Abstract
Liquid culture assays revealed a previously unreported capacity for Mycobacterium bovis BCG, M. gordonae, and M. marinum to oxidize CO and for M. smegmatis to consume molecular hydrogen. M. bovis BCG, M. gordonae, M. smegmatis, and M. tuberculosis H37Ra oxidized CO at environmentally relevant concentrations (<50 ppm); H2 oxidation by M. gordonae and M. smegmatis also occurred at environmentally relevant concentrations (<10 ppm). CO was not consumed by M. avium or M. microti, although the latter appeared to possess CO dehydrogenase (CODH) genes based on PCR results with primers designed for the CODH large subunit, coxL. M. smegmatis and M. gordonae oxidized CO under suboxic (10 and 1% atmospheric oxygen) and anoxic conditions in the presence of nitrate; no oxidation occurred under anoxic conditions without nitrate. Similar results were obtained for H2 oxidation by M. smegmatis. Phylogenetic analyses of coxL PCR products indicated that mycobacterial sequences form a subclade distinct from that of other bacterial coxL, with limited differentiation among fast- and slow-growing strains.
Mycobacteria colonize soil and aquatic environments as well as various vertebrate hosts (for examples see references3, 6, 10, 20, and 47). They have been classified as slow or fast growing based on maximal growth rates in culture (53, 57). Although many strains require organic-rich or complex medium for optimal growth (53), such conditions seldom occur in situ. Thus, slow- and fast-growing strains express traits that promote survival during nutrient starvation, when preferred substrates are otherwise lacking, or when conditions lead to imbalanced growth (for examples see references 8, 9, 12, and 51). In addition, mycobacteria undoubtedly experience hypoxic and anoxic conditions with or without nutrient limitation, which also must elicit adaptive responses.
Numerous aspects of the molecular biology, physiology, and ecology of bacterial survival responses have been explored for many taxa, including Escherichia coli and other enteric bacteria (31, 46), Bacillus subtilis (11), Pseudomonas (25, 54), Rhizobium (56), and Vibrio (39, 54), among others. Phenomena such as sporulation, encystment, reductive cell division, metabolic downshifts, and induction of various genetic systems have been well documented (30).
Mycobacterial survival mechanisms and other adaptive responses have also been investigated extensively (8, 9, 12, 23, 24, 32, 38, 45, 51, 59). The nature of latency, a form of long-term survival by the causative agents of tuberculosis, has attracted particular attention, since some estimates suggest that Mycobacterium tuberculosis and related taxa infect as much as one third of the world's population (57). These infections involve persistence under nutrient-limited suboxic or anoxic conditions within granulomas for periods of up to decades (57). Reactivation of latent mycobacteria leads to forms of tuberculosis that can be difficult to treat (57). Although significant progress has been made in understanding the mechanisms of persistence by tubercle bacilli, much remains to be learned.
Recently, Park et al. (42) have shown that several fast- and slow-growing mycobacteria (e.g., M. smegmatis and M. tuberculosis H37Ra) grow lithotrophically by using high CO concentrations. Earlier, Cho et al. (4) documented very slow growth on 100-ppm CO by Acinetobacter sp. strain JC1, which has been described subsequently as a mycobacterium (42). In contrast, growth on high H2 and CO2 concentrations has been reported for only one mycobacterial strain to date, M. gordonae (41).
The capacity to use CO and H2 is intriguing, because H2 has been proposed as a universal bacterial survival or maintenance substrate (35). CO may play a similar role. Both gases are ubiquitous, occurring in the atmosphere at concentrations of about 0.5 to 0.6 ppm and 0.1 to 0.5 ppm, respectively (44). H2 and CO are also found within lung or respiratory gases at concentrations of about 5 to 50 ppm and 2 to 5 ppm, respectively (1, 2, 17, 19, 40, 43, 47, 58). The ability of mycobacteria to consume these gases at environmentally relevant concentrations was assessed by using stationary-phase cultures grown with various media and oxic, suboxic, and anoxic conditions. In addition, sequence data for a fragment of the large subunit of CO dehydrogenase (coxL) were obtained to assess phylogenetic relationships between mycobacteria and other CO oxidizers.
MATERIALS AND METHODS
Culture growth.
M. gordonae, M. marinum, M. microti, and M. tuberculosis H37Ra were obtained from the American Type Culture Collection. M. avium, M. bovis BCG, and M. smegmatis were obtained from M. Glickam (Memorial Sloan-Kettering Cancer Center). Cultures were grown in serum bottles (60 or 160 cm3) containing one of three media: Middlebrook 7H9 with oleic acid-albumin-dextrose-catalase (OADC) supplement (4.5 ml) (53), Middlebrook 7H9 without OADC supplement (4.5 ml), or a pyruvate (25 mM), yeast extract (0.05%), mineral salts medium (PYE) (9.5 ml) (29). After sealing with butyl rubber stoppers and crimp caps, media were inoculated with 0.5-ml volumes of suitable cell suspensions. Cultures were incubated at 37°C (30°C for M. marinum and M. smegmatis) with shaking at 125 rpm.
