103:789–791. doi: 10.1161/01.cir.103.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel R, Nagueh SF, Tsybouleva N, Abdellatif M, Lutucuta S, Kopelen HA, Quinones MA, Zoghbi WA, Entman ML, Roberts R, Marian AJ. Simvastatin induces regression of cardiac hypertrophy and fibrosis and improves cardiac function in a transgenic rabbit model of human hypertrophic cardiomyopathy. Circulation. 2001;104:317–324. doi: 10.1161/hc2801.094031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsybouleva N, Zhang L, Chen S, Patel R, Lutucuta S, Nemoto S, DeFreitas G, Entman M, Carabello BA, Roberts R, Marian AJ. Aldosterone, through novel signaling proteins, is a fundamental molecular bridge between the genetic defect and the cardiac phenotype of hypertrophic cardiomyopathy. Circulation. 2004;109:1284–1291. doi: 10.1161/01.CIR.0000121426.43044.2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marian AJ, Senthil V, Chen SN, Lombardi R. Antifibrotic effects of antioxidant N-acetylcysteine in a mouse model of human hypertrophic cardiomyopathy mutation. Journal of the American College of Cardiology. 2006;47:827–834. doi: 10.1016/j.jacc.2005.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anan R, Greve G, Thierfelder L, Watkins H, McKenna WJ, Solomon S, Vecchio C, Shono H, Nakao S, Tanaka H. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy [see comments] J Clin Invest. 1994;93:280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Driest SL, Ackerman MJ, Ommen SR, Shakur R, Will ML, Nishimura RA, Tajik AJ, Gersh BJ. Prevalence and severity of “benign” mutations in the beta-myosin heavy chain, cardiac troponin T, and alpha-tropomyosin genes in hypertrophic cardiomyopathy. Circulation. 2002;106:3085–3090. doi: 10.1161/01.cir.0000042675.59901.14. [DOI] [PubMed] [Google Scholar]
- 60.Herman DS, Hovingh GK, Iartchouk O, Rehm HL, Kucherlapati R, Seidman JG, Seidman CE. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nature methods. 2009;6:507–510. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–350. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]