Abstract
Mammalian gonad differentiation involves sexually dimorphic cell-fate decisions within the bipotential gonadal primordia. Testis differentiation is initiated by a center-to-poles wave of Sry expression that induces supporting cell precursors (SCPs) to become Sertoli rather than granulosa cells. The initiation of ovary differentiation is less well understood. We identified two novel SCP markers, 1700106J16Rik and Sprr2d, whose expression is ovary-biased during early gonad development, and altered in Wnt4, Sf1, Wt1 and Fog2 mutant gonads. In XX and XY gonads, both genes were upregulated at ~E11 in a center-to-poles wave, and then rapidly downregulated in XY gonads in a center-to-poles wave, which is reminiscent of Sry expression in XY gonads. Our data suggest that 1700106J16Rik and Sprr2d may have important roles in early gonad development, and are consistent with the hypothesis that ovarian SCP differentiation occurs in a center-to-poles wave with similar timing to that of testicular SCP differentiation.
Keywords: sex determination, organogenesis, gene expression, differentiation, Sry, ovary, testis
INTRODUCTION
Mammalian ovaries and testes develop from the paired bipotential genital ridges. It is commonly thought that the genital ridges contain the precursors of the three main cell types found in adult ovaries and testes – germ cells (oocytes and spermatocytes), supporting cells (granulosa and Sertoli cells) and steroidogenic cells (theca and Leydig cells) – and that each of these cell lineages undergoes a binary cell fate decision to adopt an ovarian or a testicular cell-type fate (Gillman, 1948; Lovell-Badge, 1992). In XX and XY gonads, the supporting cell precursors (SCPs) differentiate first (Odor and Blandau, 1969; Magre and Jost, 1980) and, at least in XY gonads, it is thought that this initial commitment decision influences the fate of the other cell lineages. In XY gonads the differentiation of SCPs into Sertoli cells is initiated by the expression of Sry (Sex determining region of Chr Y) (Koopman et al., 1991). In mouse XY genital ridges Sry is transiently expressed uniquely in the SCPs between embryonic days (E) 10.5 and 12.5 (Burgoyne et al., 1988; Koopman et al., 1991; Hacker et al., 1995; Jeske et al., 1995; Albrecht and Eicher, 2001; Bullejos and Koopman, 2001). It is first expressed in cells just below the coelomic epithelium in the center of the gonad and expression spreads toward the anterior and posterior poles (Albrecht and Eicher, 2001; Bullejos and Koopman, 2001). Shortly after expression reaches the poles, it is downregulated in a center-to-poles (C-P) wave (Bullejos and Koopman, 2001).
It is not known what triggers SCPs to differentiate as granulosa cells in XX gonads. Using an Sry-EGFP transgenic reporter mouse line we previously determined that Sertoli and granulosa cells are derived from a common precursor and that upregulation of Sry-EGFP expression in XX gonads was spatially and temporally identical to that in XY gonads (Albrecht and Eicher, 2001). These results showed that SCPs in both sexes are equally competent to specifically express Sry and implied that SCPs might differentiate in a C-P wave in both sexes. However, because Sry is not expressed during normal ovary development it was unclear if endogenous granulosa cell-specific genes would be expressed in a similar way.
To further investigate when and how ovary differentiation begins we sought to identify cell-type-specific molecular markers of early ovarian development. Because the Sry-EGFP reporter transgene is an early and specific marker of pre-granulosa cells in XX gonads, we utilized these mice, FACS sorting, and microarray analysis to identify genes differentially expressed between pre-granulosa cells and other presumably undifferentiated somatic cells in E13.5 ovaries (Lee and Albrecht, in preparation). Among the genes with the highest differential expression in pre-granulosa cells were Sprr2d (small proline-rich protein 2d) and 1700106J16Rik (RIKEN cDNA 1700106J16 gene). Here, we report the characterization of the spatiotemporal expression of these two genes in normal and mutant gonads. In XY gonads, the spatiotemporal expression of both genes was strikingly similar to that of Sry in that they were expressed in SCPs, were first detected in cells just below the coelomic epithelium, and up- and downregulated in a C-P wave. In XX and XY gonads, their expression was initially similar. However, in XX gonads their expression persisted and levels were higher at stages when the genes were expressed in both sexes. In addition, 1700106J16Rik and Sprr2d expression was altered in gonads from mutants with defects in early gonadal development. Of particular interest, 1700106J16Rik expression was increased, while Sprr2d expression was decreased in E12.5 Wnt4 (wingless-related MMTV integration site 4) null XX gonads, suggesting that these genes may be regulated by WNT4, a key regulator of ovarian development (Vainio et al., 1999; Jordan et al., 2001; Jeays-Ward et al., 2003; Yao et al., 2004; Kim et al., 2006). Our data suggest that 1700106J16Rik and Sprr2d may have important roles in early gonad development. Moreover, these data are consistent with the hypothesis that ovarian SCP (pre-granulosa cell) differentiation occurs in a C-P wave with similar timing to that of testicular SCP (pre-Sertoli cell) differentiation.
RESULTS
Sprr2d and 1700106J16Rik are preferentially expressed in pre-granulosa cells
To identify cell type-specific markers during early ovary differentiation, we isolated E13.5 ovaries from mice expressing an Sry-EGFP reporter transgene, dissociated them into single cells, and labeled the endothelial and germ cells with a fluorescently conjugated CD31 antibody. FACS analysis was used to differentially isolate the GFP-marked pre-granulosa cells and unlabeled somatic cells, and the red fluorescent endothelial and germ cells were discarded. Microarray analysis was used to identify genes differentially expressed between the pre-granulosa cells and the non-endothelial somatic cells. (The details of this microarray experiment will be published elsewhere.) Among the genes with the highest differential expression were Sprr2d and 1700106J16Rik, which were expressed four and five times higher, respectively, in pre-granulosa cells.
Sprr2d is one of eleven highly homologous mouse Sprr2 genes (Sprr2a-k) clustered on Chromosome 3, which encode a family of small proline-rich proteins (SPRRs) (Song et al., 1999). SPRR2 proteins appear to be present in a variety of mammalian species (Ensembl, www.ensembl.org). SPRRs are principally expressed during the terminal differentiation of stratified squamous epithelia (Kartasova et al., 1988; Hohl et al., 1995), and are proposed to be structural components of cornified envelopes (Steven and Steinert, 1994). In adult mice, Sprr2 genes are expressed in squamous cells of the skin, footpad, vaginal epithelia, digestive tract and respiratory epithelia (Tesfaigzi and Carlson, 1999). 1700106J16Rik is an uncharacterized gene that is predicted to encode a 152 amino acid protein with no obvious motifs (Okazaki et al., 2002). Ensembl orthologue prediction by reciprocal BLAST analysis indicates that the human, chimpanzee, rhesus monkey, dog, cow and rat (mammalian) genomes contain a gene that is highly homologous to 1700106J16Rik, while putative 1700106J16Rik orthologues were not predicted to exist in chicken, frog, fish, fly, or worm (non-mammalian) genomes. In adult mice, 1700106J16Rik expression appears to be largely restricted to testis and spinal cord (Mouse Gene Prediction Database, http://mgpd.med.utoronto.ca/) (Zhang et al., 2004). To our knowledge, 1700106J16Rik and Sprr2d expression during embryonic development previously has not been characterized.
To confirm that Sprr2d and 1700106J16Rik were differentially expressed within E13.5 ovaries, their expression was analyzed by semi-quantitative RT-PCR using RNA from cells isolated independently from those used in our microarray experiments (Fig 1A). The results indicated that indeed these genes are preferentially expressed in EGFP positive pre-granulosa cells. Three Sprr2 family members, Sprr2a, Sprr2f and Sprr2g, were identified in separate experiments as the only other Sprr2 genes significantly expressed in E11.5–E13.5 gonads (data not shown). These three Sprr2 genes also were preferentially expressed in pre-granulosa cells (Fig 1A), suggesting that Sprr2a, Sprr2d, Sprr2f and Sprr2g may be coordinately regulated during fetal gonad development, as they are during epidermal differentiation (Patel et al., 2003). However, Sprr2f and Sprr2g appear to be expressed at relatively low levels.
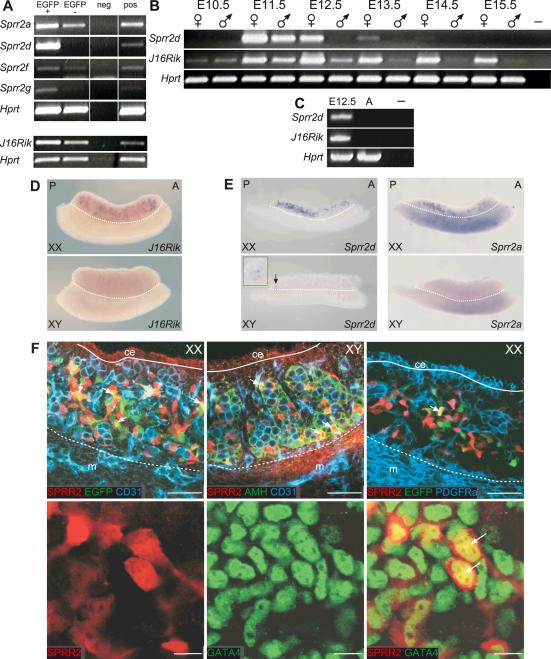
Figure 1. Expression analysis of Sprr2d and 1700106J16Rik in developing gonads.
