Abstract
The olfactory-driven blood-feeding behaviour of female Aedes aegypti mosquitoes is the primary transmission mechanism by which the arboviruses causing dengue and yellow fevers affect over 40 million individuals worldwide. Bioinformatics analysis has been used to identify 131 putative odourant receptors from the A. aegypti genome that are likely to function in chemosensory perception in this mosquito. Comparison with the Anopheles gambiae olfactory subgenome demonstrates significant divergence of the odourant receptors that reflects a high degree of evolutionary activity potentially resulting from their critical roles during the mosquito life cycle. Expression analyses in the larval and adult olfactory chemosensory organs reveal that the ratio of odourant receptors to antennal glomeruli is not necessarily one to one in mosquitoes.
Keywords: odourant receptor, olfaction, genome, Aedes aegypti, Anopheles gambiae
Introduction
Olfaction is the chemosensory modality dedicated to the detection of small airborne chemicals called odourants, which is of fundamental importance in the life cycle and life histories of insects. Aedes aegypti is responsible for the transmission of arboviral pathogens associated with dengue haemorrhagic fever, yellow fever and Rift Valley encephalitis. Olfactory signal transduction plays a critical role in female behaviours such as the search for oviposition sites or host selection and thus directly impacts their overall ability (vectorial capacity) to transmit pathogenic agents (Zwiebel & Takken, 2004).
Insect olfactory systems have been the subject of intense study in models such as Drosophila melanogaster as well as a range of both medically and economically important systems. At the molecular level, olfactory signal transduction is mediated by a complex and still enigmatic suite of proteins that are hypothesized to act through G-protein coupled receptor (GPCR) mediated pathways (van Der Goes van Naters & Carlson, 2006). These include a varied set of small molecular weight odourant binding proteins, olfactory arrestins, G-proteins, biotransformation enzymes as well as large and highly diverse families of putative odourant receptors (Ors) (reviewed in Rutzler & Zwiebel, 2005). In Drosophila, the two main insect olfactory appendages: the antennae and maxillary palps are covered by various morphological and functional classes of olfactory sensilla housing up to four primary olfactory receptor neurones (ORNs) (Shanbhag et al., 1999; Stocker, 1994). First order ORNs project to the antennal lobe (AL) where they synapse locally to interneurones and projection neurones in morphological structures called glomeruli (Shanbhag et al., 1999). With one notable exception (Goldman et al., 2005), the olfactory organization in the adult and the larva is linear: individual ORNs express one type of OR while their axons stereotypically converge to a unique glomerulus, thus giving rise to the ‘one neurone–one receptor-one receptor–one glomerulus’ paradigm (Couto et al., 2005; Fishilevich & Vosshall, 2005).
The identification of the first vertebrate Ors (Buck & Axel, 1991) led to a major effort by several groups interested in identifying insect Or genes. Nearly a decade later, the release of the D. melanogaster genome facilitated the identification of a family of Ors from D. melanogaster (Clyne et al., 1999b; Gao & Chess, 1999; Vosshall et al., 1999). The completion of the genome of the malaria mosquito, An. gambiae (Holt et al., 2002) led to the description of a second complete insect Or gene family and allowed the first comparative genomic analysis between two related insects (Fox et al., 2001; Hill et al., 2002). Multiple Ors have now been identified in species from at least four other insect orders, including Diptera, Lepidoptera, Hymenoptera and Coleoptera (Krieger et al., 2003; Robertson & Wanner, 2006; Wanner et al., 2007; Hallem et al., 2006) where a significant body of work has described insect Or genes, their patterns of expression, functions and implications for odour coding (Robertson et al., 2003; Jacquin-Joly & Merlin, 2004; Hallem et al., 2004a,b; Rutzler & Zwiebel, 2005; Ache & Young, 2005; Dahanukar et al., 2005; Jefferis, 2005). Additional studies have characterized one Or that is highly conserved across insect orders, widely coexpressed with other more diverse Ors and which is required for their function (Krieger et al., 2003; Pitts et al., 2004; Larsson et al., 2004; Jones et al., 2005; Xia & Zwiebel, 2006; Benton et al., 2006). It is apparent that with several additional insect genome projects nearly complete or about to be initiated, the future of comparative genomics is certain to yield new and exciting information.
Here we report the manual annotation and characterization of the Or gene family in the genome of Ae. aegypti (Liverpool strain). As a result of their role in olfactory signal transduction cascades, Ae. aegypti odourant receptors (AaOrs) exert significant effects throughout the mosquito life cycle and, accordingly, are expected to have a dramatic impact on the potential for the mosquito to transmit diseases.
Results
The Ae. aegypti odourant receptor family comprises 131 members
We have identified a total of 131 candidate AaOr genes in the genome of Ae. aegypti. Of these, 100 genes encode putative, complete functional proteins, 10 are incompletely annotated genes, and 21 are apparent pseudogenes (Table S1). While we believe these represent the complete Or repertoire in the Ae. aegypti genome, we acknowledge this annotation is dependent on the maturity of the current genome assembly, the inevitable refinement of which may ultimately affect changes in the complexity of the AaOr gene family.
The C-terminus is the most conserved region among AaOrs and, in most cases, includes an SYT or SYS motif near the extreme C-terminus of the conceptual translation, a feature that is conserved in both An. gambiae and D. melanogaster (Hill et al., 2002; Robertson et al., 2003; Severson et al., 2004). The complete list of AaOrs, including their predicted amino acid sequences, is available as supporting online material (Table S1). As is the hallmark of all GPCRs, all of the AaOR peptides contain multiple trans-membrane (TM) regions based on TMPred predictions (Hofmann & Stoffel, 1993). Chemosensory receptors, comprising the AaOrs and a large family of 88 gustatory receptors encoded by 79 Gr genes (Kent L.B. and Robertson H.M., unpublished data), belong to the most numerous of the GPCR classes in the Ae. aegypti genome (Hill, personal communication).
It was not possible to annotate the full-length sequence of all AaOrs, especially in 5′- and 3′-regions, and a number of incompletely annotated genes were identified. Presumably, this was due in some cases to lack of sequence conservation among insect Ors. In addition, the presence of large introns and/or incomplete assembly of genomic sequences may have complicated exon identification. AaOrs126, 127, 130 and 131 are very short incompletely annotated genes, each encoding fewer than 100 amino acids with one or two recognizable exons (Table S1). These genes may represent Ors with extremely large introns or exons that are difficult to identify for the reasons mentioned previously. Alternatively, they may be pseudogenes or may not be Ors at all, although in a couple of instances they are located near other AaOrs on the same supercontig with which they share high similarity. In any case, these short incomplete protein sequences were not used in phylogenetic analyses. Because no complete physical/cytological map is yet available in Ae. aegypti, AaOr names were assigned, when possible, based on phylogenetic relationships (Table S1, Figs 1–3).
Figure 1.
Phylogenetic relationships of the Ae. aegypti and An. gambiae odourant receptor (Or) families. Corrected distance tree generated using paup* v4 using distances corrected in TreePuzzle v5 using a maximum-likelihood model and the BLOSUM62 amino acid matrix [(Hill et al., 2002; Robertson et al., 2003; Robertson & Wanner, 2006) for methods]. The tree is rooted using the AaGPRor7and AgGPRor7 pair based on the basal position of their highly conserved ortholog DmOr83b in the Orfamily in a phylogenetic tree of the entire superfamily (Robertson et al., 2003). Bootstrap support from 1000 replications of neighbour-joining with uncorrected distances is shown on the relevant branch points. Ae. aegypti (bold branches) and An. gambiae (thin branches) gene expansions are indicated by vertical black bars. Brackets indicate apparent orthology between Ae. aegypti and An. gambiae OR proteins. Protein names are abbreviated to AaOr and AgOr, with suffixes: (P) – pseudogene; (C) (N) – lacking sequence at C- or N-terminus, respectively. Introns are named according to Fig. S3, and their presence within a lineage is indicated by bold letters. Inferred intron losses are indicated by italicized letters on the relevant branches.
Figure 3.
Intron analysis of Ae. aegypti and An. gambiae Or genes. (A) The intron positions and phases of Ae. aegypti and An. gambiae (AaOrs and AgOrs) are shown relative to a scale of the average receptor size in amino acids. The ancestral intron locations o, p, q and s (arrowheads) are shared by the two mosquito subfamilies, while idiosyncratic introns (small letters with asterisk and prime symbols) are species specific. The total number of Ors harbouring each conserved intron is indicated in (B).
