Abstract
We have shown that a xylan-degrading bacterium, W-61, excretes multiple xylanases, including xylanase 5 with a molecular mass of 140 kDa. Here, we emend the previously used classification of the bacterium (i.e., Aeromonas caviae W-61) to Paenibacillus sp. strain W-61 on the basis of the nucleotide sequence of the 16S rRNA gene, and we clone and express the xyn5 gene encoding xylanase 5 (Xyn5) in Escherichia coli and study the subcellular localization of Xyn5. xyn5 encodes 1,326 amino acid residues, including a 27-amino-acid signal sequence. Sequence analysis indicated that Xyn5 comprises two family 22 carbohydrate-binding modules (CBM), a family 10 catalytic domain of glycosyl hydrolases, a family 9 CBM, a domain similar to the lysine-rich region of Clostridium thermocellum SdbA, and three S-layer-homologous (SLH) domains. Recombinant Xyn5 bound to a crystalline cellulose, Avicel PH-101, while an N-terminal 90-kDa fragment of Xyn5, which lacks the C-terminal half of the family 9 CBM, did not bind to Avicel PH-101. Xyn5 was cell bound, and the cell-bound protein was digested by exogenous trypsin to produce immunoreactive and xylanolytic fragments with molecular masses of 80 and 60 kDa. Xyn5 was exclusively distributed in the cell envelope fraction consisting of a peptidoglycan-containing layer and an associated S layer. Thus, Paenibacillus sp. strain W-61 Xyn5 is a cell surface-anchored modular xylanase possessing a functional cellulose-binding module and SLH domains. Possible cooperative action of multiple xylanases produced by strain W-61 is discussed on the basis of the modular structure of Xyn5.
β-1,4-Xylans, the major components of hemicellulose, are heterogeneous polysaccharides consisting of a homopolymeric backbone of β-1,4-linked d-xylopyranose units and short side chains, including O-acetyl, α-l-arabinofuranosyl, and α-d-glucuronyl residues (37). It has been shown that many kinds of bacteria and fungi hydrolyze β-1,4-xylan by the use of xylanolytic enzymes, such as β-1,4-xylanases, β-xylosidases, α-glucuronidases, α-arabinofuranosidases, and esterases (13, 34, 39). β-1,4-Xylanases (EC 3.2.1.8) are the key enzymes that hydrolyze the backbone structure of β-1,4-xylans to initiate degradation of the complex polysaccharides by microorganisms. A number of β-1,4-xylanases have been purified from fungi and bacteria, and the genes encoding β-1,4-xylanases have been cloned and characterized. Several microorganisms produce multiple xylanases, implying a strategy for effective hydrolysis of β-1,4-xylan. Each enzyme may have a specialized function in the degradation of the complex polysaccharides, and specialized functions of individual xylanases may be useful for applications in the food, feed, and paper industries (13, 26, 34, 36, 39).
A gram-negative β-1,4-xylan-degrading bacterium, W-61, was previously isolated(20). The morphological, biochemical, and physiological properties of strain W-61 were characterized according to Bergey's manual of systematic bacteriology, and it was identified as Aeromonas caviae W-61 (20). However, in this study, the previously used classification of the bacterium is emended to Paenibacillus sp. strain W-61 on the basis of the nucleotide sequence of the 16S rRNA gene. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by zymography using Remazol brilliant blue-stained xylan showed that strain W-61 produced five extracellular xylanases, numbered 1, 2, 3, 4, and 5, with molecular masses of 22, 41, 58, 120, and 140 kDa, respectively (20, 21). All of the xylanases of strain W-61 have been purified and characterized (20, 21, 23, 28), and the differences among the xylanolytic products of the enzymes have been found. Briefly, xylanases 1, 2, 4, and 5 hydrolyzed oat spelt xylan to liberate xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose as the major products, whereas xylanase 3 produced xylo-oligosaccharides larger than xylohexaose from the same substrate (20, 21, 23, 28). The genes encoding xylanases 1 and 3 (i.e., xyn1 and xyn3, respectively) have been cloned and characterized: xyn1 and xyn3 encode a family 11 and a family 5 xylanase, respectively (23; V. D. Nguyen et al., unpublished results). We also showed that xylanase 2 and xylanase 4 are C-terminally truncated derivatives of xylanase 3 and xylanase 5, respectively, having enzymatic properties different from those of their original molecular species (23, 28).