Semi-quantitative RT-PCR analysis (A–C). (A) Expression of Sprr2a, d, f, g and 1700106J16Rik (J16Rik) in FACS-sorted cells from E13.5 B6 Sry-EGFP transgenic ovaries. Each gene was more highly expressed in EGFP+/CD31- cells (EGFP+) than in EGFP-/CD31- cells (EGFP-). (B) Analysis of Sprr2d and J16Rik expression in E10.5 urogenital ridges and E11.5–E15.5 gonads. In XX gonads (♀), Sprr2d expression was highest at E11.5 and rapidly decreased thereafter. In XY gonads (♂), Sprr2d was detected only at E11.5. J16Rik was expressed at very low levels in both sexes at E10.5, and expression increased at E11.5 in both sexes and was maintained in XX gonads until at least E15.5. In XY gonads, J16Rik expression decreased after E11.5, and was essentially undetectable by E13.5. Both genes were expressed more highly in XX than in XY gonads at each developmental stage analyzed. (C) Neither gene was expressed in adult ovaries. In (A–C) a reaction was performed without template as a negative control (neg or –), and Hprt served as the endogenous control. In (A) additional positive controls (pos) were performed using RNA from non-sorted cells from dissociated E13.5 B6 Sry-EGFP transgenic ovaries for the Sprr2 panel, or using RNA from intact E13.5 B6 ovaries for the J16Rik panel. In (C) E12.5 XX gonads were used as a positive control (leftmost lane). WISH analysis (D and E). (D) In E13.5 gonad/mesonephros complexes, J16Rik expression was detected in a distinct subset of cells within XX gonads, but not in XY gonads or mesonephroi of either sex. (E) In E12.5 XX gonads, Sprr2d and Sprr2a were expressed in discrete cells with a posterior bias. In E12.5 XY gonads, Sprr2d was expressed in just a few cells at the very posterior end (arrow and inset) and Sprr2a expression was not detected. A, anterior; P, posterior. Gonads are above the dotted line and mesonephroi are below. (F) Localization of SPRR2 protein(s) was analyzed using WIHC. E12.5 B6 Sry-EGFP transgenic XX gonads were labeled with antibodies to SPRR2 (red), EGFP (green) and CD31 (blue) (upper left panel). CD31 marks germ and endothelial cells and Sry-EGFP marks pre-granulosa cells. SPRR2 was expressed in a subset of CD31-negative somatic cells, some of which also expressed EGFP (yellow and arrows). E12.5 B6 XY gonads were labeled with antibodies to SPRR2 (red), AMH (green) and CD31 (blue) (upper middle panel). AMH marks Sertoli cells. SPRR2 was not expressed in CD31-positive germ cells or endothelial cells but was expressed in a subset of AMH-positive Sertoli cells (yellow and arrows). E12.5 B6 Sry-EGFP transgenic XX gonads were labeled with antibodies to SPRR2 (red) and PDGFRα (blue) (upper right panel). Sry-EGFP transgene expression (autofluorescence) is green. EGFP expressing pre-granulosa cells and SPRR2 expressing cells were located in the innermost regions of the gonad, and EGFP and SPRR2 expression overlapped in some cells (yellow and arrow). PDGFRα marks coelomic epithelium and interstitial somatic cells, and its expression did not overlap with either EGFP or SPRR2. In each upper row image, the coelomic epithelium (ce) is located above the solid line, and the mesonephros (m) below the dotted line. In the lower row, E13.25 XX gonads were labeled with antibodies to SPRR2 (red) and GATA4 (green). GATA4 marks somatic cell nuclei. SPRR2 was expressed in a subset of GATA4-positive cells, and SPRR2 protein was co-localized with GATA4 in cell nuclei (lower right panel, yellow and arrows). SPRR2 also was located in the cytoplasm. Scale bars: 50 μm (upper row), 10 μm (lower row).
When we examined Sprr2d and 1700106J16Rik expression by semi-quantitative RT-PCR in XX and XY gonads at different developmental stages, we found that these two genes displayed both sexually dimorphic and temporally dynamic expression (Fig 1B). Sprr2d was strongly expressed at E11.5 in both XX and XY gonads, and then became rapidly downregulated in XY gonads, such that no expression was detected at E12.5. In XX gonads, Sprr2d also was downregulated after E11.5, but its expression could be detected until E13.5. Sprr2d expression was not detected in E10.5, E14.5, or E15.5 gonads or in the adult ovary (Fig 1C). 1700106J16Rik was expressed at very low levels in both XX and XY E10.5 urogenital ridges (Fig 1B). By E11.5, 1700106J16Rik was upregulated in XX gonads, and also in XY gonads, but to a lesser extent. By E12.5, 1700106J16Rik was downregulated in XY gonads, while its expression continued to be upregulated in XX gonads and persisted until at least E15.5. Expression was not detected in the adult ovary (Fig 1C).
To further characterize the expression of these genes in embryonic gonads, we analyzed Sprr2d and 1700106J16Rik expression in E13.5 gonad/mesonephros complexes by whole-mount in situ hybridization (WISH) (Fig 1D and data not shown). Expression of 1700106J16Rik was not detected in the testis or mesonephros. In ovaries, expression was detected in discrete cells and was generally restricted to the central region (medulla). This medullary expression is similar to that seen for EGFP-expressing pre-granulosa cells in E13.5 Sry-EGFP transgenic ovaries (Albrecht and Eicher, 2001). These data indicate that 1700106J16Rik is expressed in a ovary-biased manner in E13.5 gonads, and preferentially, if not specifically, in pre-granulosa cells.
Expression of Sprr2d was not detected by WISH in E13.5 gonad/mesonephros complexes (data not shown), but was detected in E12.5 gonads (Fig 1E). At E12.5, expression was restricted to a subset of cells in both XX and XY gonads. In E12.5 XX gonads, Sprr2d was prominently expressed, but biased toward the posterior end. In E12.5 XY gonads, expression was detected only in a few cells at the very posterior, if at all. We also detected Sprr2a expression in E12.5 XX, but not XY, gonads by WISH, and expression was absent in the mesonephros (Fig 1E). Like Sprr2d, greater Sprr2a expression was detected in the posterior compared to the anterior half of the ovary. The similar temporal and spatial pattern of Sprr2d and Sprr2a downregulation is consistent with the idea that these genes are co-regulated during gonadal differentiation.
To further investigate whether Sprr2d is specifically expressed in the supporting cell lineage, we used an antibody that potentially recognizes multiple SPRR2 protein family members to determine the cellular localization of SPRR2 proteins in fetal gonads by whole-mount fluorescent immunohistochemistry (WIHC) and confocal microscopy (Fig 1F). In E12.5 XY gonads, SPRR2 expression was detected only within testis cords in a subset of AMH-expressing Sertoli cells. In E12.5 XX gonads, SPRR2 was expressed in a subset of somatic cells, and was not expressed in the coelomic epithelium, germ cells or endothelial cells. When we compared the expression of SPRR2 to that of PDGFRα, a marker of ovarian interstitial and coelomic epithelium cells (Lee and Albrecht, in preparation), we found that SPRR2 was not expressed in PDGFRα-expressing cells. This expression pattern is consistent with pre-granulosa cell expression, as the Sry-EGFP transgene, a specific marker of pre-granulosa cells, was expressed in a similar pattern. When we directly compared SPRR2 to Sry-EGFP expression, we found that a subset of SPRR2-expressing cells also expressed Sry-EGFP, and vice versa. Given that SPRR2 expression is restricted to a subset of pre-Sertoli cells in XY gonads, the lack of complete overlap in XX gonads might be explained by a difference in the timing of SPRR2 and Sry-EGFP expression among pre-granulosa cells. Taken together, these data indicate that Sprr2d is expressed uniquely in Sertoli cells in testes and in pre-granulosa cells in ovaries. We next investigated the subcellular localization of SPRR2s. Using GATA4 as a marker of somatic cell nuclei, we found that SPRR2 proteins were expressed in both the nucleus and cytoplasm in E13.25 XX gonads.
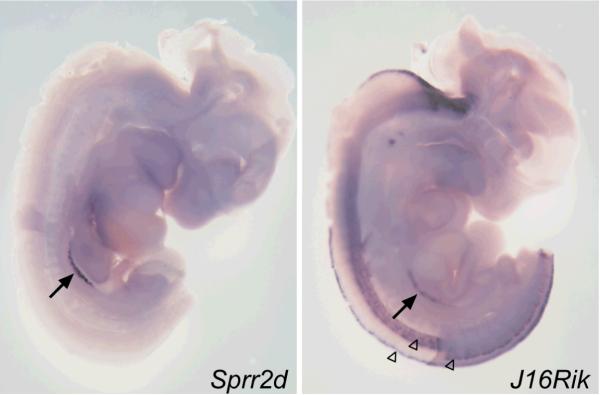
Available data suggested that the expression of Sprr2d and 1700106J16Rik in adults is restricted to a small number of tissues. To determine if the expression of these genes also was restricted during fetal development, we analyzed Sprr2d and 1700106J16Rik expression in E11.75 XX embryos by WISH (Fig 2). Sprr2d was expressed only in the gonads, while 1700106J16Rik was expressed in the gonads, and in parts of the hindbrain and spinal cord. Thus, at E11.75, Sprr2d and 1700106J16Rik have very restricted expression.
Figure 2. Spatial expression analysis of Sprr2d and 1700106J16Rik in E11.75 XX embryos.
In embryos bisected down the midline of the neural tube and analyzed by WISH, Sprr2d expression was detected only in the genital ridges (arrow) and 1700106J16Rik expression was detected in the genital ridges (arrow) and in specific regions of the developing spinal cord (open arrowheads) and hindbrain.