Pseudogenes are rare occurrences in both the D. melanogaster and An. gambiae genomes (Krzywinski et al., 2001). We identified 21 potential Or pseudogenes (Table S1), based on the occurrence of premature stop codons, mutated start codons, or small nucleotide insertion/ deletion (indels) in their open-reading frames (Table S1). Thirteen of these genes, AaOrs 22, 29, 38, 51, 54, 57, 64, 73, 77, 82, 83, 112 and 116, were detected as reverse transcription–polymerase chain reaction (RT–PCR) products in antennal tissue (Fig. 4). Sequencing the AaOr38 RT–PCR product revealed a CAG codon in the Costa Rica strain where an in-frame TAG stop-codon exists in the Liverpool strain, thus demonstrating that AaOr38 is likely to be an intact gene in the Costa Rica strain (data not shown). A similar result was observed with AaOr29 for which the Costa Rica strain contains TGG instead of a TGA stop codon at position 31 in the Liverpool strain. Further investigation is required to document whether the other potential AaOr pseudogenes show similar polymorphisms. We have also observed regular occurrences of single nucleotide polymorphisms (SNPs) in other AaOr RT–PCR amplicons in this study (data not shown). The existence of SNPs between and within strains of species has been recognized in DmOrs (Berger et al., 2001), AgOrs (Hill et al., 2002), and C. elegans chemoreceptors (Stewart et al., 2005), so their presence here is not without precedent.
Figure 4.
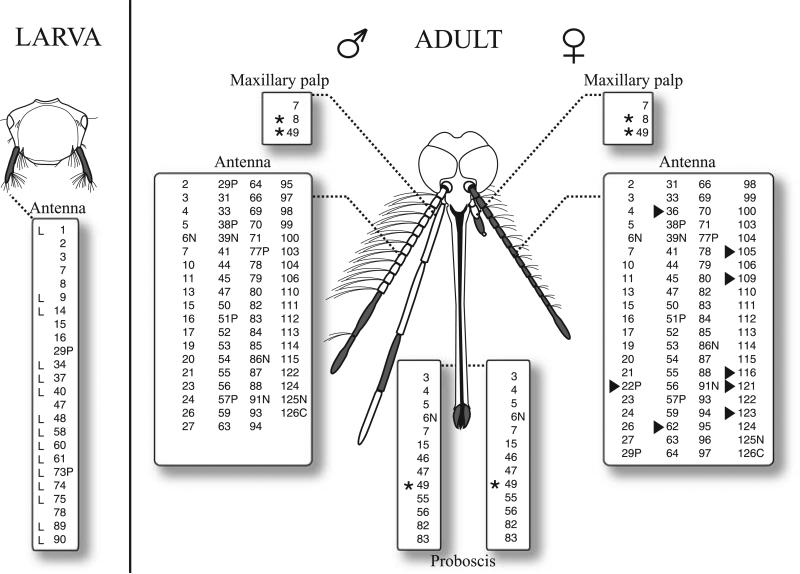
Peripheral topography of the Or transcriptome in adult and larval olfactory organs of Ae. aegypti. Schematic representation of a larval and chimeric adult Ae. aegypti heads. Left and right sides represent male and female olfactory appendages, respectively. Non-antennal expression is indicated with an asterisk. AaOr8 was detected only in the maxillary palp. Female-specific and larval-specific Ors are indicated by an arrowhead and ‘L’, respectively. Olfactory sensilla bearing segments are in shaded in grey.
AaOrs are highly divergent
Phylogenetic analysis reveals several species-specific Or expansions in Ae. aegypti and An. gambiae (Fig. 1). The AaOr family is significantly expanded relative to the Or families of An. gambiae (79 AgORs) (Hill et al., 2002) and D. melanogaster (62 DmORs) (Robertson, 2001) and is dominated by a large set of 40 AaOrs (86–125) with no closely related An. gambiae Ors (Fig. 1). The expansion and diversification of mosquito Ors reflects the apparent rapid evolution of the insect chemosensory receptors (Robertson et al., 2003). Or diversity between these two mosquitoes indicates that many species-specific differences in sensory perception have arisen in the 140–200 million years (Myr) since the Anophelinae/Culicinae divergence (Krzywinski et al., 2001). Furthermore, short branch lengths observed between many Ors within one species (Fig. 1) suggest numerous recent gene duplication events. This raises the possibility that any behaviours associated with these novel receptors have undergone recent or rapid shifts both between An. gambiae and Ae. aegypti, and probably also within these mosquito subfamilies. The diversification of the Or family is more pronounced in Ae. aegypti, suggestive of the increased potential of this mosquito to exploit a wider array of ecological niches. While it is tempting to speculate that the life history of Ae. aegypti includes a need for greater peripheral chemosensory capacity, further analyses that include AaOr deorphanization will be necessary to understand the significance of this large increase in Or complexity.
Ae. aegypti and An. gambiae share a subset of conserved Ors
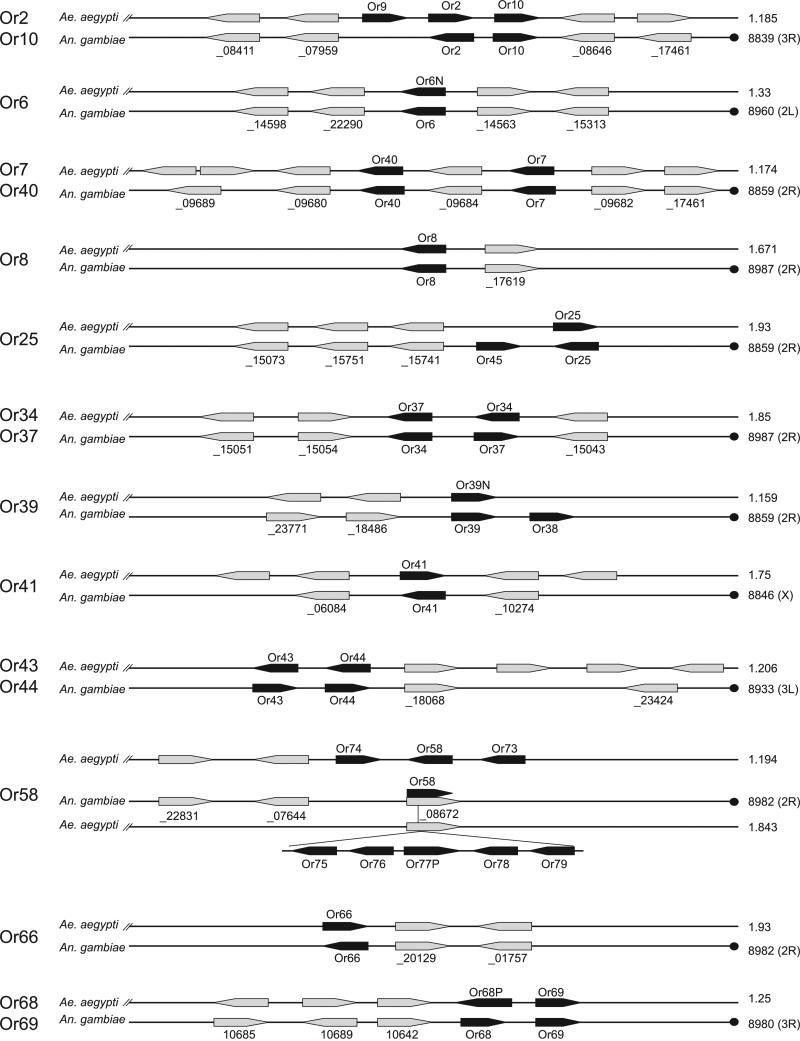
As is the case for other insect Ors, AaOrs display a high degree of divergence (Robertson et al., 2003; Clyne et al., 1999a). Indeed, the majority of AaOrs share less than 20% identity at the amino acid level with each other as well as with Or peptides from An. gambiae and D. melanogaster (data not shown). Based on phylogenetic analysis, we identified 18 Aedes/Anopheles orthologous subgroups including the highly conserved Or7 gene (Figs 1 and 2). Along with the nonconventional Drosophila DmOr83b gene (Gao & Chess, 1999; Larsson et al., 2004; Pitts et al., 2004), these genes are clear orthologs based on conserved sequence homology (Melo et al., 2004), microsynteny (Fig. 2) and, in the case of Drosophila, an essential requirement for olfactory signal transduction (Jones et al., 2005; Benton et al., 2006). The evolutionary relationships to other apparent orthologs are more complex due to lower sequence conservation and sometimes to the presence of neighbouring paralogs (Fig. 2). Another closely related group is the OR2/OR10 paralogous clade; they include AaOR10/AgOR10, AaOR9/AgOR10 and AaOR2/AgOR2, which share 71.5%, 65% and 70.5% identity, respectively. The closest Drosophila relative of AaOR2/AgOR2 is DmOR43a (< 40% identity; Hill et al., 2002); however, these receptors do not appear to share odourant ligand specificity (Wetzel et al., 2001; Hallem et al., 2004b). The remaining 20 Ae. aegypti-An. gambiae Or orthologs have no counterparts in D. melanogaster, raising the interesting possibility that these genes may be associated with conserved functions between Ae. aegypti and An. gambiae. As such, they may offer important insight into comparative studies of mosquito olfaction. Finally, it should be noted that there is potentially a second copy of AaOr7 in the Ae. aegypti genome located on supercontig 1.2048 (data not shown), which would be unique among insects to date. However, inasmuch as this very small (only 15 kb in length) supercontig only contains a partial length AaOr7 sequence and may therefore represent an anomalous sequence assembly.