In this study, we cloned and expressed the xyn5 gene in Escherichia coli and studied the subcellular localization of Xyn5. The results obtained indicate that Xyn5 is a cell surface-anchored family 10 xylanase that possesses a functional cellulose-binding module and a characteristic C-terminal part consisting of a domain similar to the lysine-rich region of Clostridium thermocellum SdbA (i.e., scaffoldin dockerin binding protein A, which anchors cellulosome to the bacterial surface) (16, 17), and three S-layer-homologous (SLH) domains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The Xyn5-producing strain W-61 was isolated and stocked by our laboratory (20). E. coli DH5 and E. coli DH5α were used as the hosts for cloning vectors. Charomid 9-36 and pUC119 were purchased from Nippon Gene (Tokyo, Japan) and TaKaRa (Kyoto, Japan), respectively. Paenibacillus sp. strain W-61 was aerobically grown at 23°C in medium I (0.7% oat spelt xylan, 0.2% yeast extract, 0.25% NaCl, 0.5% NH4Cl, 1.5% KH2PO4, 3% Na2HPO4, and 0.025% MgSO4 · 7H2O, pH 7.0) as described previously (28). The E. coli strains were cultivated in Luria-Bertani medium or 2× YT medium (29).
Analysis of 16S rRNA gene.
Chromosomal DNA was isolated from the cleared lysate of strain W-61 as described previously (23). The 16S rRNA gene was amplified by PCR essentially as described by Edwards et al. (11): 5′-AGAGTTTGATCCTGGCTCAG-3′ (primer A) and 5′-AAGGAGGTGATCCAGCCGCA-3′ (primer H) were used as the forward and reverse primers, respectively, and Ex Taq polymerase (TaKaRa) was used.
Cloning of xyn5 gene.
Standard DNA manipulations were done as described by Sambrook et al. (29). Initially, part of the xyn5 gene was amplified by PCR using chromosomal DNA from strain W-61 as the template and 5′-CARATHGTHTAYCARGAR-3′ and 5′-RACRAANCGRAGRAARAG-3′ (where R, Y, H, and N indicate A/G, T/C, A/T/C, and A/T/C/G, respectively) as the forward and reverse primers, respectively. The primers were synthesized according to the N-terminal amino acid sequences of intact Xyn5 and of fragment IV from cyanogen bromide-cleaved Xyn5 (28). PCR was done for 30 cycles with Ex Taq polymerase using the following temperature profile: 94°C for 1 min, 44°C for 1 min, and 72°C for 2 min. As a result, a 1.8-kbp fragment was amplified by PCR. The amplified 1.8-kbp fragment was inserted in the HincII site of pUC119. DNA sequencing showed that the 1.8-kbp fragment contained nucleotide sequences corresponding to the N-terminal amino acid sequences of intact Xyn5 and of fragments I, II, III, and IV from cyanogen bromide-cleaved Xyn5 (28). The cloned 1.8-kbp fragment was digested with HindIII, and the resultant 0.9-kbp fragment, which was located within the 1.8-kbp fragment, was used as a probe for DNA hybridization after being labeled with the ECL random-prime labeling system (Amersham International, Little Chalfont, Buckinghamshire, United Kingdom).