Sprr2d and 1700106J16Rik have distinctive and dynamic expression patterns
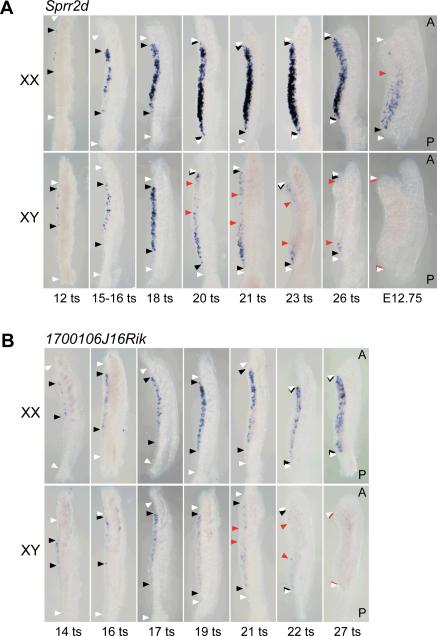
Because our initial data indicated that 1700106J16Rik and Sprr2d were preferentially expressed in SCPs in a sexually dimorphic and temporally dynamic pattern, we analyzed their expression using WISH in genital ridges from closely spaced developmental stages to determine whether these genes exhibit spatially dynamic expression. Sprr2d expression was first detected in gonads at about E11 (12 tail somite (ts) stage) in the central gonadal region that overlies the anterior mesonephric tubules (Fig 3A). Expression subsequently spread toward both the anterior and posterior poles until it was expressed throughout the length of the gonad by ~E11.6 (20 ts stage). Sprr2d expression was noticeably restricted to a distinct subset of cells located within the gonad, but was absent from the coelomic epithelium. Sprr2d was initially expressed in the same manner in both XX and XY gonads, but was then rapidly downregulated in XY gonads in a C-P wave that began after E11.5 (18 ts stage). Expression at the anterior pole was extinguished prior to expression at the posterior pole. In XX gonads at the same stages, Sprr2d expression continued to increase until ~E12.0 (25 ts stage), after which it was downregulated in an A-P wave. By E13.5, we could not detect Sprr2d expression by WISH in either ovaries or testes (data not shown).
Figure 3. Waves of Sprr2d and 1700106J16Rik expression in genital ridges.
WISH analysis of Sprr2d (A) and 1700106J16Rik (B) expression in early stage gonads. White arrowheads indicate the anterior and posterior ends of the developing gonads, black arrowheads indicate the anterior and posterior boundaries of gene upregulation, and red arrowheads indicate the anterior and posterior boundaries of gene downregulation. In each panel, the gonad is oriented to the left of the mesonephros. A, anterior; P, posterior. (A) Sprr2d was expressed initially in the central region of XX and XY gonads from 12 tail somite (ts) (~E11.0) stage fetuses, and then expression spread to the anterior and posterior poles at later stages. In XY gonads, there was a region in the center where Sprr2d expression was decreased by the 20 ts (~E11.75) stage. This region of downregulation spread both anteriorly and posteriorly until expression could not be detected in XY gonads at E12.75. In XX gonads, a region of Sprr2d downregulation was consistently detected at the anterior end by E12.75. (B) 1700106J16Rik was initially expressed in the central region of 14 ts (~E11.0) XX and XY gonads and then spread to both the anterior and posterior poles. By the 21 ts (~E11.75) stage, there was a region in the center of XY gonads where 1700106J16Rik expression was decreased. This region of downregulation spread both anteriorly and posteriorly until expression was not detected in XY gonads at the 27 ts (~E12.25) stage.
WISH analysis revealed that upregulation of 1700106J16Rik expression also occurs in a C-P wave. We detected 1700106J16Rik expression in a distinct subset of cells within the gonad just after ~E11 (14 ts stage) (Fig 3B). Like Sprr2d, 1700106J16Rik was first expressed just anterior to the central portion of the gonad, and its expression spread to both the anterior and posterior poles. It was initially expressed in roughly the same number of cells in both XX and XY gonads, but was then rapidly downregulated in E11.5 (18 ts stage) XY gonads, while it expression continued in XX gonads. Between E11.75 and E12.25 (21–26 ts stages), 1700106J16Rik expression was mostly extinguished in the central region of XY gonads. Subsequently, 1700106J16Rik expression was extinguished at the anterior and then the posterior pole by the 27 ts stage. Thus, the pattern of 1700106J16Rik expression between E10.75 and E12.25 (12–27 ts stages) was essentially identical to that of Sprr2d. However, 1700106J16Rik expression persisted longer than Sprr2d expression in ovaries, and could be detected in E13.5 and later stage ovaries by WISH (Fig 1D).
The anterior-to-posterior wave of Sprr2d downregulation in ovaries is independent of the presence of germ cells and retinoic acid signaling
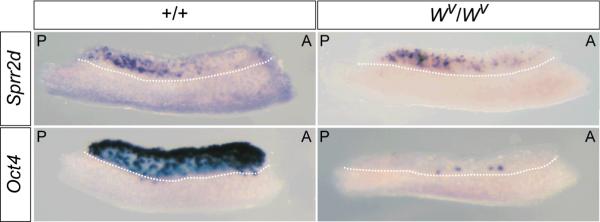
The similarity between the A-P wave of Sprr2d downregulation and the A-P wave of germ cell differentiation (Menke et al., 2003; Yao et al., 2003; Bullejos and Koopman, 2004) suggested a potential interdependence between these events. To determine if Sprr2d downregulation is dependent on the presence of germ cells, we examined its expression in E12.5 B6-Wv/Wv germ cell deficient gonads by WISH (Fig 4). Sprr2d expression did not noticeably differ between B6 wildtype and B6-Wv/Wv XX gonads. Furthermore, Sprr2d was still expressed with a posterior bias in B6-Wv/Wv E12.5 XX gonads, suggesting that in ovaries upregulation of Sprr2d, as well as the A-P wave of Sprr2d downregulation, are not dependent on germ cell signaling.
Figure 4. Sprr2d expression is not altered in germ cell-deficient ovaries.
Expression of Sprr2d and Oct4 were examined by WISH in E12.5 XX +/+ and Wv/Wv germ cell-deficient gonads. Sprr2d was expressed at equal levels, and with a posterior bias, in +/+ (n = 4) (top, left panel) and Wv/Wv (n = 4) (top, right panel) gonads. As expected, the germ cell marker Oct4 was abundantly expressed in +/+ gonads (n = 4) (bottom, left panel), but was expressed in only a few cells in Wv/Wv gonads (n = 2) (bottom, right panel). A, anterior; P, posterior.
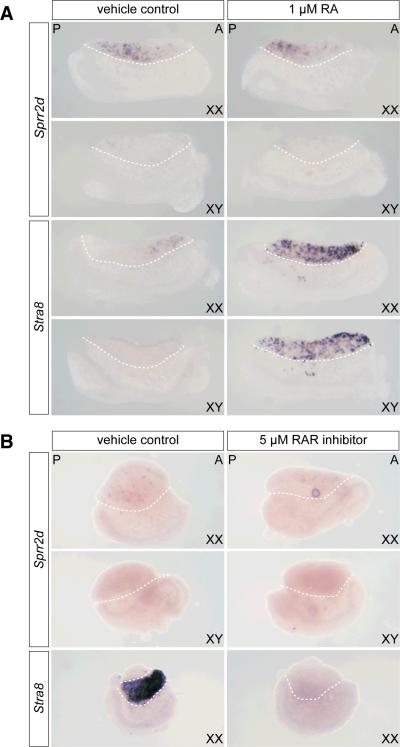
The A-P wave of Sprr2d downregulation in somatic cells was similar in timing to the A-P wave of Stra8 (stimulated by retinoic acid gene 8) upregulation in female germ cells (Menke et al., 2003). The dependence of Stra8 expression on retinoic acid (RA) signaling (Bowles et al., 2006; Koubova et al., 2006) and evidence that RA can influence SPRR expression in cultured human keratinocytes (Hohl et al., 1995; Gibbs et al., 1996) prompted us to investigate whether Sprr2d downregulation in ovaries is dependent on RA signaling. We examined the expression of Sprr2d and Stra8 by WISH in E11.5 gonads cultured with and without exogenous 1.0 μM RA for 10, 18 and 24 hrs (Figure 5A and data not shown). We found that in cultured XX gonads, Sprr2d was downregulated in an A-P wave with similar timing to the wave of Sprr2d downregulation in gonads, in vivo. We also found that this pattern of Sprr2d expression was not noticeably different in RA-treated versus untreated cultured XX gonads. In untreated cultured XY gonads, Sprr2d expression was detectable only at the posterior poles in a few cells, or not at all, which is similar to the in vivo Sprr2d expression pattern (Fig 1E). Furthermore, no difference between RA-treated and untreated XY gonads was apparent. On the other hand, Stra8 expression was induced in XX and XY gonads that had been treated with RA, confirming that the RA treatment was effective at activating RA-dependent gene expression, at least in germ cells. These results suggest that under the conditions we used, treating fetal gonads with pharmacological doses of RA for 10–24 hrs does not affect Sprr2d expression.
Figure 5. Sprr2d expression is not significantly altered in RA and RAR inhibitor treated cultured gonads.
