Abstract
Purpose
Gastric cancer may be subdivided into three distinct subtypes –proximal, diffuse, and distal gastric cancer– based on histopathologic and anatomic criteria. Each subtype is associated with unique epidemiology. Our aim is to test the hypothesis that these distinct gastric cancer subtypes may also be distinguished by gene expression analysis.
Experimental Design
Patients with localized gastric adenocarcinoma being screened for a phase II preoperative clinical trial (NCI 5917) underwent endoscopic biopsy for fresh tumor procurement. 4–6 targeted biopsies of the primary tumor were obtained. Macrodissection was performed to ensure >80% carcinoma in the sample. HG-U133A GeneChip (Affymetrix) was used for cDNA expression analysis, and all arrays were processed and analyzed using the Bioconductor R-package.
Results
Between November 2003 and January 2006, 57 patients were screened to identify 36 patients with localized gastric cancer who had adequate RNA for expression analysis. Using supervised analysis, we built a classifier to distinguish the three gastric cancer subtypes, successfully classifying each into tightly grouped clusters. Leave-one-out cross validation error was 0.14, suggesting that >85% of samples were classified correctly. Gene set analysis with the False Discovery Rate set at 0.25 identified several pathways that were differentially regulated when comparing each gastric cancer subtype to adjacent normal stomach.
Conclusions
Subtypes of gastric cancer that have epidemiologic and histologic distinction are also distinguished by gene expression data. These preliminary data suggest a new classification of gastric cancer with implications for improving our understanding of disease biology and identification of unique molecular drivers for each gastric cancer subtype.
Keywords: Gastric Cancer, cDNA Expression, classification, pathways
Introduction
Gastric cancer is the second most common cause of cancer-related mortality worldwide with 700,349 deaths annually, and is the third most common malignancy worldwide with 974,000 new cases in the year 2000(1). Gastric cancer has been considered a single heterogenous disease with several epidemiologic and histopathologic characteristics; for the purposes of medical management, gastric cancer is treated in a uniform fashion, without regard to subtype. Pathologically, gastric adenocarcinoma may be distinguished according to the Lauren’s classification as intestinal, diffuse, or mixed subtypes (2). Epidemiologically, intestinal gastric cancer, particularly of the antrum, is strongly associated with chronic inflammation (i.e. atrophic gastritis (3–4)) often as a consequence of chronic infection with H. pylori (5–6). Conversely, inflammation is characteristically absent in the development of Lauren’s diffuse type gastric cancer, particularly when as a result of a germline mutation in CDH1 (7). Anatomically, proximal gastric cancer may be classified as a third type of gastric cancer, as tumors of the gastric cardia/gastroesophageal junction (GEJ), for which inflammation of a different type (i.e. chronic gastric acid/bile reflux) may be the driving force for carcinogenesis (8–9). Proximal/GEJ tumors are also usually not diffuse in histology, similar to distal non-diffuse gastric cancer.
As noted above, at present, the histopathologic, anatomic, and epidemiologic distinctions that subdivide this disease are not taken into account in the clinical management of the disease, for either initial potentially curative treatment or in palliation of advanced disease. For patients with metastatic disease, the available cytotoxic agents are applied indiscriminately to all disease subtypes, and with only modest success (reviewed (10)). In other epithelial malignancies, such as breast (11–12) and lung adenocarcinoma (13), the identification of specific molecular phenotypes have had profound implications for treatment strategies and continued drug development (14–15). We hypothesize that gastric cancer represents at least three entirely different malignancies arising in the same organ, each with different initiating pathologic processes, and each possibly having different tumor biology. If this is true, this disease classification may lead to different treatment paradigms for individual gastric cancer subtypes.
Clinical indicators in support of this hypothesis include the suggestion that proximal gastric tumors have a worse prognosis, stage for stage, when compared to distal tumors (16), that Lauren’s diffuse gastric cancers appear to have a different pattern of spread and behaviour than intestinal gastric adenocarcioma (17), and Her2 overexpression incidence is different between intestinal and diffuse types of gastric cancer (18). We hypothesize that different tumors arising from the stomach may be distinguished at the genomic level. The implications of this new molecular classification would be significant as they would imply the presence of unique molecular drivers and unique molecular pathways for each gastric cancer subtype that may be exploited to identify prognostic and predictive biomarkers and to identify unique targets for therapy. Herein, we present our preliminary evidence as a test set supporting a molecular classification of gastric cancer into three individual diseases, supported by histopathologic and epidemiologic characteristics.
Methods
Study Population
From May 2003 to January 2006, we screened patients with gastric or gastroesophageal adenocarcinoma by endoscopic ultrasound, laparoscopy, CT scan and PET scan for enrolment in an NCI-sponsored neoadjuvant clinical trial of irinotecan and cisplatin chemotherapy followed by surgical resection (19). This protocol was reviewed and approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center and by the National Cancer Institute (NCI #5917, NCT00062374). Written informed consent was obtained from each patient. All patients without evidence of metastatic disease on CT scanning underwent pre-operative evaluation including endoscopic evaluation with ultrasound during which an endoscopic biopsy was performed for the procurement of fresh tumor tissue for RNA extraction and analysis. All endoscopic tumor biopsies were performed prior to initiation of any treatment for the malignancy.