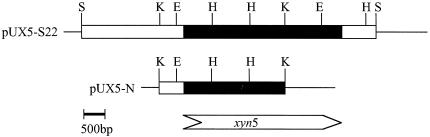
Chromosomal DNA of strain W-61 was digested with restriction enzymes, and the resultant fragments were analyzed by Southern hybridization using the labeled 0.9-kbp probe described above. Hybridization was done on Hybond N+ nylon sheets (Amersham) at 55°C overnight. As a result, the 0.9-kbp probe hybridized with bands corresponding to a 7.0-kbp SacI fragment, a 3.5-kbp EcoRI fragment, and a 3.0-kbp KpnI fragment. Because we failed to confirm that the 7.0-kbp SacI fragment contained the full-length of xyn5, we tried to clone the 3.5-kbp EcoRI fragment. The 3.5-kbp EcoRI fragments were inserted in pUC119, and colony hybridization was done on the Hybond N+ nylon sheet at 55°C overnight using the 0.9-kbp probe. A pUC119 plasmid carrying the 3.5-kbp EcoRI fragment (pE8-A1) was isolated from a positive clone. pE8-A1 was digested with KpnI or HindIII, and the resultant DNA fragments were subcloned in pUC119. DNA sequencing indicated that the 3.5-kbp EcoRI fragment lacked the nucleotides corresponding to the C-terminal region of Xyn5. Therefore, to analyze the 7.0-kbp SacI fragment, another probe for DNA hybridization was prepared on the basis of the nucleotide sequence of the 3.5-kbp EcoRI fragment: The probe corresponded to a 0.6-kbp region which is downstream of the 0.9-kbp HindIII fragment and upstream of the KpnI site on the 3.5-kbp EcoRI fragment (Fig. 1). The 0.6-kbp fragment was amplified by PCR, and the amplified 0.6-kbp fragment was labeled with the ECL random-prime labeling system. Chromosomal DNA of strain W-61 was digested with SacI, EcoRI, KpnI, and HindIII, and the resultant DNA fragments were analyzed by Southern hybridization using the 0.6-kbp probe. A restriction enzyme map for SacI, KpnI, EcoRI, and HindIII sites suggested that the 7.0-kbp SacI fragment contained the full-length xyn5 (Fig. 1). The 7.0-kbp SacI fragments were inserted into charomid 9-36 and transformed into E. coli DH5. Colony hybridization was done using the labeled 0.6-kbp probe, and four positive clones were obtained from ∼30,000 colonies. The plasmid carrying xyn5 (pC36-S8) was isolated from a positive clone. pC36-S8 was digested with SacI, and the 7-kbp fragment containing xyn5 was cloned in pUC119. A pUC119 plasmid carrying the SacI fragment in the reverse direction to the lac promoter was selected and designated pUX5-S22. pUX5-S22 was digested with EcoRI, KpnI, or HindIII, and the resultant DNA fragments were subcloned in pUC119 and sequenced.
FIG. 1.
Restriction maps of pUX5-S22 and pUX5-N. The open bars represent cloned DNA fragments in the pUC119 vector, and the solid parts correspond to the xyn5 gene in pUX5-S22 and a 5′ part of the xyn5 gene in pUX5-N. The transcriptional direction of xyn5 is shown beneath the restriction maps. S, SacI; E, EcoRI; H, HindIII; K, KpnI.
DNA sequencing and similarity search.
DNA sequencing was done using an ABI Model 377 DNA sequencer (Perkin-Elmer, Foster City, Calif.) and the ABI Prism Dye Terminator Cycle Sequencing ready-reaction kit (Perkin-Elmer). The nucleotide sequences were analyzed with the GENETYX version 10 software package (Software Development, Tokyo, Japan). A similarity search for DNA sequences was done using BLAST on the DDBJ-GenBank-EMBL nucleotide sequence databases.
SDS-PAGE, Western blotting, and zymography.
SDS-PAGE was done as described previously (14). Proteins were electroblotted onto a polyvinylidene difluoride sheet, and the blotted proteins were immunostained with specific antiserum against a 120-kDa recombinant Xyn5 [rXyn5(120k); see below]. The protein bands were visualized by using alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Promega, Madison, Wis.), nitroblue tertrazolium (Wako Pure Chemicals, Osaka, Japan), and 5-bromo-4-chloro-indolylphosphate (Wako Pure Chemicals). Antiserum was raised in female New Zealand White rabbits, which were immunized with rXyn5(120k) emulsified in Freund's complete and incomplete adjuvants (Difco Laboratory, Detroit, Mich.). Zymography for xylanases was done using Remazol brilliant blue-stained xylan (Nacalai Tesque, Kyoto, Japan), as described previously (28).