CO and H2 utilization.
M. smegmatis and M. gordonae were grown aerobically with PYE and Middlebrook 7H9 with OADC, respectively, as described above. Stationary-phase cells were harvested by centrifugation, washed with a 10 mM phosphate buffer (pH 6.5), and resuspended in buffer to an optical density (600 nm) of about 1.0. Ten milliliters of culture was transferred to 160-ml serum bottles that were subsequently sealed with butyl rubber stoppers and crimp caps. Bottle headspaces contained ambient air, nitrogen with either 0.2 or 2% oxygen (suboxic treatments), or nitrogen only (anaerobic treatments). Some suboxic and anaerobic treatments were amended with sodium nitrate to a final concentration of 10 mM. CO or hydrogen was added to bottle headspaces at final concentrations of up to 1%. Headspace subsamples were obtained at intervals by using a needle and syringe; CO concentrations were determined by gas chromatography as previously described (26-28).
CO uptake by M. bovis BCG, M. marinum, M. microti, and M. tuberculosis H37Ra was assessed by growing cultures in Middlebrook 7H9 with or without OADC supplement as described above. CO was added to culture headspaces at concentrations up to 1% 2 to 3 weeks postinoculation. Since detergents were omitted from the growth medium, cells clumped and adhered to the bottle walls, precluding optical measurements of cell density. CO uptake was monitored as described above.
PCR analyses.
Genomic DNA from 2- to 4-ml volumes of mycobacterial cultures was extracted by using a bead-beating kit (MoBio, Inc., Hayward, Calif.). Genomic extracts were used in a PCR analysis as described by King (29). Briefly, primers OMPf (5′-GGCGGCTT[C/T]GG[C/G]AA[C/G]AAGGT-3′) and O/Br (5′-[C/T]TCGA[T/C]GATCATCGG[A/G]TTGA-3′) were used in 50-μl reaction volumes with standard final concentrations for buffer, deoxynucleoside triphosphates, primers (0.1 μM each), and magnesium ion (1.5 mM). Culture extracts were used at final concentrations of approximately 1 to 10 ng. Positive controls included DNA extracts from a known CO oxidizer, Oligotropha carboxidovorans; negative controls included extracts from an organism for which no CO oxidation has been observed in vivo, Lutibacterium anuloederans (5). Reactants were denatured at 94°C in an Eppendorf Mastercycler thermocycler (Brinkmann Instruments, Inc., Westbury, N.Y.) for 3 min prior to addition of 0.5 U of MasterTaq DNA polymerase (Brinkmann Instruments, Inc.) at 80°C. Each amplification cycle consisted of a 45-s denaturation step at 94°C, a 60-s annealing step with the below temperature profile, and a 90-s extension at 72°C. Annealing temperatures varied according to a touchdown protocol, which was initiated at 62°C for two amplification cycles followed by 1°C stepwise decreases in annealing temperature (two amplification cycles for each step) to a final value of 58°C, which was used for 30 amplification cycles. A final 20-min extension at 72°C completed amplification. PCR products were visualized with ethidium bromide gel electrophoresis and were purified with MoBio PCR clean-up kits. Purified DNA was sequenced bidirectionally using the PCR primers with an ABI model 377 sequencer by the University of Maine Sequencing Facility.
Amino acid sequences were deduced from partial coxL sequences by using ExPAsy (http://us.espasy.org/tools/dna.html) and were aligned with other coxL sequences by using Clustal X, with manual adjustments as necessary. The resulting alignments were analyzed by using PAUP* 4.0b (Sinauer Associates, Inc., Sunderland, Mass.) to determine phylogenetic relationships among taxa. After excluding gapped positions, 367 residues were subjected to a neighbor-joining algorithm (1,000 bootstrap replicates) for tree construction.
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank under accession numbers AY307917 and AY333106 to AY333109.
RESULTS
CO and H2 utilization.
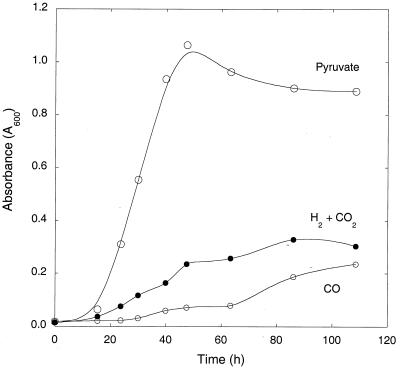
M. smegmatis grew with either 20% H2 plus 2% CO2 or 20% CO alone as carbon and energy sources (Fig. 1). Growth was slower for both of the former than for a mineral salts medium containing 25 mM pyruvate. Although the CO-based medium contained the greatest amount of substrate carbon (about 7 mmol versus 2.5 mmol for pyruvate and 0.8 mmol for H2 plus CO2), cell yields were least for CO, with intermediate yields for H2 plus CO2 based on absorbance data (Fig. 1).
FIG. 1.