Figure 2.
Analysis of microsynteny between orthologous and paralogous Ae. aegypti and An. gambiae Or genes. Or genes (black arrows) are abbreviated to AaOr and AgOr. Interspecific neighbouring homologs that share at least 50% amino acid identity are shaded in grey. The Anopheles neighbouring genes are labelled by the last five digits of their ENSANG number preceded by an underscore character (e.g. ENSANG00000008672 abbreviated to _08672). Gene orientation is indicated by arrows. Ae. aegypti supercontig number, An. gambiae scaffold number, and chromosome number are on the right side. Centromeres are indicated by a dot.
AaOrs are clustered in the genome
Although the AaOrs have not been mapped cytologically to the Ae. aegypti chromosomes, AaOr localization on super-contigs revealed that 12 AaOrs are paired in tandem arrays, seven are found as triplets and many others as larger clusters of up to 11 genes (Table S1). This is similar to the organization of the AgOrs (Hill et al., 2002) but in contrast to that of the DmOrs, which are typically dispersed as single genes throughout the genome and at most are found in triplets (Clyne et al., 1999a; Robertson et al., 2003). Overall, 18 conserved AaOrs share microsynteny with their AgOr counterparts and neighbouring genes further supporting the evolutionary relationships of these receptors (Fig. 2). Taken together, genomic clustering, conserved gene orientation, and sequence similarly provide strong evidence that these groups have arisen via gene duplication.
Ae. aegypti introns are on average larger than those of An. gambiae (Nene et al., 2007). and, not surprisingly, the gene architecture of AaOrs differs markedly from that of AgOrs. In general, AaOrs contain introns that are significantly larger, where the average is 544 bp, than those observed in An. gambiae where the average is 119 bp. The largest AaOr intron (≈ 30 kb) is found in AaOr7 and is nearly 10 times larger than the largest intron in AgOr7. We identified 18 conserved intron positions between Ae. aegypti and An. gambiae (Fig. 3). Of these 18, introns o, p, q and s are present at the base of the Or lineage and are also shared with the Drosophila Ors (Fig. 3). Intron loss is a dominant feature of the recent evolution of eukaryotes (Roy & Penny, 2006), and this phenomenon is reflected in various Or lineages where numerous instances of independent intron losses have occurred (Fig. 1). Furthermore, there are five and nine idiosyncratic intron locations specific to Ae. aegypti and An. gambiae Ors, respectively.
AaOrs are expressed during the aquatic and terrestrial stages
To validate our gene models and to explore the spatial localization and distribution of the AaOrs, we used RT–PCR to determine the expression profiles of 123 AaOrs across the olfactory apparatus of Ae. aegypti. Like most insects, the antennae of Ae. aegypti and An. gambiae constitute the principal olfactory appendages, as reflected by the sheer number of olfactory sensilla and wide olfactory response profiles (Pappenberger et al., 1996; Meijerink et al., 2000; Hallem et al., 2004b) contained within. In addition, insects manifest important olfactory responses in their maxillary palps, which display fewer olfactory sensilla and reduced olfactory response profiles (Kellogg, 1970; Grant et al., 1995). Interestingly, another population of ORNs has been identified on the proboscis (labellum) of An. gambiae (Pitts et al., 2004) that have recently been shown to harbour olfactory responses that are potentially novel to mosquitoes (Kwon et al., 2006).
Eighty-three of the 123 surveyed AaOrs (70%, excluding the previously characterized AaOr7) are expressed in the antennae (Fig. 4, Table 1). Of these, nine (AaOr22, 36, 62, 96, 105, 109, 116, 121 and 123) appeared to be expressed only in females, as they were undetectable in male antennae and other olfactory appendages. The expression of two antennal AaOrs (AaOr84 and AaOr113) was further mapped to discrete sets of Ae. aegypti ORNs using in-situ hybridization (Fig. S4). In these studies, AaOr84 and AaOr113 were expressed in a relatively small number of ORN cell bodies spanning multiple antennal segments (data not shown), where they are consistently colocalized with the AaOr7, which serves as a general ORN marker protein in Ae. aegypti (Melo et al., 2004).
Table 1.
AaOr expression survey in olfactory organs by RT–PCR. Positive expression is highlighted in grey. The number of (+) symbols represents the number of positive signals for each individual biological replicate performed; (–) negative results in any of the three biological replicates; nd, not determined. Additional information regarding these analyses may be found in the supporting text
| Gene | Adult antenna | Adult proboscis | Adult palps | Larval antenna | Forward primer | Reverse primer | |||
|---|---|---|---|---|---|---|---|---|---|
| male | female | male | female | male | female | ||||
| AaGPRor1 | - | - | - | - | - | - | +++ | ACGGGAGATCGAGAGAACAG | TGGTAGGAAGATCTGCGTC |
| AaGPRor2 | +++ | +++ | - | - | - | - | +++ | TGCTTGGCTCGATTTTGAC | AACCCTGCGGAGCAATG |
| AaGPRor3 | +++ | +++ | +++ | +++ | - | - | +++ | ACGCTGTTTGCTTGGAAC | TTCATAACGACGTAAGCCG |
| AaGPRoH | +++ | +++ | +++ | +++ | - | - | - | TCATTCTCTGCTACTACGGTG | ACGTCCGAATGGTTATGG |
| AaGPRor5 | +++ | +++ | +++ | +++ | - | - | - | TCATTCTCTGCTACTACGGTG | AAGTAGGACCATGATGTTTGC |
| AaGPRor6N | +++ | +++ | +++ | +++ | - | - | - | ACCAGTTCCGGACAATTATC | TGAAATACGACCACGATGC |
| AaGPRor7 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | AGAAAGCGATGACCATATCC | TTCAACTGCACCAACACC |
| AaGPRor8 | - | - | - | - | +++ | +++ | +++ | ACGGAAACAAATTGATTGAAGC | TGGCCAGCTCGAGTCATC |
| AaGPRor9 | - | - | - | - | - | - | +++ | TAGTGGACCATGGATTGAAC | TCATAGGATAGACGTTACCCAC |
| AaGPRor10 | +++ | +++ | - | - | - | - | - | TCCTGCTGATAATTCTACGGG | ATAAACCCGACGCAGC |
| AaGPRor11 | +++ | +++ | - | - | - | - | - | ACGACTGGTATAACTATCCCCG | AGCTTGTTTCAGGATCGTG |
| AaGPRor12C | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor13 | +++ | +++ | - | - | - | - | - | TGTTTGAACGGAACTGCTCTC | TCGTTGAGCCCGTTGAATC |
| AaGPRor14 | - | - | - | - | - | - | ++ | TGGATTGCGTACTCATATGG | TGTGATACCTTCTTTGCGTTG |
| AaGPRor15 | +++ | +++ | +++ | +++ | - | - | +++ | TCGTGGCACTTACCGG | ACTTTGCTGCTGTGATACCTTC |