Endoscopy and Endoscopic Biopsy
All patients underwent standard video endoscopy using the Olympus gastroscope GIF-160 (Olympus America, Melville, NY). Targeted biopsies of the gastric mass, ulcer edge or thickened folds were obtained using the Bard Precisor EXL coated disposable biopsy forceps (Bard International, Murray Hill, NJ). Four to 6 biopsies were performed for each patient, with each biopsy usually measuring approximately 2–3 mm in diameter. Upon receiving the biopsy tissue from the endoscopic biopsy forceps, a small sample was placed immediately into buffered formalin (for histopathologic evaluation) or saline (for immediate freezing) while still in the endoscopy suite. The specimens in saline were immediately transported to the Tumor Procurement Lab where they were individually placed in OCT media and frozen at −80°C. The time from obtaining the biopsy to OCT was less than 15 minutes. The formalin fixed samples were submitted to the Pathology Department for routine processing.
Specimen Analysis for RNA Processing
OCT embedded biopsy samples were maintained below −20°C during processing. Frozen section slides were made from OCT embedded biopsy samples and were then stained with H&E and reviewed by reference pathologists (LT, DSK). The presence or absence of invasive adenocarcinoma and the extent of malignant and non-malignant cell involvement of the sample was recorded. We defined an adequate biopsy specimen as one having a proportion of at least 80% carcinoma nuclei. Macrodissection was performed in a specimen dissection chamber maintained at −20°C, using the marked H&E slide as a guide enabling us to remove OCT and non-malignant tissue from the carcinoma. Following macrodissection, the remaining biopsy sample that was then primarily gastric adenocarcinoma was sent in liquid nitrogen to the Genome Core laboratory for RNA extraction and processing. Although samples varied in size (up to about 5 mm in diameter), most samples were approximately 2 mm in diameter. Samples that were less than 1 mm were unable to be processed by macrodissection. These minute samples were submitted for RNA extraction and processing only if they contained 100% cancer on the H&E reference slides. In cases where individual patients had several biopsy samples that were suitable for RNA processing, the samples were combined for RNA processing. Throughout these tissue handling procedures, care was taken to use RNAase free gloves and laboratory equipment to minimize contamination.
RNA isolation, probe preparation, and microarray hybridization
Total RNA was isolated from tumor specimens using RNeasy columns (Qiagen), and all samples were treated on the column with RNase-free DNase. The quality of RNA was verified before labeling by analyzing 20–50 nanograms (ng) of each sample using the RNA 6000 NanoAssay and a Bioanalyzer 2100 (Agilent). Samples with a 28S/18S ribosomal peak ratio of 1.8–2.0 were considered suitable for labeling. For samples meeting this standard, 1.2 – 2 micrograms (mcg) of total RNA (depending on availability) was used for cDNA synthesis using an oligo-dT-T7 primer and the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). Synthesis, linear amplification, and labeling of cRNA were accomplished by in vitro transcription using the Message Amp aRNA Kit (Ambion) and biotinylated nucleotides (Enzo Diagnostics). Ten mcg of labeled and fragmented cRNA were then hybridized to the Human Genome U133A GeneChip (Affymetrix), which contained 22,215 oligonucleotide-based probe sets, at 45°C for 16 hours. Post hybridization staining, washing were processed according to manufacturer (Affymetrix) guidelines. Finally, chips were scanned with a high-numerical Aperture and flying objective (FOL) lens in the GS3000 scanner (Affymetrix). The image was quantified using MAS 5.1 (MicroArray Suite, Affymetrix) with the default parameters for the statistical algorithm and all probe set scaling with a target intensity of 500.
Definition of Gastric Cancer Subtypes
Three individual types of gastric cancer are strongly suggested by clinical and epidemiologic data (20). Using these characteristics, we defined the subtypes of gastric cancer histopathologically as follows:
Proximal non-diffuse gastric cancer – The bulk of the tumor (>80%) is located in the gastric cardia which may extend up to the gastroesophageal junction and small portion of the distal esophagus. On histopathology, there is evidence of precursor glandular dysplasia or in situ carcinoma in the setting of chronic inflammation usually without atrophy. Tumor differentiation may range from well to poorly differentiated, but the pattern of tumor infiltration should not be entirely diffuse.
Diffuse Gastric Cancer – Tumor location may be anywhere in the stomach. On histopathology, there is no apparent gastritis, neither severe chronic nor atrophic. The pattern of infiltration is entirely diffuse without excessive extracellular mucin pools (colloid carcinoma is not included). There should not be any component of gland-forming intestinal-type carcinoma. The tumor is poorly differentiated signet ring cell-type either with or without intracellular mucin.
Distal non-diffuse Gastric Cancer – The bulk of the tumor is usually located in the distal stomach and may extend up to the mid body of the stomach or down to the pylorus. On histopathology, there is evidence of chronic gastritis with intestinal metaplasia and a spectrum of glandular dysplasia and in situ carcinoma. The dominant pattern is a moderately differentiated and intestinal type carcinoma without or with minor components of poorly differentiated or de-differentiated carcinoma.
Patients were assigned to an individual subtype of gastric cancer based on the histopathologic and anatomic definitions above and the expression arrays were analyzed.