Growth of M. smegmatis in mineral salts plus various carbon and energy sources: pyruvate, H2 plus CO2, and CO. Data are means of triplicates ± 1 standard error.
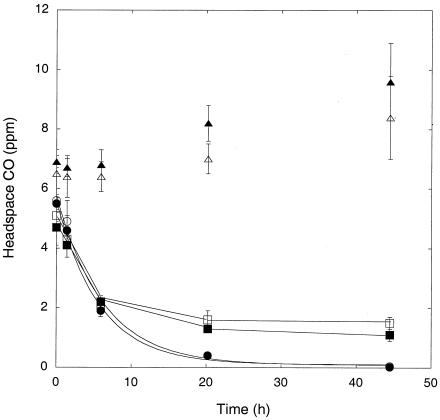
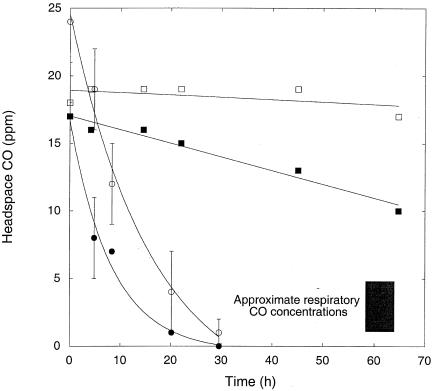
Washed, stationary-phase M. smegmatis incubated in a mineral salts buffer with oxic and suboxic headspaces or under anoxic conditions in the presence of nitrate consumed 5 ppm CO at similar initial rates (Fig. 2). CO was not consumed by cells incubated anaerobically without nitrate (Fig. 2). CO was depleted to concentrations of <0.05 ppm by cells incubated with air or 10% air; cells incubated with 1% air and under anoxic conditions with nitrate reduced CO to 1.5 ± 0.2 ppm and 1.2 ± 0.2 ppm, respectively, but these values did not differ significantly between treatments (P > 0.05). Similar results were obtained in a second trial, which showed that nitrate addition did not alter CO uptake by cells incubated with 1% air (data not shown). Nitrite concentrations of more than 1 mM were observed in anoxic plus nitrate treatments (data not shown).
FIG. 2.
CO uptake by M. smegmatis under various incubation conditions with killed controls: air (○), 10% air (•), 1% air (□), anoxic plus nitrate (▪), anoxic (Δ), autoclaved (▴). Data are means of triplicates ± 1 standard error.
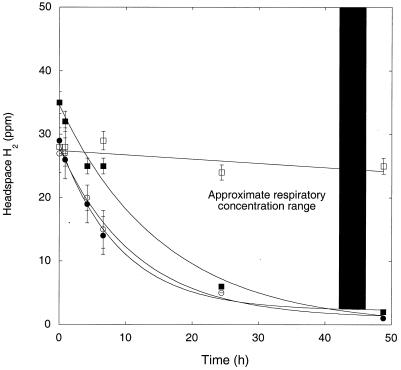
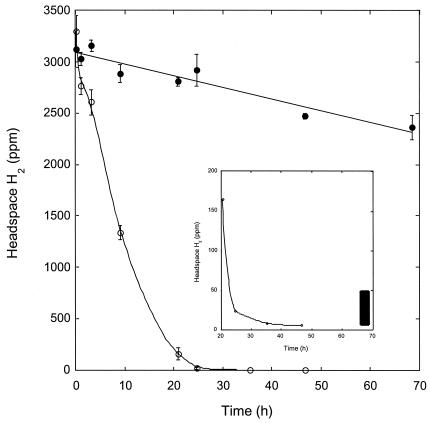
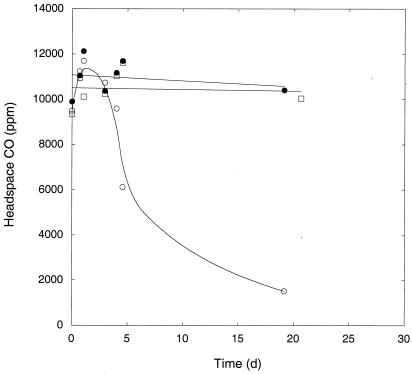
H2 was also consumed readily by washed, stationary-phase M. smegmatis incubated in a mineral salts buffer. Initial uptake rates were similar for cultures incubated under oxic and suboxic conditions and under anoxic conditions in the presence of nitrate (Fig. 3). A slight decline that did not differ statistically from zero (P > 0.05) occurred under during anoxic incubations without nitrate. H2 was reduced to levels of ≤1 ppm under oxic and suboxic conditions and to 2 ppm under anoxic conditions with nitrate. When H2 was added to stationary-phase cultures grown and maintained in PYE medium, uptake was substantially faster (>90%) than that for cultures grown and maintained in Middlebrook 7H9 medium with OADC supplement (Fig. 4). In addition, cells in PYE reduced H2 from initial concentrations of >3,500 ppm to <2 ppm in 48 h, while cells in 7H9 with OADC only reduced H2 from about 3,100 ppm to 2,400 ppm during the same interval.