| AaGPRor16 | +++ | +++ | - | - | - | - | +++ | TGGATCTACCTGTTCAACGG | TATTCCTGCGCTGTTACAAG |
| AaGPRor17 | +++ | +++ | - | - | - | - | - | AGCCTCATCCCAAATTTTC | TCTCTTTAACGTGCTGGGAC |
| AaGPRor18C | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor19 | +++ | +++ | - | - | - | - | - | AGGAACGCTCTTATTGATAAAGG | TCTCCATGTTCATAACTCCG |
| AaGPRor20 | +++ | +++ | - | - | - | - | - | TCCTTGCTACTTCTCAGTCTCCG | AGCAGCATAATTCCGGAGTAAAC |
| AaGPRor21 | +++ | +++ | - | - | - | - | - | TTTACTTCTGAACGGTGCCC | AGCATGCAAATCGAATACAGAG |
| AaGPRor22P | - | +++ | - | - | - | - | - | AGCAAAGGACATTGATGAGG | ACGCAGATGTGAGCTATGG |
| AaGPRor23 | +++ | +++ | - | - | - | - | - | ACTGGCACTCATTCTCAACTC | CTAGTTGAAACTCTGGAGCATTG |
| AaGPRor24 | +++ | +++ | - | - | - | - | - | AGGAGATTTGGAGGTGGATG | TCCCGTTGAAGTTGTTCG |
| AaGPRor25 | - | - | - | - | - | - | - | ACCGGATAAGATCTATGCTCTCG | TGGAGTAGTACGAAAATGCCG |
| AaGPRor26 | +++ | +++ | - | - | - | - | - | AGTTTTTCCACCTCAACCG | TGACCTTGGTCTCGATCTTC |
| AaGPRor27 | +++ | +++ | - | - | - | - | - | TCGTGCATATTCCAGACCTG | TCCACTGACCACTCGTACC |
| AaGPRor28 | - | - | - | - | - | - | - | TTCCAAGTGTTTGCTCAGG | TTGAGCCCGTTGTCCAC |
| AaGPRor29P | +++ | +++ | - | - | - | - | +++ | ACCGAATGCCCTCAACGTGATG | ATGGTTGTAGATGGCATCGCTAACC |
| AaGPRor30 | - | - | - | - | - | - | - | TTCGTCTACTCCATGCTCG | TAGGATGATTCGTTGTTCGC |
| AaGPRor31 | +++ | +++ | - | - | - | - | - | TGCGGGCTCAAGTTC | AGCAACGTACGAGCCAG |
| AaGPRor32P | - | - | - | - | - | - | - | TGGTCTACTCTATGTTGAGCACC | TTGCCAGTAGTAAGCTTCGC |
| AaGPRor33 | +++ | +++ | - | - | - | - | - | TGGTGCTATTATGGAAACAGAG | ACTTGAATGGCTTCTGCG |
| AaGPRor34 | - | - | - | - | - | - | +++ | AACGACCGGAAAGATCTG | ACCGAATAGGCTGTGTTTAGAG |
| AaGPRor35C | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor36 | - | +++ | - | - | - | - | - | TCGGTTGTGCGTTATTCTAC | AGGTCGCACAATTGTATCG |
| AaGPRor37 | - | - | - | - | - | - | +++ | TCCAAGATATCGAACGAGTCTAC | ACAACAGGTACATCATCATCCC |
| AaGPRor38P | +++ | +++ | - | - | - | - | - | ACGGCTTGTTCATCGGAGTTTGTC | ATGCCCAGCGAGTACCACAC |
| AaGPRor39N | +++ | +++ | - | - | - | - | - | GGTGACGGTTTGTTCATCGGAGTTT | CGGTAGCGATACCAGGGAAAATCGT |
| AaGPRor40 | - | - | - | - | - | - | +++ | ACCGCTAAAGCTAACCGC | TTAACTAGTATGGCGACTACCG |
| AaGPRor41 | +++ | +++ | - | - | - | - | - | TTGAAAGCCAACAGCGAC | ATTAAGCGTCATCATTCCTCC |
| AaGPRor42 | - | - | - | - | - | - | - | AATGACGCTGTTTGTGCTAC | ATATCGTACACCTCCTGGATG |
| AaGPRor43 | - | - | - | - | - | - | - | TGCTAGAAATGTGTTTGCTCG | ATCAAATACCAGTCCATGCC |
| AaGPRor44 | +++ | +++ | - | - | - | - | - | TGGAGTTCTGTTTGCTTGGGACAG | TGTTCAATGGTGCGAGTCCACC |
| AaGPRor45 | +++ | +++ | - | - | - | - | - | TCTAACTGTGGGAGGCTTTG | TCATCATAGCGAAGAATTGG |
| AaGPRor46 | - | - | +++ | +++ | - | - | - | AGACCTTCTCATCAAAATGGG | AACATTTCGCTGCTCGG |
| AaGPRor47 | +++ | +++ | +++ | +++ | - | - | +++ | TTGCTTGACCATGATGACC | AAGATGTTACGCTGCTCTGG |
| AaGPRor48 | - | - | - | - | - | - | +++ | TGGCACATGAGATCTACAGC | AATGAGTAAGTGGTTTGGACG |
| AaGPRor49 | - | - | ++ | - | +++ | +++ | - | TCAATGGAGATGAGCTAAGCG | TAATACCGATCTTCCGCTGC |
| AaGPRor50 | +++ | +++ | - | - | - | - | - | TGGGAACGGAACTGATAAGTG | TCATCCTCTGTGATCTTCCC |
| AaGPRor51P | +++ | +++ | - | - | - | - | - | TGGTGGCAAATGCGGTCTATGAGG | TGAAGTAAGACATCGTGGTGCGGAC |
| AaGPRor52 | +++ | +++ | - | - | - | - | - | TGCTGGAGTCTTACTGGTTCTG | ACAATCAGCAGCGTCTCC |
| AaGPRor53 | +++ | +++ | nd | nd | nd | nd | - | TTTCGATGCTGGGAGCTGAACTC | GACGGTCCGCTCTCATTTTCATG |
| AaGPRor54P | +++ | +++ | - | - | - | - | - | ACATGAGGTTTGCTTCCGATCTGC | GGCAGCCATTCGGTTGTACCATT |
| AaGPRor55 | +++ | +++ | +++ | +++ | - | - | - | TGCCGAAAGACGTTTGC | TCCGTAGAACTGCTGCATTATC |
| AaGPRor56 | +++ | +++ | +++ | +++ | - | - | - | ATGTTCGTTCTGCAATATACCG | AACAACAGCAATCGGCTCTCGGTAG |
| AaGPRor57P | +++ | +++ | - | - | - | - | - | ATGGTCGTAATCGCATCG | ATCCGATCTCAGCTTCATG |
| AaGPRor58 | - | - | - | - | - | - | +++ | TCGAAGCCAAACCCCAGGAC | ACAGCCGATGGTTGATGGATGAC |
| AaGPRor59 | +++ | +++ | - | - | - | - | - | ACTTTAAGACAAGCCCTTCGC | AGCACAGAGCATGGATCCC |
| AaGPRor60 | - | - | - | - | - | - | +++ | TTGTAACATGGTGTCGTCGTTG | ATTCCATCAGCTCGATTAGTATGC |
| AaGPRor61 | - | - | - | - | - | - | +++ | TTCACTCCAAAATTCGACAGC | TGCTGTATCTCAAATCGACTCC |
| AaGPRor62 | - | +++ | - | - | - | - | - | TGAACATCGCAGCTGGAG | AGGCCGAAAGACCGATATATTG |
| AaGPRor63 | +++ | +++ | - | - | - | - | - | TCTAGGAACAGCTTTGACGC | TGTTCAGAAGAGCCACGC |
| AaGPRor64P | +++ | +++ | - | - | - | - | - | ATCCGTGTCATGTCTCAGTATG | TGCAGTACAAATTTCAACCG |
| AaGPRor65 | - | - | - | - | - | - | - | ATTTATGCTATCAACTGGCACC | TTGTGATGAGCATCGCC |
| AaGPRor66 | +++ | +++ | - | - | - | - | - | TGACTGGTACGCAACATATG | TCGGTAATAAATGCCATGG |
| AaGPRor67 | - | - | - | - | - | - | - | TGGTCGACATTTATGCTATCAAC | ATTGTGATGAGCATCGCC |
| AaGPRor68P | - | - | - | - | - | - | - | TGTTGTGAATTTTCCGTGG | ACATGATTGGTGTTCTCGATG |
| AaGPRor69 | +++ | +++ | - | - | - | - | - | TGATGCGATGCTTTTGG | AAATTTAACGGAGCAAACCC |
| AaGPRor70 | +++ | +++ | - | - | - | - | - | TCGTCTTCACTGGCGTATTC | TTTGAGCAAATGCCACG |
| AaGPRor71 | +++ | +++ | - | - | - | - | - | GTACTTGGGGACGGAGTTGAGT | GCCGATCCAGAACATCTTTGAGCA |
| AaGPRor72 | - | - | - | - | - | - | - | TTGTGTTCATCGGTTGTGC | TGCTTCCGTGAGTTTGG |
| AaGPRor73P | - | - | - | - | - | - | +++ | CTGCTGTACGGATTCTTGTGCGA | |
| AaGPRor74 | - | - | - | - | - | - | +++ | CAAGCGGTGCGTTGATCGTC | CCGTGTAAGCGAACCACAACACC |
| AaGPRor75 | - | - | - | - | - | - | +++ | GGCATTGGTGTTTCGGGACAA | TGCCCTCATCATCACAATCAGCA |
| AaGPRor76 | - | - | - | - | - | - | - | GCGTTCGACGGCACCAG | CAGCAGCGTGAATGCCGAGT |
| AaGPRor77P | +++ | +++ | - | - | - | - | - | GCGTCATCAGAGGTTGCTGGAA | GATGCCGCCACAAGTGAATCC |
| AaGPRor78 | +++ | +++ | - | - | - | - | +++ | GCAGGCTTGTCCATCGTGTCC | TTCGCTCGTCGCATTCCACA |
| AaGPRor79 | +++ | +++ | - | - | - | - | - | CCTGCTGGTGGTTCAGCTTCG | TCGCCACGTCCCTGATCG |
| AaGPRor80 | +++ | +++ | - | - | - | - | - | GCATTCTGTTCATCGTGGGAGCA | CATTCCACCAACTCTGAACTTGATTCC |
| AaGPRor81 | - | - | - | - | - | - | - | GACGGATTGTCTGGAAGTACGAGGA | TCCGAAATATACTGGATCGCCATAA |
| AaGPRor82P | +++ | +++ | +++ | +++ | - | - | - | CGCATCCGAAACCTACCTGTGTT | TGTCATGTCGTACCGCACAAAGC |
| AaGPRor83P | +++ | +++ | +++ | +++ | - | - | - | CATCCGAAACCTACCTGTGTTGC | ACCGATTTCATCATCGTGGCAAAC |