(A) E11.5 XX and XY gonad/mesonephros complexes were cultured for 24 hr with 1.0 μM RA, or with ethanol only (vehicle control). In XX gonads, Sprr2d was expressed with a posterior bias and was almost completely absent from XY gonads, which is similar to its expression pattern in E12.5 gonads in vivo. No difference was apparent between RA-treated and control samples. Stra8 expression was induced in RA-treated, but not in control XX and XY gonads, indicating that the RA treatment was effective. (B) E11.5 XX and XY gonads were cultured with 5.0 μM RAR inhibitor (BMS204493) or ethanol only (vehicle control) for 48 hr. In XX gonads, Sprr2d was expressed in very few cells, and was absent from XY gonads. No difference in Sprr2d expression was apparent between RAR inhibitor-treated and untreated samples. (The round spot in the RAR inhibitor-treated XX gonad is a trapped air bubble.) Stra8 was expressed in untreated XX gonads, but was not expressed in RAR inhibitor-treated XX gonads, indicating that the RAR inhibitor treatment was effective. Representative gonads are shown in each panel (n ≥ 4). The gonad is located above the dotted line in each panel, and between the dotted lines in the Stra8 vehicle control panel in (B). A, anterior; P, posterior.
To further test whether the A-P wave of Sprr2d downregulation is dependent on RA signaling, we analyzed the expression of Sprr2d in gonads treated with a pan-retinoic acid receptor (RAR) inhibitor (BMS-204493), which is able to inhibit each of the RAR isotypes (α, β and γ) (Germain et al., 2002). It was previously shown that this pan-RAR inhibitor can prevent the upregulation of Stra8 in E11.5 XX gonads cultured for 48 hrs (Koubova et al., 2006). We found that culturing E11.5 gonads for 48 hrs in the presence of 5 μM BMS-204493 did not prevent the downregulation of Sprr2d (Fig 5B). In all cases analyzed, Sprr2d could only be detected in a few cells, if at all, in both RAR inhibitor-treated and untreated XX and XY gonads, and no difference in Sprr2d expression between treated and untreated gonads could be appreciated. On the other hand, Stra8 was clearly expressed in untreated XX gonads that had been cultured in parallel, and its expression was completely suppressed in XX gonads treated with RAR inhibitor, suggesting that RA signaling through RAR receptors was inhibited, at least in germ cells. Our data thus suggest that Sprr2d downregulation is not directly regulated via RAR signaling.
Sprr2d and 1700106J16Rik expression are altered in the absence of WNT4 signaling
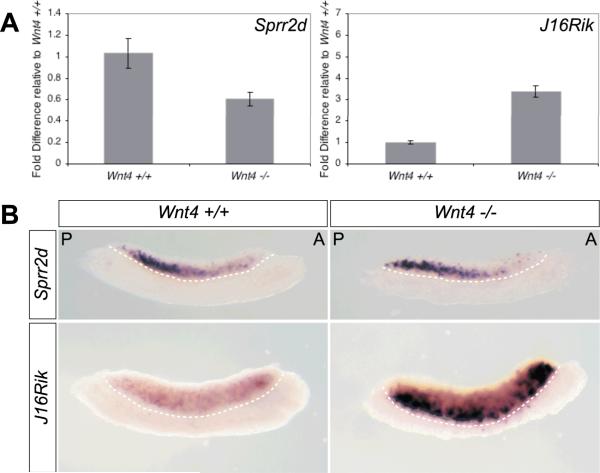
The ovary-biased expression of 1700106J16Rik and Sprr2d in fetal gonads suggests that they could be important for fetal ovary differentiation. Wnt4 plays a significant role in early ovary differentiation (Vainio et al., 1999; Jeays-Ward et al., 2003; Kim et al., 2006). To determine if Sprr2d or 1700106J16Rik are genetically downstream of Wnt4 in the ovary differentiation pathway, we examined if their expression was altered in Wnt4 mutant gonads. Quantitative real-time RT-PCR analysis revealed that Sprr2d expression was reduced in E12.5 Wnt4 −/− XX gonads to 61% of the level in E12.5 Wnt4 +/+ XX gonads (Fig 6A). When we examined Sprr2d expression by WISH, we did not detect a difference between Wnt4 +/+ and Wnt4 −/− gonads (Fig 6B), which is likely due to the differences in sensitivity between WISH and quantitative real-time RT-PCR. Notably however, the A-P wave of downregulation was evident in Wnt4 −/− XX gonads. The decrease in Sprr2d expression in Wnt4 −/− XX gonads suggests that Sprr2d expression is at least partially dependent on Wnt4 expression.
Figure 6. Sprr2d and 1700106J16Rik expression are altered in Wnt4 mutant ovaries.
(A) Quantitative real-time RT-PCR analysis of Sprr2d and 1700106J16Rik (J16Rik) expression in E12.5 Wnt4+/+ and Wnt4−/− XX gonads. Each bar represents the average fold difference in gene expression relative to E12.5 Wnt4+/+ XX gonads. Sprr2d expression in Wnt4−/− gonads was 0.6-fold of the expression in Wnt4+/+ gonads (p = 0.007). 1700106J16Rik was expressed 3.4-fold higher in Wnt4 −/− versus Wnt4+/+ gonads (p = 4E-7). Wnt4 +/+ (n = 5); Wnt4 −/− (n = 7). Error bars represent the standard error of mean. (B) WISH analysis of Sprr2d and 1700106J16Rik expression in E12.5 Wnt4+/+ and Wnt4−/− XX gonads. There was no appreciable difference in Sprr2d expression between Wnt4 +/+ (n = 2), +/− (n = 4; not shown), and −/− (n = 4) gonads. The expression of 1700106J16Rik was markedly increased in Wnt4−/−gonads (n = 6) compared to Wnt4+/+ gonads (n = 8), particularly near the gonad/mesonephros boundary (dotted line). In each panel, the gonad is located above and the mesonephros below the dotted line. A, anterior; P, posterior. For each probe, matched samples were incubated in parallel.
In contrast, quantitative real-time RT-PCR analysis revealed a 3.4-fold increase in 1700106J16Rik expression in E12.5 Wnt4 −/− compared to Wnt4 +/+ XX gonads (Fig 6A). These results were confirmed by WISH analysis (Fig 6B). Interestingly, in Wnt4 −/− gonads the cells that most strongly expressed 1700106J16Rik were located near the gonad/mesonephros boundary.
Sprr2d and 1700106J16Rik expression are decreased in Fog2, Sf1 and Wt1 mutants
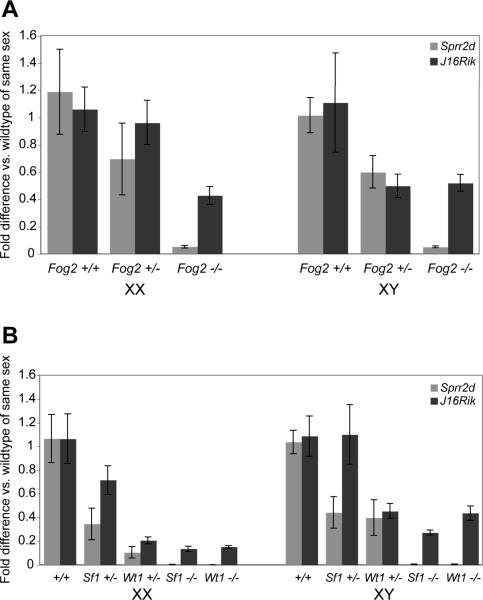
Mutations in Fog2, Sf1 and Wt1 affect the very earliest stages of gonad development in both sexes (Brennan and Capel, 2004). Because our data indicate that Sprr2d and 1700106J16Rik are expressed in SCPs at very early stages of gonad development, initially in both sexes, we examined whether Sprr2d and 1700106J16Rik expression were altered in Fog2, Wt1 and Sf1 mutant E11.5 genital ridges (Fig 7). There was a significant decrease in Sprr2d expression in XX and XY genital ridges from all three mutants, so that it was essentially undetectable in each. [Downregulation of Sprr2d in Fog2 mutants was independently seen by Manuylov et al. (2008)]. In XX genital ridges, 1700106J16Rik expression also was significantly decreased in all three mutants ranging from about 43% of wildtype expression in Fog2−/− samples to about 15% in Sf1−/− and Wt1−/− samples. In XY genital ridges, 1700106J16Rik expression also was clearly decreased, ranging from 27–53%, but the results were somewhat more variable and did not reach our threshold for statistical significance for Wt1 (p = 0.06) and Fog2 (p = 0.08). Sprr2d and 1700106J16Rik tended to be expressed at an intermediate level in Fog2, Sf1 and Wt1 heterozygous mutant genital ridges compared to either wildtype or homozygous null mutants. These data suggest that Sprr2d and 1700106J16Rik are expressed downstream of, or parallel to, Fog2, Sf1 and Wt1 in both XX and XY genital ridges.
Figure 7. Sprr2d and 1700106J16Rik expression are decreased in Fog2, Sf1, and Wt1 mutant genital ridges.
Quantitative real-time RT-PCR was used to analyze Sprr2d and 1700106J16Rik (J16Rik) expression in E11.5 XX and XY genital ridges from fetuses with null mutations in Fog2 (A), Sf1 or Wt1 (B). Each bar represents the fold difference in expression compared to wildtype samples of the same sex. Error bars represent the standard error of mean. For all comparisons between −/− and +/+ samples, p < 0.05, except for J16Rik expression in XY Wt1−/− vs. Wt1+/+ (p = 0.06) and XY Fog2−/− vs. Fog2+/+ (p = 0.08). However, all data follows a similar trend. Sample numbers are: Fog2 (XX +/+ [6], +/− [6], −/− [6]; XY +/+ [3], +/−[6], −/− [5]), Sf1 (XX +/+ [4], +/− [6], −/− [2]; XY +/+ [9], +/− [5], −/− [4]), Wt1 (XX +/+ [4], +/−[≥6], −/− [3]; XY +/+ [9], +/− [5], −/− [≥2]). Because the Sf1 and Wt1 knockout alleles are both on the B6 background, data from +/+ gonads of each sex were pooled for comparisons.