Protein assay and protein sequencing.
Protein was assayed by the method of Bradford using bovine serum albumin as a standard (5). The N-terminal amino acid sequences of the blotted protein bands were analyzed on a polyvinylidene difluoride sheet using an ABI Model 491 protein sequencer (Perkin-Elmer).
Expression and purification of recombinant Xyn5.
E. coli DH5α carrying pUX5-S22 was grown at 37°C in 2× YT medium with ampicillin (100 μg/ml). The bacteria were collected and suspended in 30 mM Tris-HCl buffer (pH 8.0) containing 2.5 mM EDTA and centrifuged at 3,500 × g for 10 min. The resultant pellet was vigorously suspended by agitation in chilled H2O containing 1 mM phenylmethylsulfonyl fluoride to give an osmotic shock to the bacterial cells. After removal of the cells by centrifugation at 15,000 × g for 10 min, the supernatant was concentrated 10-fold by ultrafiltration using a model Minitan Plate 4/PK 10,000 NMHL (Millipore Co., Bedford, Mass.). The concentrated supernatant was put onto a DEAE-Toyopearl 650 M column (Tosoh, Tokyo, Japan) equilibrated with 50 mM sodium phosphate buffer (pH 7.0). The adsorbed proteins were eluted with a 0 to 0.2 M NaCl linear gradient at a flow rate of 1 ml/min. The active fraction, eluted at 0.16 M NaCl, was dialyzed against 50 mM sodium phosphate buffer (pH 7.0) and loaded onto a DEAE-5PW column (Tosoh). The adsorbed proteins were eluted with a 0 to 0.2 M NaCl linear gradient at a flow rate of 0.5 ml/min. The active fractions, eluted at 0.16 M NaCl, were mixed with the same volume of 3 M ammonium sulfate and loaded onto a Phenyl-5PW column (Tosoh) equilibrated with 25 mM sodium phosphate buffer (pH 6.8) containing 1.5 M ammonium sulfate. The adsorbed proteins were eluted with 1.5 to 0 M ammonium sulfate at a flow rate of 0.5 ml/min. The active fractions, eluted with 0.2 to 0 M ammonium sulfate, were dialyzed against 5 mM sodium phosphate buffer (pH 7.0) and loaded onto an HA-1000 column (Tosoh). The adsorbed proteins were eluted with a 5 to 100 mM linear gradient of sodium phosphate buffer (pH 7.0) at a flow rate of 0.5 ml/min. rXyn5, eluted with 65 mM sodium phosphate buffer, was collected and dialyzed against 10 mM sodium phosphate buffer (pH 7.0) and stored at −80°C.
Preparation of an N-terminal 90-kDa fragment of Xyn5, Xyn5(90k).
The KpnI fragment obtained from pUX5-S22 was inserted into pUC119, and the plasmid possessing the KpnI fragment in the right direction (pUX5-N) (Fig. 1) was selected. E. coli DH5α harboring pUX5-N was grown at 37°C in 2× YT medium with ampicillin (100 μg/ml). The bacteria were collected and osmotically shocked as described above. After centrifugation, the supernatant was concentrated, and Xyn5(90k) was purified by high-performance column chromatography using DEAE-5PW and Phenyl-5PW columns under the conditions described above.
Analysis of hydrolytic products from oat spelt xylan.
Oat spelt xylan (1 mg) was incubated with rXyn5(120k) or rXyn5(90k) (0.2 U) in 200 μl of 50 mM sodium phosphate buffer (pH 7.0) at 37°C for 1 or 2 h. The products were analyzed by thin-layer chromatography using Silica Gel 60F 254 (E. Merck, Darmstadt, Germany) as described previously (28).