FIG. 3.
H2 uptake by M. smegmatis under varied incubation conditions: air (○), 1% air (•), anoxic (□), anoxic plus nitrate (▪). Data are means of triplicates ± 1 standard error. The bar indicates approximate respiratory H2 concentration range.
FIG. 4.
H2 uptake by M. smegmatis grown with pyruvate-mineral salts (open symbols) or Middlebrook 7H9 with OADC (closed symbols). Insert shows uptake at low concentrations. Data are means of triplicates ± 1 standard error. The bar indicates approximate respiratory H2 concentration range.
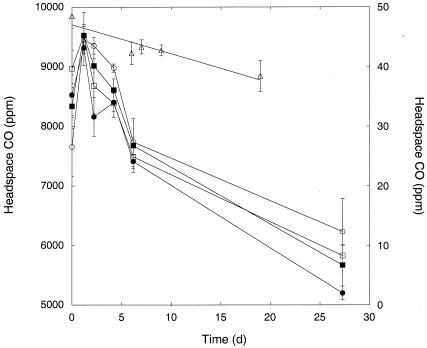
M. gordonae grown in Middlebrook 7H9 with OADC and transferred to a mineral salts buffer consumed CO present at initial headspace concentrations from 15 to 25 ppm (Fig. 5). Uptake rates were similar for oxic and suboxic conditions and were notably slower for anoxic conditions with nitrate; no uptake occurred for anoxic incubations without nitrate (Fig. 5). CO was not consumed by oxic M. gordonae grown and maintained in Middlebrook 7H9 with OADC (data not shown) or by similarly treated M. microti (Fig. 6). In contrast, after an initial lag, M. marinum grown and maintained in Middlebrook 7H9 with OADC consumed CO slowly over a period of about 3 weeks, reducing headspace concentrations from about 1% to <0.2%.
FIG. 5.
CO uptake by M. gordonae grown with pyruvate-mineral salts under various incubation conditions: air (○), 1% air (•), anoxic (□), anoxic plus nitrate (▪). Data are means of triplicates ± 1 standard error for oxic treatments, means of duplicates for anoxic treatments. The bar indicates approximate respiratory CO concentration range.
FIG. 6.
CO uptake by M. marinum (○) and M. microti (•) grown with Middlebrook 7H9 plus OADC. Data are means of duplicates. Values for sterile control are also indicated (□).
CO was also consumed by M. tuberculosis H37Ra and M. bovis BCG grown and maintained in Middlebrook 7H9 with or without OADC (Fig. 7). Uptake was slower than that for M. marinum, with a reduction in concentrations from about 1 to 0.6% over 4 weeks. M. tuberculosis H37Ra grown and maintained in Middlebrook 7H9 with OADC also consumed CO present at initial headspace concentrations of about 50 ppm; again, uptake was slow, with a 21% reduction in CO over about 2.5 weeks (Fig. 7).
FIG. 7.
CO uptake by M. tuberculosis H37Ra (squares) and M. bovis BCG (circles) under various growth conditions: Middlebrook 7H9, open symbols; Middlebrook 7H9 plus OADC, closed symbols; M. tuberculosis H37Ra grown with Middlebrook 7H9 and incubated with about 50 ppm CO (Δ). Data are means of triplicates ± 1 standard error.
PCR, sequence, and phylogenetic analyses.
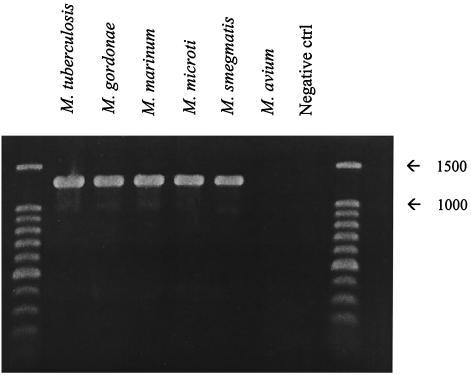
PCR products consistent with the size of the target coxL fragment (1,260 to 1,290 bp) were obtained from all mycobacterial extracts except those of M. avium (Fig. 8). Inferred amino acid sequences derived from the PCR products contained several motifs consistent with those of known coxL, including the active site, AYRCSFR, and binding sites for the molybdopterin cytosine dinucleotide cofactor (e.g., QGQ, HETT, and SRS). Mycobacterial amino acid sequences also contained several potentially unique motifs, including KTGWVYD, RLSVQTQ, and HGAGDLP, that appeared to be suitable targets for the design of genus-specific primers or probes.
FIG. 8.
PCR products from genomic extracts of selected mycobacteria using coxL primers. Expected fragment size was ca. 1,260 to 1,290 bp.