Gonad development in Fog2, Sf1 and Wt1 homozygous null mutant gonads is severely affected, and does not appear to progress much beyond the genital ridge stage. Therefore, we also examined the expression of Sprr2d and 1700106J16Rik in B6 XYPOS gonads, which develop well beyond the genital ridge stage, but are sex reversed. B6 XYPOS gonads do not develop as normal testes, but rather develop as ovaries or ovotestes (gonads with both ovarian and testicular tissue) (Eicher et al., 1982). We examined Sprr2d and 1700106J16Rik expression by WISH in E13–13.5 B6 XYPOS gonads (Supplemental Fig 1), which is the earliest timepoint at which B6 XYPOS gonads can be phenotypically distinguished as ovaries or ovotestes using a dissecting microscope. Expression of Sprr2d was undetectable in B6 XYPOS ovaries and ovotestes suggesting that Sprr2d expression is downregulated by E13.5 in B6 XYPOS sex reversed gonads, as it is in normal XX ovaries and XY testes. Expression of 1700106J16Rik was detected throughout B6 XYPOS ovaries and was essentially indistinguishable from expression in XX ovaries, although expression was perhaps slightly reduced. In B6 XYPOS ovotestes, 1700106J16Rik was expressed throughout the ovarian regions, but again the level was somewhat reduced compared to that in XX ovaries. In the testicular regions of B6 XYPOS ovotestes, 1700106J16Rik expression was greatly reduced and was detected in very few cells. Our unpublished data indicate that granulosa cell markers such as FOXL2 also are detected in a few cells within the testicular regions of B6 XYPOS ovotestes. Thus, our results are consistent with the idea that 1700106J16Rik is a ovary-biased pre-granulosa cell marker in E13.5 gonads.
DISCUSSION
Here we present the initial characterization of two novel markers of early gonadal SCPs (pregranulosa and pre-Sertoli cells). We first identified Sprr2d and 1700106J16Rik as putative pre-granulosa cell-specific genes in a microarray differential expression screen in E13.5 Sry-EGFP transgenic ovaries (Lee and Albrecht, in preparation). The following data support these initial microarray results: 1) In semi-quantitative RT-PCR experiments, both genes were preferentially expressed in pre-granulosa cells compared to non-pre-granulosa/non-vascular somatic cells isolated from independently FACS-sorted E13.5 ovaries. 2) In WISH experiments, 1700106J16Rik expression was spatially restricted in E13.5 ovaries in a pattern similar to that of the Sry-EGFP transgene. 3) SPRR2 protein expression was detected in XX gonads in a subset of cells that co-express the Sry-EGFP transgene, but not in germ cells, vascular endothelial cells, or PDGFRα-expressing interstitial somatic cells. Additionally, SPRR2 protein was detected in XY gonads in a subset of Sertoli cells, but not in any other cell type. These results suggest that SPRR2 protein(s) are transiently expressed in SCPs in XX and XY gonads. However, definitive proof that Sprr2d and 1700106J16Rik are expressed specifically in SCPs during the early stages of gonad differentiation will require additional co-localization experiments for the following reasons: First, the SPRR2 antibody we used likely recognizes multiple SPRR2-family proteins. Therefore, the SPRR2 protein expression detected with this antibody could reflect, at least in part, expression of other Sprr2-family genes such as Sprr2a. Second, some Sprr2d and 1700106J16Rik expression was detected by RT-PCR in the FACS-sorted non-pre-granulosa/non-vascular somatic cells. We think these data likely reflect the probability that the sorted non-pre-granulosa/non-vascular somatic cell population contained some pre-granulosa cells. Despite these caveats, these data and other data discussed below suggest that Sprr2d and 1700106J16Rik are predominately, if not specifically, expressed in SCPs, and that their expression is ovary-biased.
Both Sprr2d and 1700106J16Rik were expressed in a sexually dimorphic manner, with a clear ovarian bias: When both genes were expressed in XX and XY gonads, their expression was higher in XX gonads, and their expression persisted in XX gonads into later developmental stages. Recent gene expression data show that E11.5 XX gonads are transcriptionally active, and that at this stage XX and XY gonads express different subsets of genes (Nef et al., 2005; Small et al., 2005; Beverdam and Koopman, 2006). The identification of these two novel ovarian pre-granulosa cell markers further suggests that an ovarian gene expression program, particularly in SCPs, is initiated at the very earliest stages of sexually dimorphic gonad development. This expression pattern also suggests that these genes may have important roles in early ovary development, particularly in pre-granulosa cell differentiation. We note that Fst, a gene with a similar C-P spatiotemporal expression pattern and whose expression also is female-biased, is known to have a critical role in early ovary differentiation (Menke et al., 2003; Yao et al., 2004). However, it is not clear if Fst is expressed specifically in pre-granulosa cells. The roles of Sprr2d and 1700106J16Rik in mammalian gonad differentiation and sex determination remain to be determined in the future by loss- and gain-of-function experiments. However, our data lead us to make predictions about possible roles.
SPRR proteins have been proposed to be structural components of squamous epithelial cell cornified envelopes (Steven and Steinert, 1994; Hohl et al., 1995). Fetal gonad SCPs presumably do not have such structures, particularly at the very earliest stages of differentiation. In our studies, we detected SPRR2 protein(s) in both the nucleus and cytoplasm in gonadal somatic cells, suggesting that in the gonad they may have alternative functions. In mid-gestation embryos, Sprr2d expression was restricted to the genital ridges, which suggests that Sprr2d likely has a specific function during gonad development. The ovary-biased expression of Sprr2d and Sprr2a in developing gonads suggests that they could have specific roles in pre-granulosa cell differentiation, with rapid suppression in XY gonads to prevent feminization of SCPs. However, given that Sprr2d and Sprr2a were expressed transiently in both XX and XY gonads, it is also possible that they are involved in SCP specification or the initial stages of SCP differentiation in both XX and XY gonads, becoming downregulated in a C-P wave in XY gonads as testicular differentiation is initiated, or downregulated in an A-P wave in XX gonads as ovarian differentiation proceeds.
The significance of this ovarian A-P gene expression wave, which parallels the entry of female germ cells into meiosis, has yet to be determined. Interestingly, despite the similarity between the A-P wave displayed by female germ cell differentiation markers and that of Sprr2d in somatic cells, the A-P wave of Sprr2d downregulation was independent of the presence of germ cells and RA signaling. Furthermore, the A-P wave of Sprr2d downregulation was independent of WNT4 signaling. These data indicate that an A-P wave of somatic cell differentiation might facilitate the A-P wave of germ cell differentiation. Alternatively, the A-P gene expression waves in somatic cells and germ cells could be at least partially independent of each other.
1700106J16Rik is an uncharacterized gene, without any obvious functional domains. In midgestation embryos, 1700106J16Rik expression was restricted to the genital ridges and part of the developing spinal cord and hindbrain, which suggests that 1700106J16Rik may have specific functions in the development of these organs. Given that 1700106J16Rik expression was ovary-biased and persisted in ovaries until at least E15.5, we speculate that 1700106J16Rik is involved in pre-granulosa cell differentiation. Furthermore, it currently appears that both Sprr2d and 1700106J16Rik have highly conserved homologues in mammals, but do not have obvious homologues in non-mammalian genomes. These data suggest that if Sprr2d and/or 1700106J16Rik are involved in sex determination, they are involved in a mechanism that is unique to, but conserved within, mammals.
Our data show that WNT4 signaling, which is crucial for normal ovary differentiation, is necessary for the proper expression of both Sprr2d and 1700106J16Rik. However, whereas Fst expression is completely eliminated in Wnt4 −/− gonads (Jeays-Ward et al., 2004; Yao et al., 2004), Sprr2d expression was only partially decreased while, unexpectedly, 1700106J16Rik expression was increased. These data suggest that neither Sprr2d expression nor 1700106J16Rik expression are completely dependent on WNT4-mediated transcriptional activation. Reduced Sprr2d expression might be due to the fact that Wnt4 −/− XX gonads are only partially sex reversed. Alternatively, WNT4 signaling might act to upregulate Sprr2d expression in concert with other factors that have an additive effect. The reason for increased 1700106J16Rik expression in Wnt4 −/− XX gonads is difficult to explain at present, given the incomplete knowledge of the WNT4 signaling cascade in early gonad development. It could be that WNT4 partially represses 1700106J16Rik expression, or that Wnt4 and 1700106J16Rik are involved in a regulatory loop; however, further studies are necessary to distinguish between these and other possibilities. Nevertheless, the altered expression of Sprr2d and 1700106J16Rik in Wnt4 mutant ovaries is consistent with these genes having functional roles in ovary development.
We characterized Sprr2d and 1700106J16Rik expression in Sf1, Wt1 and Fog2 E11.5 mutant XX and XY gonads because these mutants exhibit decreased Sry expression in XY gonads (Capel, 1998; Hammes et al., 2001; Tevosian et al., 2002). Sprr2d and 1700106J16Rik expression were decreased in both sexes in all three mutants. Expression of Sprr2d was more severely affected in all cases, but the mechanistic significance of this finding is unclear and might simply reflect the transient nature of Sprr2d expression. Further experiments are needed to determine if reduced expression is an indirect effect cause by a reduction in the number of SCPs and/or due to direct regulation of these genes by Sf1, Wt1 and Fog2. In either case, our results suggest that Sprr2d and 1700106J16Rik are expressed downstream of, or parallel to, Sf1, Wt1, and Fog2 in both XX and XY genital ridges.