Bioinformatics
The affymetrix (HG-133A/B) dataset that contains 38 gastric tumor samples and 31 normal samples taken from stomach tissues adjacent to cancerous tissues was downloaded from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/ GSE accession number GSE13911) D’Errico et al (21). The GEO and MSKCC datasets contain 120 arrays that were processed and analyzed using the Bioconductor suite of tools in R-statistical language (www.bioconductor.org). All data were normalized using standard GCRMA functions with default parameters. Normalized data was clustered using hierarchical clustering based on Euclidian distance (hclust R-function) to verify that there is no batch effect between the two data-sets (see supplemental figure 1). Indeed, normal and tumor samples were mostly grouped together and not according to their batch. We next removed the tumor samples from the GEO dataset as there is no information on their gastric cancer type, resulting in 73 samples for further analysis (37 normal stomach and 36 gastric cancers). Probes that are present in at least half of the normal samples or half of the tumor samples were retained for further analysis (23828 probes). For analysis at the gene level, multiple probes corresponding to one gene were averaged, and probes without a gene symbol annotation were removed leaving 8740 unique genes. Differential expression analysis was done using limma R-package. Probes/genes were declared differentially expressed with fold-change cut-off greater than 2, and False Discovery Rate (FDR)=0.01 (i.e. up to 1% of the significant differences are expected to be false positives). For subtype classification only tumor samples from MSKCC were considered. To build the features (=genes) that separate different subtypes of gastric cancer, the dataset was filtered and genes with fold-changes greater than log2(1.5) in any pair-wise comparison (“proximal non-diffuse” vs “diffuse”, “proximal non-diffuse” vs “distal non-diffuse, and “diffuse” vs “distal non-diffuse”) were considered. An additional condition of (unadjusted) p-value≤0.005 in any of 3 comparison was also used. The resulting dataset used for the learning classifier has data for 785 genes and 36 tumor samples. Results are similar if probes are selected based on other criteria. To build the classifier, we opted for a supervised classification algorithm that implements regularized regression with the optimal scoring algorithm (22–23). This algorithm includes a procedure for finding gene signatures by ranking genes based on the fitted regression models. This methodology includes principal components, partial least squares, and ridge regression models. It has been applied to several microarray studies in cancer (22) and it is coded in R-package pdmclass. We used ridge regression methodology for the classification. In addition, we used Gene Set Analysis (GSA)(24) for an exploratory analysis to determine if the members of a given gene set were concordantly up or down regulated between gastric cancer subtypes and normal stomach. GSA was run with default parameters and number of permutations =500 to compute p-values, with FDR=0.25 as suggested in the GSEA manual (http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). The gene set enrichment analysis was carried out using the Molecular Signature Database (MSigDB) v2.5 released April 7, 2008. Functional analysis of differentially expressed genes was done using the DAVID tool (http://david.abcc.ncifcrf.gov/home.jsp) using all human genes as a background set.(25)
Results
Patient and Tumor Characteristics
Between November 2003 and January 2006, 57 patients with localized gastric adenocarcinoma based on CT scan imaging of the chest, abdomen, and pelvis underwent endoscopic biopsy and tissue biopsy. Of these 57 patients, tumor biopsy samples from 41 patients (72%) were adequate for RNA processing and analysis. Subsequent staging procedures (i.e. laparoscopy and FDG-PET scan) identified occult metastatic disease in 5 patients, leaving 36 patients with non-metastatic gastric cancer in the final study population (see table 1). The majority of patients had locally advanced, poorly differentiated tumors, with approximately 60% of the cases node positive on preoperative evaluation. Proximal non-diffuse gastric cancers (n=12) were more commonly Lauren’s intestinal histology. Diffuse gastric cancers (n=10) were more commonly anatomically located in the body or distal stomach, were uniformly poorly differentiated and Lauren’s diffuse histology by definition. Distal non-diffuse gastric cancers (n=14) were predominantly intestinal and mixed Lauren’s histology.
Table 1.
Patient and tumor characteristics.
| Patient Characteristic | Total population n=36 | Proximal non-Diffuse (n=12) | Diffuse (n=10) | Distalnon-diffuse (n=14) | |
|---|---|---|---|---|---|
| Age | |||||
|
| |||||
| Median (range) | 59 (25–74) | 62 (33–74) | 59 (25–70) | 58 (52–71) | |
|
| |||||
| Gender | |||||
|
| |||||
| Male/Female | 8 M/4F | 2 M/7 F | 7 M/7 F | ||
|
| |||||
| Histopathology | |||||
|
| |||||
| Differentiation | Mod-Poor | 12 | 6 | 0 | 6 |
| Poorly | 24 | 6 | 10 | 8 | |
|
| |||||
| Lauren’s | Diffuse | 14 | 0 | 10 | 0 |
| Intestinal | 19 | 9 | 0 | 10 | |
| Mixed | 8 | 3 | 0 | 4 | |
|
| |||||
| Location | |||||
|
| |||||
| GEJ/Proximal | 15 | 12 | 3 | 0 | |
| Body/Distal | 21 | 0 | 7 | 14 | |
|
| |||||
| Stage at EGD | |||||
|
| |||||
| Node positive | 23(64%) | 9(69%) | 5 (56%) | 9(64%) | |
| Early (T1–T2, N0, M0) | 7 | 0 | 2 | 5 | |
| Locally Advanced (T3–4 or N+) | 29 | 12 | 8 | 9 | |
Gastric Cancer Subtypes and Normal Stomach
Our aim was to examine whether genomic signatures would significantly differentiate gastric cancer subtypes that were assigned solely based on anatomic and histopathologic knowledge. We first compared gastric cancer versus normal stomach. We examined expression data from two independent data sets (MSKCC data 36 tumor, 10 adjacent normal stomach and D’Errico data (21) 38 tumor, 31 adjacent normal stomach). (21)Tumor samples and normals from both datasets cluster primarily according to their malignancy status (normal, tumor), and not according to the dataset (see supplemental Figure 1). Then, when evaluating MSKCC gastric tumors (i.e. study population that was annotated according to gastric cancer subtype) versus normal adjacent stomach, we identified a large number of genes that were differentially expressed in each type of gastric cancer versus normal (limma analysis, FC cut-off=2, FDR=0.01; see Figure 2). We noted that while there is a large overlap in the genes differentially expressed between gastric cancer types and normal, a significant number of genes uniquely differentiate each subtype of gastric cancer from normal stomach. Direct comparison of tumor subtypes (full dataset) yielded 3 genes (with FDR=0.05) that are differentially expressed between proximal non-diffuse and diffuse gastric cancer subtypes, including PSCA (prostate stem cell antigen) and PGA3 (pepsinogen A) which were both down-regulated >20 fold in proximal non-diffuse gastric cancer when compared with diffuse gastric cancer, and TRIM32 which was upregulated (>2 fold) in proximal non-diffuse gastric cancer.