Binding of rXyn5(120k) and rXyn5(90k) to crystalline cellulose and insoluble xylan.
Ten micrograms of purified rXyn5(120k) or rXyn5(90k) was mixed with 10 to 40 mg of Avicel PH-101 (Fluka, Neu-Ulm, Germany) or 10 mg of oat spelt xylan (Nacalai Tesque) in 300 μl of 50 mM potassium phosphate buffer (pH 7.0). The mixtures were incubated at 0°C for 30 min with occasional stirring and then centrifuged at 13,000 × g and 4°C for 10 min. The supernatants obtained were stored at 4°C, and the precipitates were suspended in the same buffer and centrifuged. The precipitates obtained were suspended in 300 μl of the same buffer. Ten microliters of the supernatants and the suspended precipitates were withdrawn and analyzed using Western immunoblotting.
Preparation of subcellular fractions of strain W-61.
Strain W-61 was grown at 23°C for 12, 24, 36, or 48 h in the presence of 0.7% oat spelt xylan. Bacteria were collected by centrifugation at 5,000 × g and 4°C for 15 min, and the supernatant was stored. The cells were suspended in 10 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid- NaOH buffer (pH 7.4) and disrupted by being passed through a French pressure cell (SLM Instruments, Rochester, N.Y.) at 1,200 kg/cm2 two times. The passed fraction was centrifuged at 5,000 × g to remove unbroken cells, and the supernatant obtained was centrifuged at 140,000 × g and 4°C for 2 h. The supernatant obtained was stored as a cytosol fraction. The precipitated membrane fraction was suspended in the same buffer, layered onto a discontinuous sucrose gradient (2.02, 1.44, and 0.77 M sucrose), and centrifuged using an SW40Ti rotor (Beckman Instruments, Palo Alto, Calif.) at 52,000 × g and 4°C for 15 h, as described by Smit et al. (32). Fractions, collected by puncturing the bottom of the centrifugal tube, were assayed for absorbance at 280 nm, for Xyn5, and for succinate dehydrogenase activity (24).
Electron microscopy.
Specimens on a carbon-coated grid were negatively stained with 1% (wt/vol) sodium phosphotungstic acid (pH 7.2) and examined under an electron microscope (H-8100; Hitachi Co., Tokyo, Japan) at an acceleration voltage of 100 kV.
Susceptibility of cell-bound Xyn5 to trypsin digestion.
Strain W-61 was grown at 23°C for 24 h, and the bacteria were collected by centrifugation and washed with 10 mM sodium phosphate buffer, pH 7.0, containing 0.85% NaCl. The washed cells were suspended in 200 μl of the same buffer and treated with trypsin from bovine pancreas (Sigma Chemicals, St. Louis, Mo.) at a final concentration of 0, 1, 10, or 100 μg/ml at 37°C for 30 min. After the addition of 0.1 mM N-tosyl-l-lysyl chloromethyl ketone (Sigma Chemicals), the cell suspensions were centrifuged at 8,000 × g and 4°C for 10 min. Portions of the trypsin-treated cell suspensions and the supernatants (1.6 and 16 μl, respectively) were withdrawn and analyzed by Western immunoblotting and zymography.
Nucleotide sequence accession numbers.
The sequence of the 16S rRNA gene of strain W-61 was deposited under DDBJ-GenBank-EMBL accession number AB110989. The nucleotide sequence of xyn5 and its flanking regions was deposited under DDBJ-GenBank-EMBL accession number AB098080.
RESULTS
Analysis of 16S rRNA gene of strain W-61.