| AaGPRor84 | +++ | +++ | - | - | - | - | - | TCGGGTTGTTGCTATCGTTGATGA | ACGGAACCAAAAACCCTGTTGAG |
| AaGPRor85 | +++ | +++ | - | - | - | - | - | GACGCTTTGGGACTGTTGGTT | TCCAAATCCCTGCTGAGCTTGT |
| AaGPRor86N | +++ | +++ | - | - | - | - | - | TGCGAGCAGAGCTTACAATAGCAT | TTCTCGTGTGGGGAATAATCGTGA |
| AaGPRor87 | +++ | +++ | - | - | - | - | - | AATCATGCGTCACATTGAGTTGCTT | TGATAAACCCCCGCAGCTAAATTG |
| AaGPRor88 | +++ | +++ | - | - | - | - | - | TGGGGATAACGGTGTTCTTCATCG | TCGGGTGCCAATCTTCATTATCATCA |
| AaGPRor89 | - | - | - | - | - | - | +++ | ATCTGGATGGACGAGCGATATTCAA | GCGTAGGATTTCTGGAGGATCTGC |
| AaGPRor90 | - | - | - | - | - | - | +++ | TCAACAAGGTATTCGGACCAACGA | CGAGGAAAGCGAACAGCGTGTA |
| AaGPRor91N | +++ | +++ | - | - | - | - | - | GATTTTCGCCCAGTAGCCCTGTA | CATCAGGTCCGTCAAATGTTGCTG |
| AaGPRor92 | - | - | - | - | - | - | - | CGTATGTCGCACTTGTGGAGACC | CACCACCAGCATTCAGCGAAAAA |
| AaGPRor93 | +++ | +++ | - | - | - | - | - | TGCTAACGGGATGACTCGCTTTG | GCTGCTGATAGTCGGAATGGTTCTC |
| AaGPRor94 | ++ | +++ | - | - | - | - | - | AAGGATTTTCGTTCGGCTCGGTA | GCAGCATTGACCTGTACGAGCA |
| AaGPRor95 | ++ | +++ | - | - | - | - | - | GCTATTGGGTTCATGGTGGAATGC | TCCGAATGATGTTTTGGCGAATGT |
| AaGPRor96 | - | +++ | - | - | - | - | - | ATGCGTTTGCCGTGACATACTACT | GAGATCAGAAATACTCCAGCACAGC |
| AaGPRor97 | +++ | +++ | - | - | - | - | - | GTTCTGGAGTGCTATTGGTGGTG | CGATCATCAGCGAAGTACGCATCT |
| AaGPRor98 | +++ | +++ | - | - | - | - | - | CGGAGGAATCGGAGGACTGTTT | ACATTTGATTGGTGGTGGGTTGGA |
| AaGPRor99 | ++ | ++ | - | - | - | - | - | ATACGAAGCGGGTCTGGGTAATG | TTCCGCTCGATGTTGTTGATCGTA |
| AaGPRor100 | +++ | +++ | - | - | - | - | - | TGGCAACTACTCCGATATTCCTGGTG | TCGAACAGCCCCAGACATCG |
| AaGPRor101 | - | - | - | - | - | - | - | ATGGGAATCAACGCAACTTCTGGA | GTGTTCAGCAAACGAACGAAAGCA |
| AaGPRor102 | - | - | - | - | - | - | - | AGACGCTTGCTCATCTTGTGCT | TCTGAAACAATCCCAGGCAGTCCA |
| AaGPRor103 | +++ | +++ | - | - | - | - | - | CTCCTGGGAATGCGAATGAAGTTGA | CGAAAATGATGTTCCCGATGGAAGCA |
| AaGPRor104 | +++ | +++ | - | - | - | - | - | GGAATGCGGATGAAACTCGGAATGA | AACAGCCCAAAGCAATCCACTGAA |
| AaGPRor105 | - | +++ | - | - | - | - | - | TGTGCTCGTTGCCTTTATTTCTGC | AATCCGTTGAATCCCATGTTTTGC |
| AaGPRor106 | +++ | +++ | - | - | - | - | - | TGGGAAATCATCAGTCGGGAAATCA | TGAGCGATGAACAGAAATGCTCCA |
| AaGPRor107 | - | - | - | - | - | - | - | CAGGATGTTATCGCACCGTTTCACA | TGCGGTGTTAAGCAATTCAACGAA |
| AaGPRor108P | - | - | - | - | - | - | - | TGTGCCGCTACTTTGCTGTTAGGA | AGTTCATCAGCATTCCTCGTCGTG |
| AaGPRor109 | - | ++ | - | - | - | - | - | TTTCAGCATTGGAACGAGTGGCTA | CATCAGCATCGTGTAGGCAGTGTC |
| AaGPRor110 | +++ | +++ | - | - | - | - | - | GTTGCCCAAGATGTTTGAAATACGC | GTGGGATGTCAGTTCACTTCGGATT |
| AaGPRor111 | +++ | +++ | - | - | - | - | - | GCCTTGGAACATGCTTTTACCTATCC | CGCATTGAGGAAGCTGACAAACG |
| AaGPRor112P | +++ | +++ | - | - | - | - | - | TCGGGAGGTGAGGACTGTTGTG | CGGCTGTAAGGCATCTGTGC |
| AaGPRor113 | +++ | +++ | - | - | - | - | - | TGCATCCGCATTTGCCTACTTG | CACTCCGTTGATGGTGTTGTGC |
| AaGPRor114 | +++ | +++ | - | - | - | - | - | ATTTTAGCCAACCGCTTTCGTTCG | CGGTTGTGGGGCATAAGAATAGCA |
| AaGPRor115 | +++ | +++ | - | - | - | - | - | AAAGTTTTAGCCAACCGCTTTCATTC | CGGTTGTGGGGCATAAGAATAGCA |
| AaGPRor116P | - | +++ | - | - | - | - | - | GCCGCTGTTTCGTTCTTTCTATTGG | TGTTGATGTCGAATATGCCCATGC |
| AaGPRor117 | - | - | - | - | - | - | - | TCCAATTTCCCGTCCCATCATTCT | CGATAGAAAGAACGAAACAGCGGCT |
| AaGPRor118P | - | - | - | - | - | - | - | TGTGTTGGAGTTACCCTCGTTGGA | CATCAAAACGGAGTACGCAACATCG |
| AaGPRor119 | - | - | - | - | - | - | - | TCTTCTGGCGTTGACTGGAATGTTTC | GGGTTACGAAGGTGAATGTGGTGA |
| AaGPRor120P | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor121 | - | +++ | - | - | - | - | - | TCCTGTAGCCAAGTTCTGCTCGT | AACAAAGCCGACAAAAGCAATCGT |
| AaGPRor122 | +++ | +++ | - | - | - | - | - | TGGTGCCATCTGGTTGATTCGT | CAGAAACATCAGCACCGAGTAAGCA |
| AaGPRor123 | - | +++ | - | - | - | - | - | AGCATGATATTGGCGGACTGTTGAA | TGTTCGCATTGCCACAAAGTCAGA |
| AaGPRor124 | +++ | +++ | - | - | - | - | - | TGTCGTCGCTTGGAACTGGAAT | GCTGTGACGGGCGTTAATCCA |
| AaGPRor125N | +++ | +++ | - | - | - | - | - | CTGGATGCCGCTGATTTGGT | CATTCCCAAAAGTGGACGGATCAA |
| AaGPRor126C | +++ | +++ | - | - | - | - | - | ATGAAAGCGATTGTTAGCTTCTGGAA | GCGACTAATGCCAAAGGTTTCCAT |
| AaGPRor127 | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor128P | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor129P | nd | nd | nd | nd | nd | nd | nd | ||
| AaGPRor130C | - | - | - | - | - | - | - | GGACCTGCTGATGATTGCTTTTGG | TTTTGCGGTAACACCTTCCCACAG |
| AaGPRor131P | nd | nd | nd | nd | nd | nd | nd | ||
| Summary | 75/123 | 84/123 | 13/123 | 13/123 | 3/123 | 3/123 | 24/123 | ||
The maxillary palps of Ae. aegypti mediate carbon dioxide and other chemical responses (Kellogg, 1970; Grant et al., 1995), are sexually dimorphic, and harbour a small subset of olfactory sensilla basiconica on the fourth segment (McIver, 1971). In addition to the widely expressed nonconventional AaOr7 gene, only the two non-antennal AaOr9 and AaOr49 (Fig. 4, asterisks) were found in the palpi of both sexes (Fig. 4, Table 1). Of these, only AaOr8, which has a potential ortholog in An. gambiae, is palp specific. AaOr49 is also expressed in the proboscis (Fig. 4, Table 1). In-situ hybridization showed that AaOr49 was found to be expressed throughout the fourth maxillary palpomere of females, which has been reported to be the sole olfactory sensilla-bearing segment (McIver, 1982). The proboscises of Ae. aegypti and An. gambiae are homologous structures and the expression of multiple AgOrs and robust olfactory responses, perhaps involved in mediating the penultimate stages of blood feeding behaviours, have been described on the labellum of An. gambiae (Kwon et al., 2006). Similarly, we have identified a subset of 12 AaOrs (excluding AaOr7) expressed in the proboscis of Ae. aegypti. It is interesting to note that, of these, only AaOr6 has a potential AgOr ortholog that displays labellum expression (Kwon et al., 2006). This conservation of mosquito proboscis Or expression and presumptive olfactory function is, to date, unique among dipterans and further underscores important differences in the olfactory processes between fruit flies and mosquitoes. Indeed, such differences may be associated with the anautogenous requirement for blood meals that underlies the vectorial capacity of these mosquitoes. Not including AaOr7, we found 23 AaOrs in the larval antenna. Fifteen of these (1, 9, 14, 34, 37, 40, 48, 58, 60, 61, 73P, 74, 75, 89 and 90) are larval specific (Fig. 4). Most larval Or genes form monophyletic clusters and/or are linked on the genome (Figs 1 and 2). Such is the case for the AaOr1/34/37, AaOr2/9, AaOr7/40, AaOr3/14/15/48, AaOr60/61, AaOr89/90, AaOr1/34/37 and AaOr58/73P/74/75/78.