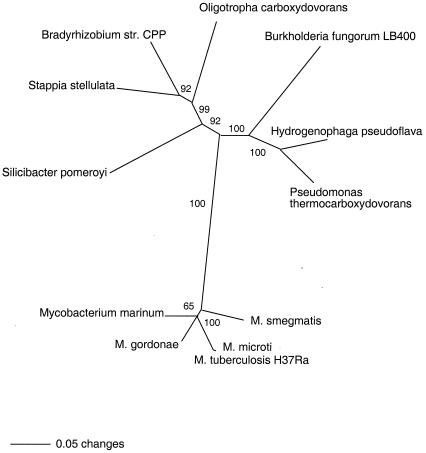
Phylogenetic analyses indicated that mycobacterial coxL sequences were related to but distinct from those of most known CO oxidizers (Fig. 9). Mycobacterial sequences clustered together and formed a subclade with high bootstrap support (100%) by using neighbor-joining (Fig. 9) and parsimony algorithms (data not shown). Sequences for M. tuberculosis H37Ra, M. tuberculosis H37Rv (data not shown), M. bovis (data not shown), and M. microti were essentially identical and were not phylogenetically resolvable. However, coxL from mammalian pathogens appeared distinct from that of other taxa.
FIG. 9.
Unrooted neighbor-joining tree from a phylogenetic analysis of an approximately 1,100-bp coxL sequence with 1,000 bootstrap replicates. Bootstrap support of >50% is indicated at nodes.
DISCUSSION
Results presented here and reported by others document growth by several mycobacteria, including M. smegmatis and M. tuberculosis H37Ra (Fig. 1) (42), on gas phase CO at concentrations up 50%. A mycobacterium originally described as Acinetobacter sp. strain JC1 also grows with CO concentrations as low as 0.01% (100 ppm), albeit very slowly (4). Similar observations described here and published by others demonstrate growth on high H2 concentrations by M. gordonae and M. smegmatis (reference 41 and Fig. 1, respectively).
However, growth with high substrate concentrations does not necessarily predict responses to environmentally relevant concentrations or conditions. For example, Acinetobacter sp. strain JC1 appears unable to grow by using ambient laboratory CO concentrations (4); indeed, its biomass decreases markedly over time when incubated with ambient CO. A comparable phenomenon has been described for methane-oxidizing bacteria, most of which cannot grow on or even consume ambient methane concentrations under typical culture conditions (18).
In this study, uptake assays (Fig. 2 to 7) revealed that oxic stationary-phase cultures of fast- and slow-growing mycobacteria utilize CO and H2 at concentrations observed in natural systems (i.e., <1 to 5 ppm for CO and H2 [21, 22, 26 to 28, 60]), including ambient air and human respiratory gases (<2 to 5 ppm for CO [1, 2, 19, 40, 44, 58] and <2 to 50 ppm for H2 [17, 43]). CO and H2 uptake also occur during incubations with suboxic conditions and anoxic conditions with nitrate (Fig. 2, 3, and 5). Results from the latter incubations suggest that ambient atmospheric CO and H2 may not support mycobacterial uptake but that activity occurs at higher levels found naturally in respiratory gases and some aquatic systems.
Although nitrate dissimilation has been observed for only a few CO oxidizers incubated with high substrate concentrations (15), results presented here suggest that nitrate-dependent anaerobic CO and H2 uptake at low concentrations represent potentially important adaptive responses. Clearly, anoxic conditions occur frequently in many natural systems (13). Moreover, anaerobic metabolism, and nitrate respiration in particular, appears to be especially significant for mycobacterial persistence in granuloma tissues. For example, Fritz et al. (14) have shown that M. bovis BCG deficient in nitrate respiration could not persist in mice used as a tuberculosis model. Accordingly, CO- and H2-dependent nitrate dissimilation may contribute to mycobacterial persistence during periods of anaerobiosis in natural systems and animal hosts.
CO and H2 uptake at low, naturally occurring concentrations may also contribute to mycobacterial maintenance metabolism under oxic conditions during periods when supplies of preferred organic substrates are limited. Substrate limitation typifies aquatic and soil systems (30, 34) from which mycobacteria are routinely isolated. Substrate deprivation also characterizes conditions that occur within granuloma tissues (8). Accordingly, mycobacteria possess several mechanisms for coping with substrate limitation, including the stringent response (45). CO and H2 uptake during stationary phase and decreased CO and H2 uptake by M. smegmatis cultures grown with a relatively rich medium (e.g., see Fig. 4) are consistent with known responses to substrate availability and represent a potentially important but previously unrecognized adaptive response.
Due to certain similarities with virulent M. tuberculosis (e.g., strain H37Rv), M. bovis BCG, M. marinum, M. smegmatis, and M. tuberculosis H37Ra have been proposed as model organisms for understanding aspects of latency and other phenomena (38, 49, 52, 55, 59). For example, responses of M. tuberculosis H37Rv to anaerobiosis and substrate deprivation are similar to those of M. bovis BCG and M. smegmatis, all of which share a number of genes involved in adaptation to stress (38).