The xylan-degrading bacterium W-61 was previously identified as A. caviae W-61 on the basis of morphological and biochemical properties (20). The strain W-61 used in this study revealed the same gram-negative rod-shaped morphology and the same zymographic profile of xylanases as the original bacterial stock did. However, because we failed to detect 2-keto-3-deoxyoctonate in the membrane fractions of strain W-61, we tried to verify the bacterial species by sequencing the 16S rRNA gene. As a result, the 16S rRNA gene of strain W-61 revealed 99, 97, 96, and 94% identity with those of Paenibacillus illinoisensis, Paenibacillus pabuli, Paenibacillus amylolyticus, and Paenibacillus macquariensis, respectively. Previous reports said that P. illinoisensis, P. pabuli, and P. amylolyticus were gram positive, whereas P. macquariensis and Paenibacillus glucanolyticus were gram negative (1, 9, 19, 31). Furthermore, no sporulating W-61 cell was observed under a phase-contrast microscope after a 3-week cultivation on Luria-Bertani plates, while P. illinoisensis and P. amylolyticus have been shown to form spores (31). Taking this evidence together, we classified W-61 as Paenibacillus sp. strain W-61.
Cloning and nucleotide sequence of the xyn5 gene of Paenibacillus sp. strain W-61.
xyn5 of strain W-61 was cloned, and the nucleotide sequences of xyn5 and its flanking regions were determined. The open reading frame of xyn5 consisted of 3,981 nucleotides, encoding a protein consisting of 1,326 amino acids with a predicted molecular weight of 142,760. Putative −35 (GTGAGA) and −10 (TAAATT) sequences were found 83 and 60 bp upstream of the translation initiation codon of xyn5, respectively. A potential Shine-Dalgarno sequence (GAAGG) was also present 12 bp upstream of the gene. No termination loop was found downstream of the TAA stop codon. The nucleotide sequence corresponding to a typical signal peptide was present at the 5′ end of the open reading frame. Taken together with the N-terminal amino acid sequence determined for purified Xyn5 (28), this indicates that nascent Xyn5 would be cleaved by a signal peptidase at the position between Ala27 and Val28 to produce mature Xyn5 comprising 1,299 amino acid residues with a predicted molecular mass of 140,008 Da.
Amino acid sequence and modular structure of Xyn5.
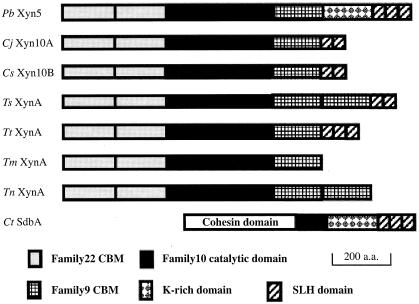
A similarity search on the SWISS-PROT database for the amino acid sequence of Xyn5 showed that mature Xyn5 is a multidomain enzyme consisting of eight discrete domains: two family 22 carbohydrate binding modules (CBMs), a family 10 catalytic domain of glycosyl hydrolases, a family 9 CBM, a domain which has 36% identity with the lysine-rich region of C. thermocellum SdbA (17), and three SLH domains, in order from the N terminus. Figure 2 shows the modular structure of Xyn5 schematically, along with those of several family 10 xylanases and C. thermocellum SdbA. The N-terminal part of Xyn5 has the same domain organization as several family 10 xylanases, and this part of Xyn5 has 51.0% amino acid sequence identity with the N-terminal part of Clostridium josui Xyn10A. In contrast, the C-terminal part of Xyn5 has the same domain organization as C. thermocellum SdbA, a cell surface-anchored protein which binds cellulosome at its N-terminal cohesin domain (16, 17).
FIG. 2.
Modular structures of Paenibacillus sp. strain W-61 Xyn5 and several family 10 xylanases, along with C. thermocellum SdbA. Modular structures, including family 22 CBMs, family 10 catalytic domains, family 9 CBMs, lysine-rich domains, and SLH domains, are illustrated for Paenibacillus sp. strain W-61 (Pb) Xyn5, C. josui (Cj) Xyn10A, C. sterocorarium (Cs) Xyn10B, T. saccharolyticum (Ts) XynA, T. thermosulfurigenes (Tt) XynA, T. maritima (Tm) Xyn10A, T. neapolitana (Tn) XynA, and C. thermocellum (Ct). a.a., amino acid residues.