Both Sprr2d and 1700106J16Rik were upregulated in a C-P wave in E11.0 – E11.5 XX and XY gonads, and downregulated in a C-P wave in E11.5 – E12.0 XY gonads. Sprr2d was downregulated in an A-P wave in XX gonads beginning around E12.5, while 1700106J16Rik expression persisted in XX gonads until at least E15.5. These data indicate that the initiation of Sprr2d and 1700106J16Rik expression was not sex-specific, whereas their downregulation was sex-specific. We are very intrigued by the dynamic temporal and spatial patterns of Sprr2d and 17000106J16Rik expression because they mimic the expression pattern of the testis determining gene, Sry (Albrecht and Eicher, 2001; Bullejos and Koopman, 2001). Given these similarities, it is tempting to speculate that these three genes may be regulated by common mechanisms. It is not clear how Sry expression is upregulated, although an epigenetic mechanism mediated by DNA methylation has been proposed (Nishino et al., 2004). It is thought that Sry expression is downregulated by SOX9 (Chaboissier et al., 2004). Therefore, we predict that Sprr2d and 1700106J16Rik expression are downregulated by SOX9 in XY gonads, and future studies will be aimed at testing this hypothesis.
A C-P wave of gene expression during mouse gonad differentiation has previously been described for Sry, Sox9 and Fst {Albrecht, 2001 #1; Bullejos, 2001 #81; Bullejos, 2005 #108; Menke, 2003 #11; Sekido, 2004 #109; Wilhelm, 2005 #130}. We now add Sprr2d and 1700106J16Rik to this list. Whether Sprr2d and 1700106J16Rik have critical roles in pre-granulosa cell or ovary differentiation has yet to be determined. However, we note that Sry and Sox9 are necessary for testicular SCP differentiation into Sertoli cells [reviewed in{Brennan, 2004 #3}] and that Fst is necessary for early ovary differentiation {Yao, 2004 #125}. Nevertheless, the data presented here suggest that SCPs in both XX and XY gonads differentiate in a C-P wave and are consistent with the hypothesis that SCP differentiation begins at the same stage in both sexes.
EXPERIMENTAL PROCEDURES
Mouse strains
C57BL/6J (B6) mice homozygous for an Sry-EGFP transgene (B6-Tg92/Tg92) were previously described (Albrecht and Eicher, 2001) as were the B6 XYPOS Tg4 mice [Tg(Sry)4Ei] (Eicher et al., 1995). The Wnt4 knockout mice on the 129S1/SvImJ background (129S1-Wnt4) were a generous gift from Dr. Andy McMahon (Stark et al., 1994). The Fog2 knockout mice (Tevosian et al., 2002), a generous gift from Dr. Sergei Tevosian, Ph.D. were received on a mixed B6/129 background and then backcrossed to B6. The samples used here were from N4 and N5 backcross generation animals. The Sf1 (Luo et al., 1994) and Wt1 (Kreidberg et al., 1993) knockout mice, generous gifts from Dr. Keith Parker and Dr. Jordan Kreidberg, respectively, also were received on a mixed B6/129 background and then backcrossed to B6. The samples used here were from >N20 backcross generation animals. Mice with the KitW-v mutation on the B6 background (B6-Wv) were purchased from The Jackson Laboratory. CD-1 mice were purchased from Charles River Laboratories. The Boston University Laboratory Animal Science Center is AAALAC accredited and all animal procedures were approved by the BUMC Institutional Animal Care and Use Committee.
Timed matings were used for all experiments. Noon on the day of vaginal plug detection was designated as E0.5. Embryos between E10.5 and 12.5 were more precisely staged by counting the number of tail somites (E10.5 ~ 8ts, E11.5 ~ 18ts, E12.5 ~ 30ts) (Hacker et al., 1995) and embryos older than E12.5 were staged by limb morphology (Kaufman, 2003).
Genotyping
The sex of all embryos from 129S1-Wnt4 and B6 XYPOS Tg4 matings, and of all embryos younger than E12.5, was determined using a PCR-based assay (Capel et al., 1999). The sex of all other E12.5 or older embryos was determined by gonad morphology.
The genotype of B6-Wv embryos was determined using a PCR and restriction enzyme digest-based assay [adapted from (Cable et al., 1995)]. Primers flanking the Wv mutation were used to amplify the Kit gene (5'-ATGACCCTGCCTTCTTCCTT and 5'-GTCTGAAGGCTCCCAGCATA; 538bp product). The PCR product was digested with NsiI giving a 538bp uncut fragment from the wildtype allele and, 347bp and 191bp fragments from the Wv allele. The genotype of Sf1 embryos was determined using a multiplex PCR assay to amplify both the wildtype (410bp) and the targeted (~700bp) alleles (KO-F: 5'-ACAAGCATTACACGTGCACC, KO-R: 5'-TGACTAGCAACCACCTTGCC, and Neo2: 5'-AGGTGAGATGACAGGAGATC). The genotype of Fog2 embryos was determined using PCR primers to detect the wildtype allele (P10: 5'-TTGGCTTTCAGACCCAGAG and P12: 5'-ATTTGGCCATCTGCTGCCA; 614bp product, S. Tevosian, pers. comm.), and the Neo cassette in the targeted allele (Neo1: 5'-CTTGGGTGGAGAGGCTATTC and Neo2 as listed above; 260bp product). Previously described PCR-based assays were used to genotype embryos from B6 XYPOS Tg4 (Capel et al., 1999), Wnt4 (Stark et al., 1994) and Wt1 matings (Kreidberg et al., 1993).
Whole-mount in situ hybridization
Standard protocols for whole-mount in situ hybridization were used with minor modification (Wilkinson, 1992; Henrique et al., 1995). Briefly, gonad/mesonephros complexes or whole embryos were dissected and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4°C. The tissues were hybridized with digoxigenin (DIG)-labeled RNA probes, and the riboprobes were detected using an alkaline phosphatase-conjugated antibody and NBT/BCIP chromogen. Antisense and sense probes were tested in parallel, and only signal specific to the antisense probes was reported as gene-specific expression. Unless otherwise noted, each result shown was replicated in a minimum of four samples. Riboprobes were produced by in vitro transcription in the presence of DIG-labeled dUTP (Roche). Sprr2d probes were generated by PCR using the following primers (5'-ACTTTGGAGAACCCGATCCT and 5'-CAGGAAATGAAACCATGCTG; 637 bp product) and cloned into pCRII-TOPO (Invitrogen). 1700106J16Rik probes were generated by PCR using the following primers (5'-TTTCTTGGCTTCCTCTCTGC and 5'-GCCTCTCTGCATGGTGTTATT; 629 bp product) and cloned into pCRII-TOPO. Sprr2a probes were generated from a full-length cDNA IMAGE clone (4126389), and the Oct4 and Stra8 probes were generated from plasmids kindly provided by Drs. David Page and Jana Koubova (Menke et al., 2003).
Whole-mount indirect fluorescent immunohistochemistry
Gonad/mesonephros complexes were fixed in 4% PFA in PBS overnight at 4°C. Whole-mount indirect fluorescent immunohistochemistry was essentially performed as previously described (Albrecht and Eicher, 2001), except that a different blocking buffer (3% BSA, 10% donkey serum, 0.1% Triton-X, 0.02% sodium azide in PBS) was used when the anti-PDGFRα or anti-MIS (AMH) antibodies were employed. Primary antibodies and dilutions were as follows: rabbit polyclonal anti-SPRR2 (1:500, Apotech), goat polyclonal anti-PDGFRα (1:500, R&D Systems), rat monoclonal anti-CD31 (PECAM) (1:300, BD Pharmingen), goat polyclonal anti-GATA4 (C-20) (1:400, Santa Cruz Biotechnology), rabbit polyclonal anti-GFP (1:2000, Molecular Probes) and goat polyclonal anti-MIS (AMH) (C-20) (1:200, Santa Cruz Biotechnology). Secondary antibodies were donkey anti-IgG conjugated with Cy3 or Cy5 (Jackson ImmunoResearch, 1:500) or AF488 (Molecular Probes, 1:750). Samples were imaged using a Zeiss LSM510 confocal microscope.
The rabbit polyclonal anti-SPRR2 antibody (Apotech) was raised against a synthetic peptide whose sequence is highly conserved among human and mouse SPRR2 proteins (but not among SPRR1 or SPRR3 proteins), and has previously been used to detect SPRR2 expression in mouse tissues (Hohl et al., 1995; Nozaki et al., 2005).
RNA isolation and cDNA synthesis
Total RNA was isolated using the RNeasy mini RNA isolation kit (Qiagen) according to the manufacturer's instructions. Samples were treated with DNase I (Ambion) for 2 hrs at 37°C, and subsequently treated with DNase inactivation reagent (Ambion). All RNA samples used for RTPCR analysis were determined to be free of DNA by PCR using primers specific for Myog (5'-TTACGTCCATCGTGGACAGCAT and 5'-TGGGCTGGGTGTTAGTCTTAT) (Capel et al., 1999), Sprr2d (5'-CCCATCTTCCTTCCAAATCC and 5'-AACATGGAGGGTGAAAGGTG) (Beverdam and Koopman, 2006), or Lhx1 (5'-GGCGAGGAGCTCTACATCATAG and 5'-CTTGGGAATCCGGAGATAAAC) (Albrecht et al., 2003). cDNA was synthesized from equal amounts of each RNA sample to be compared using oligo-dT primers and SuperScript II reverse transcriptase (RT) (Invitrogen). Samples prepared without RT served as negative controls (-RT control).