Several of the strains in this study may also serve as model organisms for understanding CO and H2 metabolism. Although M. bovis BCG and M. tuberculosis H37Ra consume CO more slowly than other strains (for example, compare Fig. 2 to Fig. 7), which is consistent with relatively slow growth on CO as reported for M. tuberculosis H37Ra (42), both consume high and environmentally relevant CO concentrations when grown with Middlebrook 7H9 containing or lacking OADC supplement. M. marinum uses CO similarly, albeit with faster uptake rates (Fig. 6), while in contrast, M. gordonae and M. smegmatis CO utilization appears more sensitive to culture conditions. Collectively these organisms encompass a range of responses to CO, H2, and culture conditions that provide a framework for understanding activity by pathogenic M. tuberculosis.
Neither M. avium, which causes a tuberculosis-like disease in immunocompromised individuals (6), nor M. microti, which causes a tuberculosis-like disease in rodents and humans (37), appear to oxidize CO under the conditions used in this study (Fig. 6; results for M. avium not shown). Results for M. avium are consistent with the absence of CODH genes from itsgenome (http://ncbi.nlm.nig.gov/cgi-bin/Entrez/genom_table_cgi) and the lack of a coxL product from PCR of genomic extracts (Fig. 8). Results for M. microti are somewhat enigmatic, since it yields a coxL PCR product (Fig. 8), the sequence of which is virtually identical to that of the coxL PCR product from M. tuberculosis H37Ra (Fig. 9). Assuming M. microti contains a complete cox operon, lack of CO uptake indicates that its controls of expression differ from controls for other mycobacteria. Further analysis of the differences between M. microti and other mycobacteria may prove useful for understanding the diversity of mycobacterial responses to substrate limitation and the environmental factors that govern those responses.
At present the range of physiological responses to CO and the taxonomic diversity of mycobacterial CO oxidizers are unknown. Evidence from genome sequences (7, 16) and enzymatic assays (42) indicate that M. bovis and M. tuberculosis do not contain ribulose-1,5-bisphosphate carboxylase (rubisco), a key enzyme involved in lithotrophic CO2 fixation. This may account in part for differences in CO oxidation (e.g., compare Fig. 2 to Fig. 7) and growth rates (42). However, it is not yet clear whether other slow-growing mycobacteria also lack rubisco or differ in other respects from fast-growing mycobacteria.
Results presented here suggest that CO utilization occurs commonly among slow-growing mycobacteria, with some exceptions (e.g., M. avium and perhaps M. microti). A recent report suggests that many fast-growing strains also oxidize CO (42). Collectively, these taxa form a subgroup distinct from proteobacterial CO oxidizers, which in turn form groups generally consistent with 16S phylogeny (Fig. 9). Whether mycobacterial CO oxidizers as a group can be differentiated further from their proteobacterial counterparts based on physiological traits remains to be seen. Genomic evidence suggests that the cox operon in M. tuberculosis is organized similarly to that of a well-known α-proteobacterial carboxydotroph, Oligotropha carboxidovorans (33, 48, 50), but this does not imply physiological similarities. For example, O. carboxidovorans fixes CO2 lithotrophically by using rubisco (36), an enzyme absent from M. tuberculosis. Interestingly, rubisco is also absent from at least one α-proteobacterial CO oxidizer, Silicibacter pomeroyi (M. A. Moran, personal communication).
In summary, this is the first report of CO utilization by slow- and fast-growing mycobacteria at ambient or near-ambient concentrations and is the first report of mycobacterial CO oxidation under suboxic and anoxic conditions. It is also the first report of H2 utilization by M. smegmatis and of CO utilization by M. bovis BCG and M. marinum, both of which serve as model organisms for aspects of tuberculosis. The results establish CO and H2 as substrates that may contribute to mycobacterial maintenance metabolism during periods of limited heterotrophic substrate availability.
Acknowledgments
I thank M. Glickman (Memorial Sloan-Kettering Cancer Center) for gifts of M. bovis BCG, M. avium, and M. smegmatis. I also thank M. Glickman and C. Nathan (Cornell University) for helpful input.
This study was supported in part by the C. S. Darling Endowment and NSF LExEn grant 00-85495.
Contribution 385 from the Darling Marine Center.