The N-terminal region of Xyn5 containing family 22 CBMs (i.e., CBM22-1 and CBM22-2), extending from positions 43 to 365 (numbered from the initial Met of the polypeptide encoded by xyn5), have sequence similarity with the family 22 CBMs of C. josui Xyn10A, Clostridium stercorarium Xyn10B, C. thermocellum Xyn10B, Clostridium fini XynC, Thermotoga maritima Xyn10A, and Thermoanaerobacterium saccharolyticum XynA (i.e., the sequence identities between the family 22 CBMs of Xyn5 and those of the other xylanases are 46.8 to 21.9%). It has been shown that the family 22 CBM of T. maritima Xyn10A and several other family 10 xylanases have the ability to bind soluble xylan (3, 8, 10, 18) and that the family 22 CBMs of T. maritima Xyn10A and T. saccharolyticum XynA contribute to the heat stability of these enzymes (15, 18, 35). The family 10 catalytic domain of Xyn5, extending from positions 370 to 712 (Fig. 2), has extensive sequence similarity with the catalytic domains of several family 10 xylanases (i.e., 61.6, 52.7, 38.5, 37.9, 37.3, and 36.8% sequence identity with C. josui Xyn10A, C. stercorarium Xyn10B, Thermotoga neapolitana XynA, T. maritima Xyn10A, C. thermocellum XynX, and Thermoanaerobacterium thermosulfurigenes XynA, respectively). The Glu residues (Glu498 and Glu621), which are considered to be involved in general acid-base catalysis, are conserved in Xyn5. CBM9 of Xyn5, extending from positions 713 to 898, has high sequence similarity with family 9 CBMs of other family 10 xylanases (i.e., 67.6, 57.8, 50.0, 49.5, 49.2, 45.9, and 45.4% sequence identity with those of C. josui Xyn10A, C. stercorarium Xyn10B, C. thermocellum XynX, T. thermosulfurigenes XynA, T. saccharolyticum XynA, T. neapolitana XynA, and T. maritima Xyn10A, respectively). The family 9 CBMs of C. stercorarium Xyn10B and T. maritima Xyn10A have been shown to bind to crystalline cellulose, noncrystalline cellulose, and xylo-oligosaccharides (2, 38). The region, which ranges from positions 939 to 1147 and is located between CBM9 and the SLH domains, has 36% identity with the lysine-rich region of C. thermocellum SdbA, although the lysine content (5.9%) of this region is less than that (12.5%) of the lysine-rich region of C. thermocellum SdbA. The SLH domains of Xyn5, ranging from positions 1148 to 1326, have 47.5 to 29.8% sequence identity with those of C. thermocellum SdbA, T. saccharolyticum XynA, T. thermosulfurigenes AmyB, C. josui CelA, C. thermocellum OlpB, Bacillus anthracis Sap, and B. anthracis EA1.
Expression of xyn5 in E. coli and properties of recombinant Xyn5 and an N-terminal 90-kDa fragment of Xyn5.
Whole cells of E. coli 5α harboring pUX5-S22 gave a major and a minor immunoreactive and xylanolytic band corresponding to 120 and 140 kDa, respectively, on Western immunoblotting and zymography (results not shown). Therefore, xyn5 was expressed in the recombinant E. coli DH5α, and the major part of rXyn5 was present as a truncated form with a molecular mass of 120 kDa. Because xyn5 was inserted into pUC119 in the reverse orientation to the lac promoter, the putative promoter of xyn5 would be functional in the recombinant E. coli. Furthermore, Western immunoblotting showed that rXyn5 was present in the periplasmic fraction of the recombinant E. coli (results not shown), suggesting that the putative signal peptide of Xyn5 is recognizable by the secretory system of E. coli.