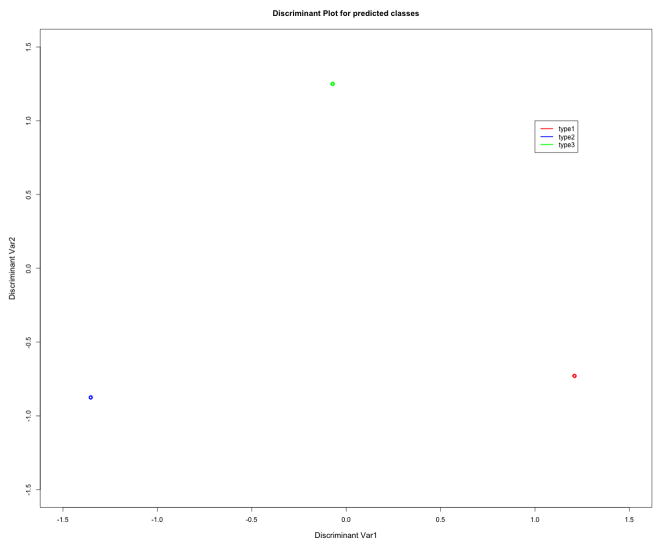
Figure 2.
Discriminant Plot for Gene Signatures showing that the RNA expression signatures of samples from different gastric cancers are quite well separated and very tightly grouped into 3 types of gastric cancer (supplemental table 1): Proximal non-diffuse (type 1), Diffuse (type 2), and Distal non-diffuse (type 3). This sample classification was done using pdmclass R-function, using ridge regression. The leave-one-out cross validation error is 0.14, which implies that >85% samples are classified correctly. The output from pdmClass cross-validation confusion matrix is provided to the right of the discriminant plot.
We then performed functional categories analysis on groups of differentially expressed genes using the DAVID tool. Genes that were upregulated in all three gastric cancer types versus normal are significantly enriched in many typical cancer-related categories including cell cycle, cell proliferation, cell adhesion, platelet derived growth factor binding, and EGF-domain, while genes that are downregulated in all gastric cancer types versus normal are enriched in digestion, disease mutation, and lipid metabolism. Similarly, genes that are up regulated in proximal and distal non-diffuse gastric cancer (but not diffuse gastric cancer) are enriched in numerous cell cycle and mitosis related categories, as well as p53 signaling pathways, while genes downregulated in both non-diffuse gastric cancer subtypes are enriched in digestion, drug metabolism, and response to various stimuli (nutrient levels, hormone stimulus, organic substance).
Gene Expression Analysis and Development of Molecular Classification
We next focused on identifying gene signatures that can be used to classify gastric cancer subtypes. We used ridge regression method on a smaller set of genes (785 genes; see Methods). (22–23)
The classifier built on these genes separates the three subtypes quite well and the samples are tightly grouped into three distinct groups (see Figure 2). The leave-one-out cross validation error is 0.14 which implies that >85% samples are classified correctly. In our patient population of 36 patients, 29 tumors were locally advanced (T3 or greater, or N+). When limiting the analysis to these 29 cases, the leave-one-out cross validation error is 0.13, implying that still > 85% of locally advanced samples are classified correctly. The top genes that separate proximal non-diffuse and diffuse gastric cancer include PGA3, PSCA, XIST, SST, ABCA8 (down regulated in proximal non-diffuse versus diffuse by over 2-fold), and PRF1, CXCL9, CXCL10, IF144L, PLA2G2A (upregulated). Prostate stem cell antigen (PSCA), for example, was over 20 fold diminished in proximal non-diffuse relative to diffuse gastric cancer subtypes. The top genes that separate proximal non-diffuse from distal non-diffuse gastric cancer include MSLN, IGJ, ENPP4, PLA2G2A (downregulated in proximal non-diffuse vs distal non-diffuse) and PF4V1, HMBO1, CYP2J2, DSC3, and S100a12 (upregulated). Notably, PLA2G2A is nearly 7-fold upregulated in proximal non-diffuse versus diffuse gastric cancer (mean fold-change) and nearly 12 fold upregulated in distal non-diffuse versus diffuse gastric cancer. The top genes that separate diffuse gastric cancer from distal non-diffuse gastric cancer include ABCA8 (≥ 4 fold), HMBOX1, COCH, S100A12, CYP2J2 (up regulated in diffuse vs. distal non-diffuse), and IFI44L (4 fold), HOXA9, MSLN, and ENPP4 (down regulated in diffuse vs. distal non-diffuse).