REFERENCES
- 1.Andersson, J. A., R. Uddman and L.-O. Cardell. 2000. Carbon monoxide is endogenously produced in the human nose and paranasal sinuses. J. Allergy Clin. Immunol. 105:269-273. [DOI] [PubMed] [Google Scholar]
- 2.Biernacki, W. A., S. A. Kharitonov, and P. J. Barnes. 2001. Exhaled carbon monoxide in patients with lower respiratory tract infection. Respir. Med. 95:1003-1005. [DOI] [PubMed] [Google Scholar]
- 3.Boldrin, B., A. Tiehm, and C. Fritzsche. 1993. Degradation of phenanthrene, fluorine, fluoranthene, and pyrene by a Mycobacterium sp. Appl. Environ. Microbiol. 59:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, J. W., H. S. Yim, and Y. M. Kim. 1985. Acinetobacter isolates growing with carbon monoxide. Korean J. Microbiol. 23:1-8. [Google Scholar]
- 5.Chung, W.-K., and G. M. King. 2001. Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensis sp. nov. Appl. Environ. Microbiol. 67:5585-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Conner, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Couture, M., S.-R. Yeh, B. A. Wittenberg, J. B. Wittenberg, Y. Ouellet, D. L. Rosseau, and M. Guertin. 1999. A cooperative oxygen-binding hemoglobin from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 96:11223-11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham, A. F., and C. L. Spreadbury. 1998. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J. Bacteriol. 180:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dailloux, M., C. Laurain, R. Weber, and P. Hartemann. 1999. Water and nontuberculous mycobacteria. Water Res. 33:2219-2228. [Google Scholar]
- 11.Dauner, M., T. Storni, and U. Sauer. 2001. Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 183:7308-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick, T., B. H. Lee, and B. Murugasu-Oei. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163:159-164. [DOI] [PubMed] [Google Scholar]
- 13.Fenchel, T., G. M. King, and T. H. Blackburn. 1998. Bacterial biogeochemistry: the ecophysiology of mineral cycling. Academic Press, New York, N.Y.
- 14.Fritz, C., S. Maass, A. Kreft, and F.-C. Bange. 2002. Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect. Immun. 70:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frunzke, K., and O. Meyer. 1990. Nitrate respiration, denitrification and utilization of nitrogen sources by aerobic carbon monoxide-oxidizing bacteria. Arch. Microbiol. 154:168-174. [Google Scholar]
- 16.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallfrisch, J., and K. M. Behall. 1999. Breath hydrogen and methane responses of men and women to breads made with white flour or whole wheat flours of different particle sizes. J. Am. Coll. Nutr. 18:296-302. [DOI] [PubMed] [Google Scholar]
- 18.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath, I., L. E. Donnelly, A. Kiss, P. Paredi, S. A. Kharitonov, and P. J. Barnes. 1998. Elevated levels of carbon monoxide are associated with an increased expression of heme oxygenase-1 in airway macrophages in asthma: a new marker of chronic inflammation. Thorax 53:668-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iivanainen, E., T. Sallantaus, M. L. Katila, and P. J. Martikainen. 1999. Mycobacteria in runoff waters from natural and drained peatlands. J. Environ. Qual. 28:1226-1234. [Google Scholar]
- 21.Johnson, J. E., and T. S. Bates. 1996. Sources and sinks of carbon monoxide in the mixed layer of the tropical South Pacific Ocean. Global Biogeochem. Cyc. 10:347-359. [Google Scholar]
- 22.Jones, R. D. 1991. Carbon monoxide and methane distribution and consumption in the photic zone of the Sargasso Sea. Deep-Sea Res. 38:625-635. [Google Scholar]
- 23.Keer, J., M. J. Smeulders, and H. D. Williams. 2001. A purF mutant of Mycobacterium smegmatis has impaired survival during oxygen-starved stationary phase. Microbiology 147:473-481. [DOI] [PubMed] [Google Scholar]
- 24.Keer, J., M. J. Smeulders, K. M. Gray, and H. D. Williams. 2000. Mutants of Mycobacterium smegmatis impaired in stationary-phase survival. Microbiology 146:2209-2217. [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y., L. S. Watrud, and A. Matin. 1995. A carbon starvation survival gene of Pseudomonas putida is regulated by σ54. J. Bacteriol. 177:1850-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, G. M. 1999. Attributes of atmospheric carbon monoxide oxidation by Maine forest soils. Appl. Environ. Microbiol. 65:5257-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, G. M. 2000. Impacts of land use on atmospheric carbon monoxide consumption by soils. Glob. Biogeochem. Cyc. 14:1161-1172. [Google Scholar]
- 28.King, G. M. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69:4067-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King, G. M. 2003. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69:7257-7265. [DOI] [PMC free article] [PubMed]
- 30.Kjelleberg, S. (ed.) 1993. Starvation in bacteria. Plenum Press, New York, N.Y.