Discussion
The Aedes Or gene repertoire is 60% larger than its Anopheles counterpart. Phylogenetic comparison demonstrates that the vast majority of ORs in each subgenome is species-specific, indicating a higher degree of evolutionary activity in Ors that is likely to reflect their critical roles during the mosquito life cycle. At the other end of the spectrum, the observed conservation of a subset of ORs in these two distant species may also reflect common biological constraints. One caveat in this, or indeed any, comparison of the two mosquito genomes is that future refinements in the overall assemblies may necessitate the re-evaluation of the Or annotations used in this study. Errors of assembly such as false duplications or simple gaps in sequence data may obscure the actual number of Ors in either mosquito genome. This may be especially relevant for the Ae. aegypti WGS assembly, which lacks a corresponding physical map, is much larger in size and considerably more difficult to assemble because of the large amounts of repetitive DNA in the genome (Nene et al., 2007).
Expression of 20 AaOrs (≈ 16%) is undetectable in adult and larva mosquitoes. This may in part reflect low levels of transcripts distributed across a limited number of ORNs, thereby confounding amplification by means of RT–PCR. In other instances, it is likely that a subset of AaOrs are specifically expressed in wings or legs as observed in the legs of An. gambiae (Pitts et al., 2004). We cannot exclude the possibility that these genes have been rendered nonfunctional (pseudogenes) through mutations outside of their open-reading frame or through mis-sense mutations. Taken together, the differential AaOr expression patterns within the peripheral olfactory apparatus provide a molecular basis for the response profiles of these tissues, as has been demonstrated for D. melanogaster (Krzywinski et al., 2001; Hallem et al., 2004b) in which the odour space of each olfactory tissue is determined by the spectrum of Ors expressed. Moreover, if one accepts that the peripheral segregation of AaOrs into the antennal and non-antennal structures observed here is nonrandom, and may support broad biological functions for each appendage (e.g. host, mate or oviposition site recognition), then it reflects a biologically constrained topography for AaOr expression in the periphery.
It is reasonable to suggest that the observed female-specific expression, detected by RT–PCR, may indicate the involvement of these AaOrs in female-specific, olfactory-mediated behaviours. Moreover, these data provide an additional departure from the accepted ‘one receptor–one neurone’ and ‘one receptor-one glomerulus’ paradigms of olfactory organization in that one might also expect this sexual dimorphism to be manifest in AL anatomy. However, the only apparent AL sexual dimorphism is restricted to a single additional glomerulus in male Ae. aegypti (Ignell et al., 2005). Alternatively, the observed difference in antennal PCR signals between female and male may just be a reflection of the lower number of olfactory sensilla and ORNs in males (McIver, 1982). The absence of male-specific Ors suggests that both male and female mosquito olfactory-mediated behaviours are able to utilize a common Or array as shown in Drosophila (Ejima et al., 2007). While Ors expression has been reported in the larva of Drosophila (Fishilevich & Vosshall, 2005; Kreher et al., 2005), this is the first time that Ors have been shown to express in aquatic insects. The larval antenna of Diptera harbours two chemosensory organs. The peg organ is assumed to be a contact chemosensory sensillum based on the presence of a single terminal pore while the sensory cone is multiporous consistent with olfaction (Nicastro et al., 1998). The cone of Ae. aegypti is innervated by approximately 13 receptor neurones (Zacharuk et al., 1971). This amount does not account for the number of expressed AaOrs in the larval antenna. However, mosquito Ors are also expressed in the adult proboscis, which has been considered a gustatory organ (Pitts et al., 2004; Kwon et al., 2006). It is therefore possible that AaOrs may be found in the larval peg organ as well or that more than one AaOr per neurone is expressed in the cone. We found a direct correlation between larval Or expression, sequence conservation and genomic proximity suggesting that larval Or genes may be under the control of larval-specific regulatory elements. A recent study demonstrated that the expression of Ors in adult Drosophila is under a hierarchical code of regulatory elements and transcription factors dictating organ and neurone specificity (Ray et al., 2007).
In Drosophila, transgenic reporter lines have facilitated systematic analyses of the anatomical relationships between discrete ORNs and their synaptic targets within AL glomeruli. The results of these studies have largely adhered to the one receptor–one neurone and one glomerulus–one receptor principles that are characteristic of mammalian olfactory systems (Couto et al., 2005; Fishilevich & Vosshall, 2005; Kreher et al., 2005). This paradigm is also likely to be valid in the honeybee Apis mellifera, where between 156 and 166 AL glomeruli (Galizia et al., 1999) and 163 intact Or genes have recently been identified (Robertson & Wanner, 2006). In contrast, mosquito olfactory networks seem to be paradoxical in this regard as the Or repertoires of both An. gambiae and Ae. aegypti are significantly greater than the approximately 50 AL glomeruli present in both vector species (Ignell et al., 2005). This raises the intriguing possibility that there may be significant differences across insect taxa in how chemosensory information is encoded.