Gene Set Analysis
In addition, we applied the Gene Set Analysis tool to perform exploratory pathway analysis by comparing each gastric cancer subtype to normal, as well as by performing direct subtype comparisons. Pathway analysis may be indicative of underlying biological processes Table 2 provides the list of pathways that were either upregulated or downregulated with FDR=0.25. We observed that proximal and distal non-diffuse gastric cancers had a number of upregulated pathways including glycolysis and gluconeogenesis when compared to normal stomach, whereas no pathways were identified to be upregulated in diffuse gastric tumors. Conversely, each gastric tumor subtype shows downregulation of the KEGG HSA05222 (small lung cancer) pathway versus normal stomach. This pathway contains several tumor suppressors, including p53, PTEN, RB and FHIT.
Table 2.
Pathways and a priori defined sets of genes that show statistically significant, concordant differences between each gastric cancer subtype and normal stomach (using Gene Set analysis, GSA with FDR= 0.25).
| Significant pathways between “Proximal non-diffuse” vs Normal Stomach | |
|---|---|
| Upregulated in “Proximal non-diffuse” | Downregulated in “Proximal non-diffuse” |
| Bile acid biosynthesis HSA00624_1_AND_2_METHYLNAPHTHALENE_DEGRADATION HSA00641_3_CHLOROACRYLIC_ACID_DEGRADATION TYROSINE_METABOLISM GLUCONEOGENESIS GLYCEROLIPID_METABOL GLYCOLYSIS HSA00561_GLYCEROLIPID_METABOLISM HSA00980_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450 HSA00010_GLYCOLYSIS__AND_GLUCONEOGENESIS |
HSA05222_SMALL_CELL_lung cancer |
| Molecular Functions | |
| ATPASE_ACTIVITY__COU HYDROLASE_ACTIVITY PRIMARY_ACTIVE_TRANS ACTIVE_TRANSMEMBRANE HORMONE_ACTIVITY |
METALLOENDOPEPTIDASE_ACTIVITY” RNA_DEPENDENT_ATPASE_ACTIVITY ATP_DEPENDENT_RNA__HELICASE_ACTIVITY |
| Significant pathways differentiate “Diffuse” vs Normal Stomach | |
| Upregulated in “Diffuse” | Downregulated in “Diffuse” |
| HSA04512_ECM_RECEPTOR_INTERACTION HSA05222_SMALL_CELL_lung cancer |
|
| Significant pathways that differentiate “Distal non-diffuse” vs Normal Stomach | |
| Upregulated in “Distal non-diffuse” | Downregulated in “Distal non-diffuse” |
| GLYCEROLIPID_METABOLISM HSA00010_GLYCOLYSIS__AND_GLUCONEOGENESIS HSA00624_1_AND_2_METHYLNAPHTHALENE_DEGRADATION BILE_ACID_BIOSYNTHES”IS GLUCONEOGENESIS GLYCOLYSIS PROPANOATE_METABOLISM TYROSINE_METABOLISM VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION HSA00280_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION HSA00640_PROPANOATE__METABOLISM HSA00641_3_CHLOROACRYLIC_ACID_DEGRADATION |
HSA05222_SMALL_CELL_ |
| Biologic Processes | |
| COVALENT_CHROMATIN_MODIFICATION HISTONE_MODIFICATION |
REGULATION_OF_DNA_REPLICATION REGULATION_OF_MITOTIC_CELL_CYCLE |
| Molecular Functions | |
| ATPASE_ACTIVITY__COUPLED_TO_MOVEMENT_OF_SUBSTANCES HYDROLASE_ACTIVITYACTING_ON_ACID_ANHYDRIDES__CATALYZING_TR ANSMEMBRANE_MOVEMENT_OF_SUBSTANCES PRIMARY_ACTIVE_TRANSMEMBRANE_TRANSPORTER_ACTIVITY ATPASE_ACTIVITY__COULED_TO_TRANSMEMBRANE_MOVEMENT_OF_IONS COENZYME_BINDING STEROID_BINDING LIPID_BINDING |
|
Discussion
Gastric cancer is a heterogeneous disease with differences in epidemiology and histopathology that, when coupled with anatomic location, may be distinguished into at least 3 different cancers (20). We explored this hypothesis by examining the gene expression of individual gastric cancer subtypes in a training set, performing a comparison with the expression analysis of adjacent normal stomach as well as amongst individual gastric subtypes. We found that individual diseases arising from the stomach defined a priori, namely “proximal non-diffuse”, “diffuse”, and “distal non-diffuse” gastric cancer, have distinct gene expression. These findings from our training set are both clinically consistent and have significance in the context of current clinical management of gastric cancer. For example, two large global studies evaluating new cytotoxic and biologic therapies have failed to demonstrate a survival benefit over the standard of care (26–27). The investigators suggested that this may be partially due to failure to appreciate biological differences in gastric cancer and how these differences may affect response to treatment.