- 31.Li, C., Y. P. Tao, and L. D. Simon. 2000. Expression of different-size transcripts from clpP-clpX operon of Escherichia coli during carbon deprivation. J. Bacteriol. 182:6630-6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim, A., M. Eleuterio, B. Hutter, B. Murugasu-Oei, and T. Dick. 1999. Oxygen depletion-induced dormancy in Mycobacterium bovis BCG. J. Bacteriol. 181:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer, O., K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck, and M. Kraut. 1990. Utilization of carbon monoxide by aerobes: recent advances. FEMS Microbiol. Rev. 87:253-260. [Google Scholar]
- 34.Morita, R. Y. 1993. Bioavailability of energy and the starvation state, p. 1-23. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 35.Morita, R. Y. 2000. Is H2 the universal energy source for long-term survival? Microb. Ecol. 38:307-320. [DOI] [PubMed] [Google Scholar]
- 36.Morsdorf, G., K. Frunzke, D. Gadkari, and O. Meyer. 1992. Microbial growth on carbon monoxide. Biodegredation 3:61-82. [Google Scholar]
- 37.Niemann, S., E. Richter, H. Dalügge-Tamm, H. Schlesinger, D. Graupner, B. Königstein, G. Gurath, U. Greinert, and S. Rüsch-Gerdes. 2000. Two cases of Mycobacterium microti-derived tuberculosis in HIV-negative immunocompetent patients. Emerg. Infect. Dis. 6:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Toole, R., M. J. Smeulders, M. C. Blokpoel, E. J. Kay, K. Lougheed, and H. W. Williams. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 185:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Östling, J., L. Holmquist, and S. Kjelleberg. 1996. Global analysis of the carbon starvation response of a marine Vibrio species with disruptions in genes homologous to relA and spoT. J. Bacteriol. 178:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paredi, P., M. J. Leckie, I. Horvath, L. Allegra, S. A. Kharitonov, and P. J. Barnes. 1999. Changes in exhaled carbon monoxide and nitric oxide levels following allergen challenge in patients with asthma. Eur. Respir. J. 13:48-52. [DOI] [PubMed] [Google Scholar]
- 41.Park, S. S., and B. T. DeCicco. 1974. Autotrophic growth with hydrogen of Mycobacterium gordonae and scotochromogenic Mycobacterium. Int. J. Syst. Bacteriol. 24:338-345. [Google Scholar]
- 42.Park, S. W., E. H. Hwang, H. Park, J. A. Kim, J. Heo, K. H. Lee, T. Song, E. Kim, Y. T. Ro, S. W. Kim, and Y. M. Kim. 2003. Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 185:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peukhuri, K., T. Poussa, and R. Korpela. 1998. Comparison of a portable breath hydrogen analyzer (Micro H2) with a Quintron MicroLyzer in measuring lactose maldigestion, and the evaluation of a Micro H2 for diagnosing hypolactasia. Scand. J. Clin. Lab. Investig. 58:217-224. [DOI] [PubMed] [Google Scholar]
- 44.Prather, M. J., R. Derwent, D. Ehhalt, P. Fraser, E. Sanhueza, and X. Zhou. 1995. Other trace gases and atmospheric chemistry, p. 73-126. In J. T. Houghton, L. G. Meira Filho, J. Bruce, H. Lee, B. A. Callender, E. Haites, N. Harris, and K. Maskell (ed.), Climate change 1994. Cambridge University Press, Cambridge, United Kingdom.
- 45.Primm, T. P., S. J. Andersen, V. Mizrahl, D. Avarbock, H. Rubin, and C. E. Barry III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182:4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross, B. C., P. D. Johnson, F. Oppedisano, L. Marino, A. Sievers, T. Stinear, J. A. Hayman, M. G. Veitch, and R. M. Robins-Browne. 1997. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl. Environ. Microbiol. 63:4135-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santiago, B., U. Schübel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115-124. [DOI] [PubMed] [Google Scholar]
- 49.Schafer, M. P. 1999. Detection and characterization of airborne Mycobacterium tuberculosis H37Ra particles, a surrogate for airborne pathogenic M. tuberculosis. Aerosol Sci. Technol. 30:161-173. [Google Scholar]
- 50.Schübel, U., M. Kraut, G. Mörsdorf, and O. Meyer. 1995. Molecular characterization of the gene cluster coxMSL encoding the molybdenum-containing carbon monoxide dehydrogenase of Oligotropha carboxydovorans. J. Bacteriol. 177:2197-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomon, J. M., G. S. Leung, and R. R. Isberg. 2003. Intracellular replication of Mycobacterium marinum within Dictyhostelium discoideum: efficient replication in the absence of host corin. Infect. Immun. 71:3578-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sommers, H. M., and R. C. Good. 1985. Mycobacterium, p. 216-248. In E. H. Lennette, A. Balows, W. J. Hausler, Jr., and H. J. Shadomy (ed.), Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, D.C.
- 54.Srinivasan, S., J. Östling, T. Charlton, R. de Nys, K. Takayama, and S. Kjelleberg. 1998. Extracellular signal molecule(s) involved in the carbon starvation response of marine Vibrio sp. strain S14. J. Bacteriol. 180:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stinear, T. P., G. A. Jenkin, P. D. R. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorne, S. H., and H. D. Williams. 1997. Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 179:6894-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Volk, W. A., B. M. Gebhardt, M.-L. Hammarskjold, and R. J. Kadner. 1996. Essentials of medical microbiology, 5th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 58.Vreman, H. J., J. J. Mahoney, and D. K. Stevenson. 1993. Electrochemical measurement of carbon monoxide in breath: interference by hydrogen. Atmos. Environ. 27A:2193-2198. [Google Scholar]
- 59.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zafiriou, O. C., S. S. Andrews, and W. Wang. 2003. Concordant estimates of oceanic carbon monoxide source and sink processes in the Pacific yield a balanced global “blue-water” CO budget. Glob. Biogeochem. Cycles 17:1015. [Google Scholar]