Semi-quantitative RT-PCR
For temporal gene expression analysis by semi-quantitative RT-PCR, RNA was extracted from genital ridges attached to mesonephroi (E10.5) or genital ridges/gonads separated from mesonephroi (E11.5–E15.5). Embryos were from B6 or B6D2F1 mice. For EGFP+/CD31- vs. EGFP-/CD31- RT-PCR, RNA was isolated from E13.5 B6-Tg92/Tg92 ovarian cells that had been sorted by FACS (Lee and Albrecht, in preparation).
Sprr2d, Sprr2f and Sprr2g expression was assessed using previously described gene-specific primers (Song et al., 1999). Primers for Sprr2a (5'-CTGAGACTCAAGTACGATGTCTTACTACC and 5'-TTCTCTGTGAGGAGCCATCATAAGCAC) were modified from those described by Song et al. (1999) to account for a B6 strain polymorphism. 1700106J16Rik expression was assessed using the primers described previously. Hprt expression served as an endogenous control for total RNA levels using the following primers (5'-CCTGCTGGATTACATTAAAGCACTG and 5'-GTCAAGGGCATATCCAACAACAAAC) (Koopman et al., 1989).
Quantitative real-time RT-PCR
For analysis of gene expression in E11.5 samples (Sf1, Wt1 and Fog2 mutants), both urogenital ridges (gonads with mesonephroi) were collected as a pair from each embryo; and for E12.5 samples (Wnt4), gonads were separated from mesonephroi and collected as a pair from each embryo. The number of samples of each genotype analyzed is indicated in the legends for Figs. 6 and 7.
Quantitative real-time PCR reactions were performed on a ABI PRISM 7900HT Sequence Detection System, using Power SYBR Green PCR Master Mix (Applied Biosystems) and the following primers: 1700106J16Rik (5'-AGGGTCCATTCTTATGAAGGTGA and 5'-GTAGGAGGGCCATGACTGAGA: 102 bp product), Sprr2d (5'-CCCATCTTCCTTCCAAATCC and 5'-AACATGGAGGGTGAAAGGTG: 65 bp product) (Beverdam and Koopman, 2006), and Hprt (5'-CATTATGCCGAGGATTTGGAA and 5'-CACACAGAGGGCCACAATGT: 123 bp product) (Bouma et al., 2004). Primers were used at a concentration of 75 nM. All primer pairs produced single products of the expected size, without the formation of primer dimers, as assessed by agarose gel electrophoresis and dissociation curve analysis. Validation experiments were performed according to Applied Biosystems guidelines (AppliedBiosystems, 2004), and the PCR efficiencies of the endogenous control (Hprt) and each of the other genes were approximately equal, yielding validation slopes less than 0.1. The total reaction volume was 10 μL, and each cDNA sample was measured in triplicate, in parallel with single -RT controls and no template controls. The PCR cycle conditions were: two initial steps at 60°C for 2 min and 95°C for 15 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min.
For each sample and gene analyzed, the cycle threshold (Ct) was measured as the number of cycles to reach a threshold value of 0.2 from each of the three replicate sample wells. The baseline was set from 3 to 15. The SDS2.2 or SDS2.3 programs (Applied Biosystems) were used to determine the average cycle threshold (avg Ct) value resulting from the three technical replicate reactions from each sample. Hprt (endogenous control) avg Ct values were subtracted from the avg Ct value for each sample to give the normalized average Ct value (ΔCt). For each gene analyzed, the mean ΔCt value of the XX +/+ and XY +/+ was calculated. The fold difference between each sample and the average +/+ expression level was calculated by (1) subtracting the mean +/+ ΔCt from each ΔCt value to arrive at the ΔΔCt value and (2) using the formula 2−ΔΔCt to calculate the fold difference (FD) (Livak and Schmittgen, 2001). Average FD and standard error of mean (SEM) values were then determined.
Statistical analysis
Two-tailed unpaired Student t-tests were performed. In this study, we consider a comparison with p < 0.05 to be statistically significant.
Gonad culture and treatments
Gonad/mesonephros complexes were dissected from E11.25 – E12.0 (15 – 23 tail somites) CD-1 embryos. From each embryo, one gonad was treated, while the other served as an untreated control. 5 μm TMTP Isopore membrane filters (Millipore) were floated on 0.5 mL of culture medium [Dulbecco's Modified Eagle Medium (Invitrogen), 10% fetal bovine serum and 50 μg/mL ampicillin] that did or did not contain retinoic acid (RA) or RA receptor (RAR) inhibitor, as described below. Three to five gonad/mesonephros complexes were placed on each filter, and the gonad cultures were incubated at 37°C and 5% CO2.
For the RA treatment experiments, gonads were cultured for 24 hr with 1.0 μM all-trans-RA (Sigma) (1.0 μL of 1 mM all-trans-RA dissolved in ethanol per mL of culture medium) or in the presence of a vehicle control (1.0 μL ethanol/mL culture medium). For the RAR inhibitor treatment experiments, gonads were cultured for 48 hr with 5.0 μM of a pan-RAR inhibitor (BMS-204493, Bristol-Myers-Squibb) (1.0 μL of a 5 mM solution of BMS-204493 dissolved in ethanol per mL of culture medium) or in the presence of a vehicle control (Germain et al., 2002; Koubova et al., 2006). The culture medium was refreshed every 24 hr.
Supplementary Material
Sprr2d and 1700106J16Rik expression in E13.25–13.5 gonad/mesonephros complexes was analyzed by WISH. Sprr2d (left panels) was not detected in any gonads at this stage, which is similar to wildtype. 1700106J16Rik expression (right panels) was detected throughout XYPOS ovaries and was essentially indistinguishable from wildtype XX ovaries. In XYPOS ovotestes, 1700106J16Rik was expressed similar to wildtype XX ovaries in the ovarian regions lacking testis cords, but was greatly reduced in testicular regions with cords (within the black dotted circle). 1700106J16Rik was not detected in XYPOS Tg4 “rescued” testes. In each panel, the gonad is located above, and the mesonephros below, the white dotted line. A, anterior; P, posterior. For XYPOS ovotestes analyzed for Sprr2d expression n=3, for all others n ≥4.
ACKNOWLEDGEMENTS
We are grateful to Caryn Navarro, Joe St.George, Karen Symes and Sergei Tevosian for reading a previous version of the manuscript and for providing helpful insights, to Ihn Young Song for help with in situ hybridizations, to Karen Schlauch for statistical advice, and to members of the Albrecht and Page labs for helpful discussions. We thank David Page and Jana Kubova for providing the Oct4 and Stra8 plasmids; Wellington Cardoso, Christopher Zusi and Bristol-Myers Squibb for providing the BMS-204493 RAR antagonist; and Andy McMahon, Sergei Tevosian, Keith Parker, and Jordan Kreidberg for providing mice. This research is supported by grants from the Department of Medicine, BUSM (Pilot Project), the National Institute of Child Health and Human Development (HD042779), and the American Cancer Society (Research Scholar Grant 06-033-01-DDC).