Defining and spatially characterizing the complete repertoire of the AaOr transcriptome is a critical step toward the functional characterization of these receptors. Further study of the highly divergent Or gene families will reveal ways in which these receptors influence chemosensory-driven behaviours, some of which are likely to be unique to certain mosquito species and may have a bearing on their ability to transmit disease. Accordingly, the characterization of this gene family in Ae. aegypti has potentially far-reaching implications for basic research, vector control and global health.
Experimental procedures
Gene identification and annotation
The Ae. aegypti (Liverpool strain) whole genome sequencing database at the Broad Institute (www.broad.mit.edu/annotation/disease_vector/aedes_aegypti/) was queried using previously described OR sequences from An. gambiae (Hill et al., 2002), D. melanogaster (Clyne et al., 1999b; Vosshall et al., 1999; Zwiebel & Takken, 2004) and Heliothis virescens (Krieger et al., 2003) by tBLASTn (Altschul et al., 1990). Recently, trace amine-associated receptors (TAARs) have been implicated in odour recognition in mice (Liberles & Buck, 2006) and we used these sequences from vertebrates (Lindemann & Hoener, 2005; Gloriam et al., 2005) to query the Ae. aegypti genome. Our searches resulted in the identification of several class-A biogenic amine receptors (data not shown; Nene et al. 2007). Putative Or sequences were manually annotated using Sequencher v4.5 (Gene Codes, Inc., Ann Arbor, MI) and refined using the Softberry Splice Site Prediction program (www.softberry.com/berry.phtml?topic=fsplice&group=programs &subgroup=gfind) and TMPred (Hofmann & Stoffel, 1993). All Ae. aegypti ORs identified in this manner were used in successive tBLASTn searches to identify other candidate sequences, which were annotated as above. Final AaOR annotations were compared with the AaegL1.1 Genebuild at Vectorbase (http://www.vectorbase.org). The ClustalW algorithm (Thompson et al., 1994) was used to align protein sequences and predict the exon/intron structures (see ‘Intron/exon analysis’ below) to further support and refine the final annotation predictions. In some cases, it was possible to confirm whole or partial Or gene predictions using tentative consensus sequences identified from the TIGR Ae. aegypti Gene Index (AeGI Release 5.0, 29 June 2005; http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=a_aegyptii).
Intron/exon analysis
The predicted structure of each gene was reconstructed as described above. We first compared intron locations for each monophyletic clade within a species and then representatives of individual clades. Species comparisons were carried out in a similar fashion between Ae. aegypti and An. gambiae, and only the most parsimonious intron locations were considered to be conserved. All other were considered idiosyncratic.
Phylogenetic analysis
Deduced amino acid sequences of AaOrs were aligned with AgORs using ClustalW (Thompson et al., 1994) and phylogenetic analysis was performed using distance methods in paup* v4.0b10 (Swofford, 1998). For the details of corrected distance phylogenetic methods, see Fig. S1 legend and (Hill et al., 2002). Bootstrap analysis was used to assess statistical support for relationships via uncorrected neighbour-joining analysis of 1000 pseudo-replicated data sets.
Expression analysis using RT–PCR
Twenty-five antennae, proboscises, and maxillary palps each from 5-, 6-, 7- and 8-day-old Ae. aegypti were hand dissected. Three separate collections were done per sex. We also hand dissected three separate collections of larval antennae (larval stages 3 and 4). Total RNA was prepared using the RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized from RNA samples using SuperScript III RT according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). To control for genomic DNA contamination, each cDNA synthesis was carried out in duplicate: one with RT (experimental) and one without (no RT control). We used the Ae. aegypti housekeeping gene for Lysosomal Aspartic Protease (AaLAP: (13)) to control the quantity of each cDNA sample. PCR cycling conditions were as follows: 94 °C (2 min) followed by 35 cycles (40 cycles for proboscis, palp and larval antenna cDNAs) at 94 °C (25 s), 55 °C (20 s), and 72 °C (1 min). PCR products were then visualized with ethidium bromide on 2% agarose gels.
Confirmation of Or sequence
Aedes aegypti Or PCR amplicons were sequenced to confirm predicted coding regions for a subset of genes. Twenty-seven AaOr PCR products corresponding to predicted cDNA amplicons were gel-extracted (QiaexII, Qiagen) and directly sequenced at Vanderbilt University.
In situ hybridization and immunolabelling
cDNA fragments were amplified utilizing the oligonucleotides 5′-CTC AGG GAT TTT TGT AGC AGG AGC A-3′ and CAC TCC GTT GAT GGT GTT GTG C-3′ (AaOr113), 5′-GTG GAA AGG ATG ACG CAA AAG CA-3′, and 5′-ACG GAA CCA AAA ACC CTG TTG AG-3′ (AaOr84), 5′-GC GAG TTG ATC TAC TTT ACC G-3′ and 5′-GGT TCC AAG TAC CAG GGA TAG C-3′ (AaOr49). For synthesis of digoxigenin-labelled sense and anti-sense RNA probes, amplification products were cloned into the pCRII-TOPO vector (Invitrogen). In situ hybridization and probe preparation were carried out as described previously (Kwon et al., 2006). AaOr signals were visualized by peroxidase-coupled antidigoxigenin antibodies (Roche, Indianapolis, IN, USA) at 1 : 200 dilution. Peroxidase signals were detected using tyramide signal amplification plus cyanine 3 system (Perkin-Elmer, Wellesley, MA, USA) according to the manufacturer's instructions. AaOR7 and nervana/HRP immuno-labelling, respectively, was carried out using anti-AgOR7 antibodies and anti-HRP antibodies, respectively, as previously described (Melo et al., 2004; Rutzler et al., 2006). We used anti-HRP instead of anti-AgOR7 as it has been previously reported to weakly labels the cell body of the capitate pegs in the maxillary palp of Ae. aegypti (Melo et al., 2004). All presented images were captured within the dynamic signal range and have not been adjusted in contrast. Optical sections of 1.2 μm are shown.
Mosquito rearing
Aedes aegypti (Costa Rica strain), originally established in 2001 (Perich et al., 2003) was kindly provided by Mark Q. Benedict (Centers for Disease Control and Prevention, Atlanta, GA, USA). Embryos were hatched in flat plastic pans with distilled water. Costa Rica strain larvae were reared on a 2 : 1 mixture of ground fish food (Aquaricare Koi floating blend, Victor, NY, USA) and bakers yeast, and Liverpool larvae were reared on ground guinea pig food (Nutriphase products, Phoenix, AZ, USA). Pupae were transferred to plastic cups within 4-litre plastic containers, from which newly emerged adults were collected the next morning. Adult mosquitoes were maintained in 4 litre plastic containers at relative humidity under a 12 h/12 h photoperiod and provided with a 10% dextrose solution.
Supplementary Material
Acknowledgements
We acknowledge the insights and contributions regarding Aedes GPCRs of Drs C. Hill and D. Hogenkamp (Purdue University), Dr H. Schioth for providing zebrafish TAAR sequences. We also thank Z. Li and P. Russell for mosquito rearing, L. Chretien, L. Johns and D. Rinker for technical assistance, Dr A. McAinsh for editorial comments and Dr P. Abbot (Vanderbilt University) for critical reading of the manuscript. This work was supported by NIH grants AI56081 (to H.M.R.), A1056402 and DC04692 (to L.J.Z.).