When comparing gastric cancer subtypes to adjacent normal, we identified a significant number of genes that uniquely distinguished individual gastric cancer subtypes from normal stomach. These data provide support for the hypothesis that gastric cancer subtypes may be distinguished molecularly.
Then, using supervised classification, we demonstrate a greater than 85% ability to successfully distinguish gastric cancer subtypes by gene expression. This was the case when examining all cases (both early and advanced) and also when limiting the cases to advanced disease. One gene of interest that significantly distinguished proximal non-diffuse gastric cancer from diffuse gastric cancer, prostate stem cell antigen (PSCA), has already been implicated in gastric cancer. An intronic polymorphism (rs2294008) in PSCA, resulting in reduced PSCA expression, is significantly associated with increased risk for diffuse gastric cancer compared with an intestinal subtype (virtually all distal non-diffuse) in a Japanese population (28–29). Expression of another gene, PLA2G2A, a secreted phospholipase, has prognostic significance in gastric cancer. Specifically, tumors expressing high levels of PLA2GA2 have improved survival compared with patients with low PLA2G2A expressing tumors (30) Recently, it was demonstrated that PLA2G2A is a target of Wnt/β-catenin signalling in gastric cancer cells, and is associated with negative regulation of genes associated with invasion and metastasis (31). Using gene set analysis, a number of pathways were either up- or downregulated when individual gastric cancer subtypes were compared to normal stomach. For example, the glycoloysis pathway is upregulated in proximal and distal non-diffuse gastric cancers relative to diffuse gastric cancer. Glycolysis is the process of converting glucose into pyruvate and generating small amounts of ATP. It is a central pathway that produces important precursor metabolites, and may explain the increased glucose metabolism of many cancers (i.e. Warburg effect (32)), and is commonly linked with FDG-avidity for PET scanning. Consistent with this finding, diffuse gastric cancers (i.e. those that did not demonstrate upregulation of glyocolysis on GSA) are commonly FDG non-Avid, unlike their non-diffuse counterparts.
Oncogenic pathways have been used previously to identify pathway signatures in malignancy, and similar to this report, to identify pathway signatures in subtypes of malignancy such as breast cancer (33–34). Ooi and colleagues examined a subset of oncogenic and tumor suppressor pathways in gastric cancer, including the RAS pathway (35–36) which was identified in our gene set analysis as downregulated in proximal non-diffuse gastric cancer when compared with diffuse gastric cancer. These investigators suggested that combinations of several pathways may provide greater predictive value for patient outcomes than individual pathways themselves (35). Specifically, they noted three pathways were dysregulated in > 70% of gastric cancers: proliferation/stem cell; NF-kB, and Wnt/Beta catenin. They validated the pathways in gastric cancer derived cell lines (35), however the location of the primary tumor was not included in their analysis – that is, whether or not the different pathways were associated with proximal, diffuse, or distal gastric cancer subtypes. Similarly, in another gene expression analysis, investigators identified three subgroups of cancer as “tumorigenic”, “reactive”, and “gastric-like”(37). In their analysis, there was no association between intestinal and diffuse Lauren classification, or between tumor sites. Notably, both studies suggest gastric cancer may be subdivided genomically, with different prognoses, independent of stage (35, 37). However, no study to date has incorporated epidemiological and histopathologic data together with anatomic location, as we have done, to define subtypes of gastric cancer. Based on epidemiology and pathology, we proposed a division of gastric cancer into three distinct types of gastric cancer (20). Chronic inflammation (e.g. from H. pylori infection) is required for the development of distal gastric cancer, usually intestinal type (38), and a diet high in fruits or vegetables is protective for this type of gastric cancer. Proximal gastric cancer is most associated with obesity and gastroeophageal reflux disease (9), perhaps causing inflammation via different pathways than in distal non-diffuse gastric cancer, whereas diffuse gastric cancer does not currently have established environmental or clinical risk factors (20). Our genomic analysis data confirm a clear molecular distinction in these types of gastric cancer. The value of this classification may be demonstrated even with currently defined biomarkers in gastric cancer, namely Her-2-neu. This gene is amplified or overexpressed in approximately 25% of gastric cancer cases, and overexpression convers sensitivity to Her-2 targeted therapy, and importantly a significant survival advantage when patients with Her2 positive tumors are treated with trastuzumab and chemotherapy (39). Her2 amplification or overexpression is not uniform across gastric cancer subtypes, most prevalent in proximal or GEJ gastric cancer (~30% Her2 positivity rate) and least prevalent in diffuse type gastric cancer (~5% Her2 positivity rate). Assessment of Her2 positivity rates, therefore, depend entirely on the constituent population studied, and will be higher in areas were proximal gastric cancers prevail, and less frequent where diffuse gastric cancers prevail.
These data are parallel to emerging analyses in other malignancies. For example, recent approaches of molecular classification of breast cancer have identified three distinct subclasses of breast cancer with both biologic and prognostic significance. These subclasses are defined as estrogen receptor (ER) and/or progesterone receptor (PR) positive tumors, HER-2 amplified tumors, and ER/PR/HER2 (triple) negative tumors. The three breast cancer subtypes have been reproducibly identified by gene expression profiling in multiple breast cancer cohorts and exhibit consistent prognostic significance (11–12). Clinical implications of this subclassification of breast cancer include the development of therapeutic strategies, such as the use of PARP inhibition for triple negative disease (14, 40), as well as potentially significant racial and ethnic ramifications, such as the identification of triple negative breast tumors as more prevalent in premenopausal African American woman (39%)(41).