Grant Information: National Institute of Child Health and Human Development: HD042779 American Cancer Society Research Scholar Grant: 06-033-01-DDC
REFERENCES
- Albrecht KH, Eicher EM. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev Biol. 2001;240:92–107. doi: 10.1006/dbio.2001.0438. [DOI] [PubMed] [Google Scholar]
- Albrecht KH, Young M, Washburn LL, Eicher EM. Sry expression level and protein isoform differences play a role in abnormal testis development in C57BL/6J mice carrying certain Sry alleles. Genetics. 2003;164:277–288. doi: 10.1093/genetics/164.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AppliedBiosystems Guide to performing relative quantitation of gene expression using real-time quantitative PCR. 2004 [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet. 2006;38:1430–1434. doi: 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Koopman P. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet. 2006;15:417–431. doi: 10.1093/hmg/ddi463. [DOI] [PubMed] [Google Scholar]
- Bouma GJ, Hart GT, Washburn LL, Recknagel AK, Eicher EM. Using real time RT-PCR analysis to determine multiple gene expression patterns during XX and XY mouse fetal gonad development. Gene Expr Patterns. 2004;5:141–149. doi: 10.1016/j.modgep.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, Hamada H, Koopman P. Retinoid Signaling Determines Germ Cell Fate in Mice. Science. 2006 doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Brennan J, Capel B. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat Rev Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- Bullejos M, Koopman P. Spatially dynamic expression of Sry in mouse genital ridges. Dev Dyn. 2001;221:201–205. doi: 10.1002/dvdy.1134. [DOI] [PubMed] [Google Scholar]
- Bullejos M, Koopman P. Germ cells enter meiosis in a rostro-caudal wave during development of the mouse ovary. Mol Reprod Dev. 2004;68:422–428. doi: 10.1002/mrd.20105. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Buehr M, Koopman P, Rossant J, McLaren A. Cell-autonomous action of the testis-determining gene: Sertoli cells are exclusively XY in XX <--> XY chimeric mouse testis. Development. 1988;102:443–450. doi: 10.1242/dev.102.2.443. [DOI] [PubMed] [Google Scholar]
- Cable J, Jackson IJ, Steel KP. Mutations at the W locus affect survival of neural crest-derived melanocytes in the mouse. Mech Dev. 1995;50:139–150. doi: 10.1016/0925-4773(94)00331-g. [DOI] [PubMed] [Google Scholar]
- Capel B. Sex in the 90s: SRY and the switch to the male pathway. Annu Rev Physiol. 1998;60:497–523. doi: 10.1146/annurev.physiol.60.1.497. [DOI] [PubMed] [Google Scholar]
- Capel B, Albrecht KH, Washburn LL, Eicher EM. Migration of mesonephric cells into the mammalian gonad depends on Sry. Mech Dev. 1999;84:127–131. doi: 10.1016/s0925-4773(99)00047-7. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Shown EP, Washburn LL. Sex reversal in C57BL/6J-YPOS mice corrected by a Sry transgene. Philos Trans R Soc Lond B Biol Sci. 1995;350:263–269. doi: 10.1098/rstb.1995.0160. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Washburn LL, Whitney JB, III, Morrow KE. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science. 1982;217:535–537. doi: 10.1126/science.7089579. [DOI] [PubMed] [Google Scholar]
- Finkelman FD, Yang M, Perkins C, Schleifer K, Sproles A, Santeliz J, Bernstein JA, Rothenberg ME, Morris SC, Wills-Karp M. Suppressive effect of IL-4 on IL-13-induced genes in mouse lung. J Immunol. 2005;174:4630–4638. doi: 10.4049/jimmunol.174.8.4630. [DOI] [PubMed] [Google Scholar]
- Germain P, Iyer J, Zechel C, Gronemeyer H. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature. 2002;415:187–192. doi: 10.1038/415187a. [DOI] [PubMed] [Google Scholar]
- Gibbs S, Backendorf C, Ponec M. Regulation of keratinocyte proliferation and differentiation by all-trans-retinoic acid, 9-cis-retinoic acid and 1,25-dihydroxy vitamin D3. Arch Dermatol Res. 1996;288:729–738. doi: 10.1007/BF02505289. [DOI] [PubMed] [Google Scholar]
- Gillman J. Contributions to Embryology. Carnegie Institution of Washington; Washington, D.C.: 1948. The development of the gonads in man, with a consideration of the role of fetal endocrines and the histogenesis of ovarian tumors; pp. 83–131. [Google Scholar]
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- Hammes A, Guo J-K, Lutsch G, Leheste J-R, Landrock D, Ziegler U, Gubler M-C, Schedl A. Two splice variants of the Wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Hohl D, de Viragh PA, Amiguet-Barras F, Gibbs S, Backendorf C, Huber M. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursor proteins. J Invest Dermatol. 1995;104:902–909. doi: 10.1111/1523-1747.ep12606176. [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K, Dandonneau M, Swain A. Wnt4 is required for proper male as well as female sexual development. Dev Biol. 2004;276:431–440. doi: 10.1016/j.ydbio.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K, Hoyle C, Brennan J, Dandonneau M, Alldus G, Capel B, Swain A. Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development. 2003;130:3663–3670. doi: 10.1242/dev.00591. [DOI] [PubMed] [Google Scholar]
- Jeske YW, Bowles J, Greenfield A, Koopman P. Expression of a linear Sry transcript in the mouse genital ridge. Nat Genet. 1995;10:480–482. doi: 10.1038/ng0895-480. [DOI] [PubMed] [Google Scholar]
- Jordan BK, Mohammed M, Ching ST, Delot E, Chen XN, Dewing P, Swain A, Rao PN, Elejalde BR, Vilain E. Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am J Hum Genet. 2001;68:1102–1109. doi: 10.1086/320125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartasova T, van Muijen GN, van Pelt-Heerschap H, vandePutte P. Novel protein in human epidermal keratinocytes: regulation of expression during differentiation. Mol Cell Biol. 1988;8:2204–2210. doi: 10.1128/mcb.8.5.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman M. The Atlas of Mouse Development. Academic Press; London: 2003. [Google Scholar]
- Kim Y, Kobayashi A, Sekido R, Dinapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. Fgf9 and Wnt4 Act as Antagonistic Signals to Regulate Mammalian Sex Determination. PLoS Biol. 2006;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Collignon J, Lovell-Badge R. Zfy gene expression patterns are not compatible with a primary role in mouse sex determination. Nature. 1989;342:940–942. doi: 10.1038/342940a0. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lovell-Badge R. The role of Sry in mammalian sex determination. Ciba Found Symp. 1992;165:162–179. doi: 10.1002/9780470514221.ch10. discussion 179–182. [DOI] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- Magre S, Jost A. The initial phases of testicular organogenesis in the rat. An electron microscopy study. Arch Anat Microsc Morphol Exp. 1980;69:297–318. [PubMed] [Google Scholar]
- Manuylov NL, Smagulova FO, Leach L, Tevosian SG. Ovarian development in mice requires the GATA4-FOG2 transcription complex. 2008 doi: 10.1242/dev.024653. Development in press. [DOI] [PubMed] [Google Scholar]
- Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol. 2003;262:303–312. doi: 10.1016/s0012-1606(03)00391-9. [DOI] [PubMed] [Google Scholar]
- Morris JS, Stein T, Pringle MA, Davies CR, Weber-Hall S, Ferrier RK, Bell AK, Heath VJ, Gusterson BA. Involvement of axonal guidance proteins and their signaling partners in the developing mouse mammary gland. J Cell Physiol. 2006;206:16–24. doi: 10.1002/jcp.20427. [DOI] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Nishino K, Hattori N, Tanaka S, Shiota K. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J Biol Chem. 2004;279:22306–22313. doi: 10.1074/jbc.M309513200. [DOI] [PubMed] [Google Scholar]
- Nozaki I, Lunz JG, 3rd, Specht S, Stolz DB, Taguchi K, Subbotin VM, Murase N, Demetris AJ. Small proline-rich proteins 2 are noncoordinately upregulated by IL-6/STAT3 signaling after bile duct ligation. Lab Invest. 2005;85:109–123. doi: 10.1038/labinvest.3700213. [DOI] [PubMed] [Google Scholar]
- Odor DL, Blandau RJ. Ultrastructural studies on fetal and early postnatal mouse ovaries. II. Cytodifferentiation. Am J Anat. 1969;125:177–215. doi: 10.1002/aja.1001250205. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Patel S, Kartasova T, Segre JA. Mouse Sprr locus: a tandem array of coordinately regulated genes. Mamm Genome. 2003;14:140–148. doi: 10.1007/s00335-002-2205-4. [DOI] [PubMed] [Google Scholar]
- Pradervand S, Yasukawa H, Muller OG, Kjekshus H, Nakamura T, St Amand TR, Yajima T, Matsumura K, Duplain H, Iwatate M, Woodard S, Pedrazzini T, Ross J, Firsov D, Rossier BC, Hoshijima M, Chien KR. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. Embo J. 2004;23:4517–4525. doi: 10.1038/sj.emboj.7600454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HJ, Poy G, Darwiche N, Lichti U, Kuroki T, Steinert PM, Kartasova T. Mouse Sprr2 genes: a clustered family of genes showing differential expression in epithelial tissues. Genomics. 1999;55:28–42. doi: 10.1006/geno.1998.5607. [DOI] [PubMed] [Google Scholar]
- Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- Steven AC, Steinert PM. Protein composition of cornified cell envelopes of epidermal keratinocytes. J Cell Sci. 1994;107(Pt 2):693–700. [PubMed] [Google Scholar]
- Tesfaigzi J, An G, Wu R, Carlson DM. Two nuclear proteins in tracheal epithelial cells are recognized by antibodies specific to a squamous differentiation marker, sprI. J Cell Physiol. 1995;164:571–578. doi: 10.1002/jcp.1041640315. [DOI] [PubMed] [Google Scholar]
- Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins. A review. Cell Biochem Biophys. 1999;30:243–265. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development. 2002;129:4627–4634. doi: 10.1242/dev.129.19.4627. [DOI] [PubMed] [Google Scholar]
- Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. In situ hybridization: A Practical Approach. IRL Press; Oxford: 1992. pp. 75–83. [Google Scholar]
- Yao HH, DiNapoli L, Capel B. Meiotic germ cells antagonize mesonephric cell migration and testis cord formation in mouse gonads. Development. 2003;130:5895–5902. doi: 10.1242/dev.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao HH, Matzuk MM, Jorgez CJ, Menke DB, Page DC, Swain A, Capel B. Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev Dyn. 2004;230:210–215. doi: 10.1002/dvdy.20042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, Laurin N, Eftekharpour E, Sat E, Grigull J, Pan Q, Peng WT, Krogan N, Greenblatt J, Fehlings M, van der Kooy D, Aubin J, Bruneau BG, Rossant J, Blencowe BJ, Frey BJ, Hughes TR. The functional landscape of mouse gene expression. J Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann N, Doepker MP, Witte DP, Stringer KF, Fulkerson PC, Pope SM, Brandt EB, Mishra A, King NE, Nikolaidis NM, Wills-Karp M, Finkelman FD, Rothenberg ME. Expression and regulation of small proline rich protein (SPRR)2 in allergic inflammation. Am J Respir Cell Mol Biol. 2005 doi: 10.1165/rcmb.2004-0269OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sprr2d and 1700106J16Rik expression in E13.25–13.5 gonad/mesonephros complexes was analyzed by WISH. Sprr2d (left panels) was not detected in any gonads at this stage, which is similar to wildtype. 1700106J16Rik expression (right panels) was detected throughout XYPOS ovaries and was essentially indistinguishable from wildtype XX ovaries. In XYPOS ovotestes, 1700106J16Rik was expressed similar to wildtype XX ovaries in the ovarian regions lacking testis cords, but was greatly reduced in testicular regions with cords (within the black dotted circle). 1700106J16Rik was not detected in XYPOS Tg4 “rescued” testes. In each panel, the gonad is located above, and the mesonephros below, the white dotted line. A, anterior; P, posterior. For XYPOS ovotestes analyzed for Sprr2d expression n=3, for all others n ≥4.