References
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Suzuki T, Senti KA, Stubbs J, Schaffner G, Dickson BJ. Genetic mapping with snp markers in Drosophila. Nat Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel mulitgene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Certel SJ, De Bruyne M, Zaslavsky L, Johnson WA, Carlson JR. The odor specificities of a subset of olfactory receptor neurons are governed by acj6, a pou-domain transcription factor. Neuron. 1999a;22:339–347. doi: 10.1016/s0896-6273(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999b;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Hallem EA, Carlson JR. Insect chemoreception. Curr Opin Neurobiol. 2005;15:423–430. doi: 10.1016/j.conb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Ejima A, Smith BP, Lucas C, Van Der Goes Van Naters W, Miller CJ, Carlson JR, et al. Generalization of courtship learning in Drosophila is mediated by cis-vaccenyl acetate. Curr Biol. 2007;17:599–605. doi: 10.1016/j.cub.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci USA. 2001;98:14693–14697. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizia CG, Mcilwrath SL, Menzel R. A digital three-dimensional atlas of the honeybee antennal lobe based on optical sections acquired by confocal microscopy. Cell Tissue Res. 1999;295:383–394. doi: 10.1007/s004410051245. [DOI] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Gloriam DE, Bjarnadottir TK, Yan YL, Postlethwait JH, Schioth HB, Fredriksson R. The repertoire of trace amine G-protein-coupled receptors: Large expansion in zebrafish. Mol Phylogenet Evol. 2005;35:470–482. doi: 10.1016/j.ympev.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Goldman AL, Van Der Goes Van Naters W, Lessing D, Warr CG, Carlson JR. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Grant AJ, Wigton BE, Aghajanian JG, O'Connell RJ. Electrophysiological responses of receptor neurons in mosquito maxillary palp sensilla to carbon dioxide. J Comp Physiol A. 1995;177:389–396. doi: 10.1007/BF00187475. [DOI] [PubMed] [Google Scholar]
- Hallem E, Fox AN, Zwiebel LJ, Carlson JR. A mosquito odorant receptor tuned to a component of human sweat. Nature. 2004a;427:212–213. doi: 10.1038/427212a. [DOI] [PubMed] [Google Scholar]
- Hallem E, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004b;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu Rev Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. Tmbase – a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Ignell R, Dekker T, Ghaninia M, Hansson BS. Neuronal architecture of the mosquito deutocerebrum. J Comp Neurol. 2005;493:207–240. doi: 10.1002/cne.20800. [DOI] [PubMed] [Google Scholar]
- Jacquin-Joly E, Merlin C. Insect olfactory receptors: Contributions of molecular biology to chemical ecology. J Chem Ecol. 2004;30:2359–2397. doi: 10.1007/s10886-004-7941-3. [DOI] [PubMed] [Google Scholar]
- Jefferis GSXE. Insect olfaction: a map of smell in the brain. Curr Biol. 2005;15:R668–R670. doi: 10.1016/j.cub.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Jones WD, Nguyen TA, Kloss B, Lee KJ, Vosshall LB. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr Biol. 2005;15:R119–R121. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kellogg FE. Water vapour and carbon dioxide receptors in Aedes aegyptii. J Insect Physiol. 1970;16:99–108. doi: 10.1016/0022-1910(70)90117-4. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46:445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Krieger J, Klink O, Mohl C, Raming K, Breer H. A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189:519–526. doi: 10.1007/s00359-003-0427-x. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding anophelinae (diptera, culicidae) phylogeny: insights from nuclear single-copy genes and the weight of evidence. Syst Biol. 2001;50:540–556. [PubMed] [Google Scholar]
- Kwon HW, Lu T, Rutzler M, Zwiebel LJ. Olfactory responses in a gustatory organ of the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2006;103:13526–13531. doi: 10.1073/pnas.0601107103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Lindemann L, Hoener MC. A renaissance in trace amines inspired by a novel GPCR family. Trends Pharmacol Sci. 2005;26:274–281. doi: 10.1016/j.tips.2005.03.007. [DOI] [PubMed] [Google Scholar]
- McIver S. Comparative studies on the sense organs on the antennae and maxillary palps of selected male culicine mosquitoes. Can J Zool. 1971;49:235–239. doi: 10.1139/z71-033. [DOI] [PubMed] [Google Scholar]
- McIver SB. Sensilla of mosquitoes (diptera: Culicidae). J Med Entomol. 1982;19:489–535. doi: 10.1093/jmedent/19.5.489. [DOI] [PubMed] [Google Scholar]
- Meijerink J, Braks MA, Braak AA, Adam W, Dekker T, Posthumus MA, Beek TA, Van Loon JJA. Identification of olfactory stimulants for Anopheles gambiae from human sweat samples. J Chem Ecol. 2000;26:1367–1382. [Google Scholar]
- Melo AC, Rutzler M, Pitts RJ, Zwiebel LJ. Identification of a chemosensory receptor from the yellow fever mosquito, Aedes aegypti, that is highly conserved and expressed in olfactory and gustatory organs. Chem Senses. 2004;29:403–410. doi: 10.1093/chemse/bjh041. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Melzer RR, Hruschka H, Smola U. Evolution of small sense organs: sensilla on the larval antennae traced back to origin of the diptera. Naturwissenschaften. 1998;85:501–505. [Google Scholar]
- Pappenberger B, Geier M, Boeckh J. Responses of antennal olfactory receptors in the yellow fever mosquito Aedes aegypti to human body odours. Ciba Found Symp. 1996;200:254–63. doi: 10.1002/9780470514948.ch18. Discussion 263–6, 281–284. [DOI] [PubMed] [Google Scholar]
- Perich MJ, R.N.O., Castro AL, Alfaro AW, Platt KB, Solano T, Rowley WA. Evaluation of the efficacy of lambdacyhalothrin applied by three spray application methods for emergency control of Aedes aegypti in Costa Rica. J Am Mosq Control Assoc. 2003;19:58–62. [PubMed] [Google Scholar]
- Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector, Anopheles gambiae. Proc Natl Acad Sci USA. 2004;101:5058–5063. doi: 10.1073/pnas.0308146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Van Der Goes Van Naters W, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. Taste: independent origins of chemoreception coding systems? Curr Biol. 2001;11:R560–R562. doi: 10.1016/s0960-9822(01)00343-8. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100(Suppl. 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Penny D. Large-scale intron conservation and order-of-magnitude variation in intron loss/gain rates in apicomplexan evolution. Genome Res. 2006;16:1270–1275. doi: 10.1101/gr.5410606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutzler M, Zwiebel L. Molecular biology of insect olfaction: Recent progress and conceptual models. J Comp Physiol A Neuroethol Sens Neural Beh Physiol. 2005:1–14. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- Rutzler M, Lu T, Zwiebel LJ. Galpha encoding gene family of the malaria vector mosquito Anopheles gambiae. Expression analysis and immunolocalization of agalphaq and agalphao in female antennae. J Comp Neurol. 2006;499:533–545. doi: 10.1002/cne.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson DW, Debruyn B, Lovin DD, Brown SE, Knudson DL, Morlais I. Comparative genome analysis of the yellow fever mosquito Aedes aegypti with Drosophila melanogaster and the malaria vector mosquito Anopheles gambiae. J Hered. 2004;95:103–113. doi: 10.1093/jhered/esh023. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Müller B, Steinbrech RA. Atlas of olfactory organs of Drosophila melanogaster. 1. Types and external organization, innervation and distribution of olfactory sensilla. Int J Insect Morphol Embryol. 1999;28:377–397. [Google Scholar]
- Stewart MK, Clark NL, Merrihew G, Galloway EM, Thomas JH. High genetic diversity in the chemoreceptor superfamily of Caenorhabditis elegans. Genetics. 2005;169:1985–96. doi: 10.1534/genetics.104.035329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Paup*: Phylogenetic Analysis Using Parsimony and Other Methods. 4th edn. Sinauer Press; New York: 1998. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Goes van Naters W, Carlson JR. Insects as chemosensors of humans and crops. Nature. 2006;444:302–307. doi: 10.1038/nature05403. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wanner KW, Anderson AR, Trowell SC, Theilmann DA, Robertson HM, Newcomb RD. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol Biol. 2007;16:107–119. doi: 10.1111/j.1365-2583.2007.00708.x. [DOI] [PubMed] [Google Scholar]
- Wetzel CH, Behrendt H, Gisselmann G, Storkuhl KF, Hovemann B, Hatt H. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proc Natl Acad Sci USA. 2001;98:9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Zwiebel LJ. Identification and characterization of an odorant receptor from the West Nile virus mosquito, Culex quinquefasciatus. Insect Biochem Mol Biol. 2006;36:169–176. doi: 10.1016/j.ibmb.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharuk RY, Yin LR-S, Blue SG. Fine structure of the antenna and its sensory cone in the larvae of Aedes aegypti (1.). J Morphol. 1971;135:273–298. doi: 10.1002/jmor.1051350303. [DOI] [PubMed] [Google Scholar]
- Zwiebel LJ, Takken W. Olfactory regulation of mosquito–host interactions. Insect Biochem Mol Biol. 2004;34:645–652. doi: 10.1016/j.ibmb.2004.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.