In summary, for the first time to our knowledge, we have demonstrated that malignancies arising from the stomach that have epidemiologic and histologic distinction can also be distinguished by genomic/molecular analysis. These data have significant ramifications. Our analysis suggests that (1) different types of cancers arise from the stomach, (2) there likely exist unique molecular drivers that may be identified amongst specific genetic pathways that distinguish each disease, and (3) the presence of different biomarkers and therapeutic targets for each disease is also likely. We are performing a separate validation study to confirm the classification error estimate of our classifier. However, these data provide corroborating molecular evidence of a new classification for gastric cancer. Ultimately, such distinction will allow us to begin to manage each of these diseases differently and uniquely. As we improve our understanding of gastric cancer heterogeneity and its clinical consequences, our hope is to improve patient outcomes with improved prevention, screening and treatment options, using distinct biologic subtypes for improved application of targeted therapies.
Supplementary Material
Figure 1.
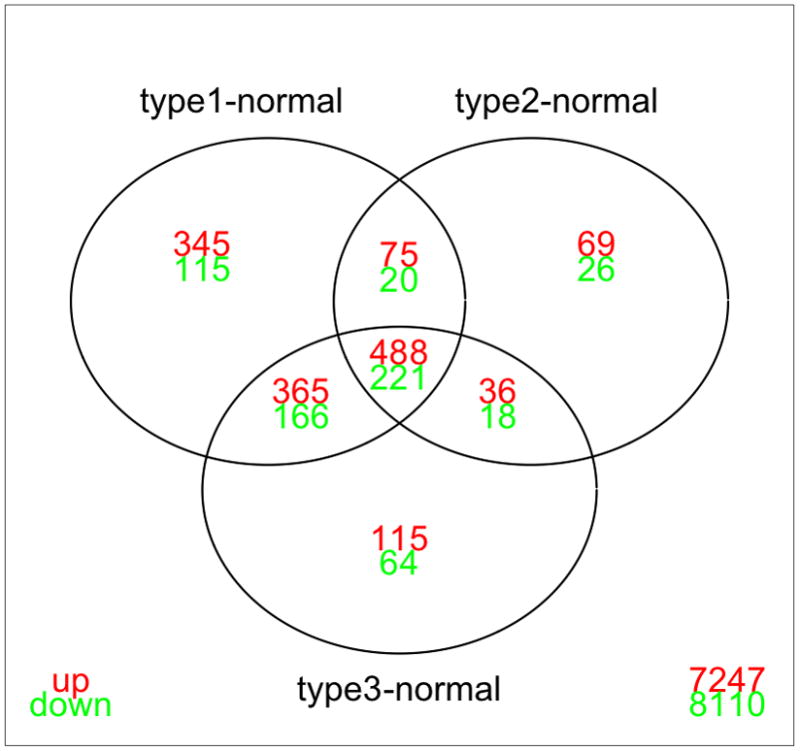
Venn Diagram demonstrating the distribution of probes that are significantly different between gastric cancer subtypes and normal stomach. Although there is a large overlap between differentially expressed genes in each subgroup comparison versus normal, a large number of genes uniquely differentiate each individual gastric cancer subtype and normal stomach. Analysis is done using limma (with fold-change cut-off 2, and FDR=0.01; values for multiple probe values for the same gene were averaged). Type 1 = proximal non diffuse, type 2 = diffuse, and type 3 = distal non-diffuse gastric cancer.
Acknowledgments
Supported by ASCO Career Development Award (Manish A. Shah, MD), NCI/CTEP (NCI# 5917), and NCI Contract for Early Drug Development (N01 CM 62206, David P. Kelsen, MD)
Footnotes
Individual Author Awards:
Manish A Shah: ASCO Career Development Award, FDA Orphan Disease Product Grant 1R01FD003755-01A1
David P Kelsen: NCI Contract N01 CM62206
References
- 1.Parkin DM, Bray FI, Ferlay J, Pisani P. Global cancer statistics: 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Lauren T. The two histologic main types of gastric carcinoma. Acta Pathol Microbiol Scand. 1965;64:34. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 3.Yuo WC, Blot WJ, Li JY, et al. Precancerous gastric lesions in a population at high risk of stomach cancer. Cancer Res. 1993;53:1317–21. [PubMed] [Google Scholar]
- 4.Correa P, Haenszel W, Cuello C, et al. Gastric precancerous process in a high risk population: cross-sectional studies. Cancer Res. 1990;50:4731–6. [PubMed] [Google Scholar]
- 5.Peek RMJ, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature RevCancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 6.Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 7.Carneiro F, Huntsman DG, Smyrk TC, et al. Model of the early development of diffuse gastric cancer in E-cadherin mutation carriers and it’s implications for patient screening. J Pathol. 2004;203:681–7. doi: 10.1002/path.1564. [DOI] [PubMed] [Google Scholar]
- 8.Blot WJ, DeVesa SS, Kneller RW, Fraumeni JF. Rising incidence of adenocarcinoma of the esophagus and gastric cardia. Journal of the American Medical Association. 1991:1287–9. [PubMed] [Google Scholar]
- 9.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–62. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Power DG, Kelsen DP, Shah MA. Advanced gastric cancer -Slow but steady progress. Cancer Treat Rev. 2010 doi: 10.1016/j.ctrv.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000:406. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 12.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Shaughnessy JO, Osborne C, Pippen J, Yoffe M. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized phase II trial. J Clin Oncol. 2009;27 abs 3. [Google Scholar]
- 15.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakaguchi T, Watanabe A, Sawada H, et al. Characteristics and clinical outcome of proximal-third gastric cancer. J Am Coll Surg. 1998;187:352–57. doi: 10.1016/s1072-7515(98)00191-4. [DOI] [PubMed] [Google Scholar]
- 17.Marrelli D, Roviello F, de Manzoni G, et al. Different patterns of recurrence in gastric cancer depending on Lauren’s histologic type: longitudinal study. World J Surg. 2002;26:1160–5. doi: 10.1007/s00268-002-6344-2. [DOI] [PubMed] [Google Scholar]
- 18.Hofman M, Stoss O, Shi D, Buttner R, van de Vijver M, et al. Assessment of a Her2 scoring system for gastric cancer: results from a validation study. Histopathology. 2008;52:797–805. doi: 10.1111/j.1365-2559.2008.03028.x. [DOI] [PubMed] [Google Scholar]
- 19.Shah MA, Yeung H, Coit D, et al. A phase II study ofpreoperative chemotherapy with irinotecan (CPT) and cisplatin (CIS) for gastric cancer (NCI 5917): FDG-PET/CT predicts patient outcome. Amercan Society of Clinical Oncology, Annual Proceedings; Orlando, FL. 2007. Anstract #4502. [Google Scholar]
- 20.Shah MA, Kelsen DP. Gastriccancer: A primer on the epidemiology and biology of the disease and an overview of the medical management of advanced disease. J Natl Compr Canc Netw. 2010;8:437–47. doi: 10.6004/jnccn.2010.0033. [DOI] [PubMed] [Google Scholar]
- 21.D’Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–69. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh D. Penalized discriminant methods for the classification of tumors from gene expression data. Biometrics. 2003:59. doi: 10.1111/j.0006-341x.2003.00114.x. [DOI] [PubMed] [Google Scholar]
- 23.Hastie T, Tibshirani R, Buja A. Flexible discriminant analysis by optimal scoring. J Am Statist Assoc. 1994;89:1255–70. [Google Scholar]
- 24.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107–29. [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Ajani JA, Rodriguez W, Bodoky G, et al. Multicenter phase III comparison of cisplatin/S-1 with cisplatin/infusional fluorouracil in advanced gastric or gastroesophageal adenocarcinoma study: the FLAGS trial. J Clin Oncol. 2010;28:1547–53. doi: 10.1200/JCO.2009.25.4706. [DOI] [PubMed] [Google Scholar]
- 27.Kang YK, Ohtsu A, Van Cutsem E, et al. AVAGAST: A randomized, double-blind, placebo-controlled, phase III study of first-line capecitabine and cisplatin plus bevacizumab or placebo in patients with advanced gastric cancer (AGC) J Clin Oncol. 2010;28:absLBA4007. [Google Scholar]
- 28.Cancer SGoMGPf. Sakamoto H, Yoshimura K, Saeki N, Katai H, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–40. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 29.Saeki N, Gu J, Yoshida T, Wu X. Prostate stem cell antigen: A Jekyll and Hyde molecule? Clin Cancer Res. 2010;16:3533–8. doi: 10.1158/1078-0432.CCR-09-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung SY, Chen XE, Chu KM, et al. Phospholipase A2 group IIA expression in gastric adenocarcinoma is associated with prolonged survival and less frequent metastases. Proc Natl Acad Sci U S A. 2002;99:16203–8. doi: 10.1073/pnas.212646299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganesan K, Ivanova T, Wu Y, et al. Inhibition of gastric cancer invasion and metastases by PLA2G2A, a novel beta-catenin/TCF target gene. Cancer Res. 2008;68:4277–86. doi: 10.1158/0008-5472.CAN-07-6517. [DOI] [PubMed] [Google Scholar]
- 32.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker M, Sommer A, Kratzschmar JR, Seidel H, Pohlenz HD, et al. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther. 2005;4:151–68. [PubMed] [Google Scholar]
- 35.Ooi CH, Ivanova T, Wu J, Lee MH, Tan IB, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLos Genet. 2009;5:e1000676. doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiyama T, Haruma K, Kitadai Y, Masuda H, Miyamoto M, et al. K-ras mutation in helicobacter pylori-associated chronic gastritis in patietns with and without gastric cancer. Int J Cancer. 2002;97:562–66. doi: 10.1002/ijc.1644. [DOI] [PubMed] [Google Scholar]
- 37.Tay ST, Leong SH, Yu K, et al. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309–16. [PubMed] [Google Scholar]
- 38.Correa P, Shiao YH. Phenotypic and genotypic events in gastric carcinogenesis. Cancer Res. 1994;54 (7 Suppl):1941s–43s. [PubMed] [Google Scholar]
- 39.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of Her2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010 doi: 10.1016/S0140-6736(10)61121-X. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 41.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.