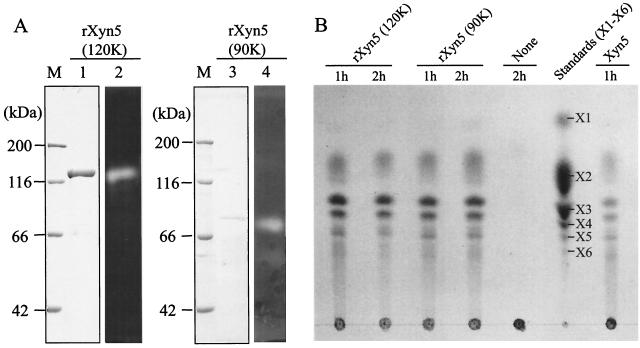
rXyn5 was purified as a xylan-degrading 120-kDa protein from the periplasmic fraction of E. coli harboring pUX5-S22 (Fig. 3A, lanes 1 and 2). The N-terminal amino acid sequence of the purified Xyn5 was determined to be Ala-Asp-Pro-Glu-Ala-Ser-Asn-Ser-Gln-Ile-Val-Tyr-, indicating that rXyn5 was purified as a truncated form lacking the three N-terminal amino acid residues (i.e., Val-Ser-Ala) and the C-terminal 20-kDa region. The three N-terminal amino acids of Xyn5 would have been cleaved by the signal peptidase of E. coli DH5α or by limited proteolysis in the periplasm of the bacterium. Truncation of the C-terminal 20-kDa fragment would occur mainly in the periplasm of E. coli DH5α, because Western immunoblotting for whole cells of the recombinant E. coli DH5α revealed a major immunoreactive band corresponding to 120 kDa, as stated above. The truncated form of rXyn5 was designated rXyn5(120k), and the cleaved C-terminal 20-kDa fragment may correspond to the SLH domains on the basis of molecular size. To characterize the enzymatic and substrate-binding properties of the N-terminal part of Xyn5, we also constructed a plasmid carrying the 5′ part of xyn5 (i.e., pUX5-N) (Fig. 1), which encodes the N-terminal part of Xyn5, including CBM22, the family 10 catalytic domain, and the N-terminal half of CBM9. The recombinant N-terminal fragment of Xyn5 was expressed in E. coli DH5α and purified from the periplasmic fraction of the bacterium. The purified N-terminal fragment of Xyn5 gave a xylan-degrading protein band corresponding to 90 kDa and was designated rXyn5(90k) (Fig. 3A, lanes 3 and 4). Protein sequencing showed that the N-terminal amino acid sequence of rXyn5(90k) was the same as that of rXyn5(120k). Both rXyn5(120k) and rXyn5(90k) hydrolyzed oat spelt xylan to liberate xylobiose, xylotriose, xylotetraose, and xylopentaose and traces of xylose and xylohexaose (Fig. 3B), indicating that rXyn5(120k) and rXyn5(90k) produced the same hydrolytic products as did xylanase 5 purified from the culture supernatants of strain W-61 (28).
FIG. 3.
(A) SDS-PAGE and zymography of purified rXyn5(120k) and rXyn5(90k). Lanes 1 and 3, protein bands stained with Coomassie brilliant blue R-250; lanes 2 and 4, zymography using Remazol brilliant blue-stained xylan; lanes M, molecular markers. (B) Thin-layer chromatography of hydrolytic products from oat spelt xylan by recombinant enzymes. Thin-layer chromatography was done as described in Materials and Methods. None, without enzyme. Xylose (X1) and xylo-oligosaccharides (X2 to X6) were used as standards.
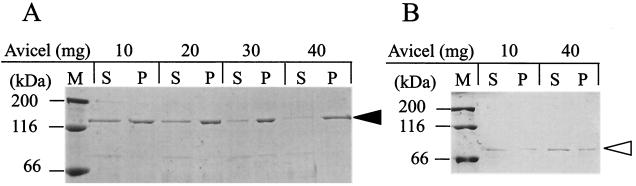
The family 10 xylanases, such as T. maritima Xyn10A and C. stercorarium Xyn10B, have cellulose-binding CBM9 (2, 38). Therefore, we tested the binding of rXyn5(120k) and rXyn5(90k) to a crystalline cellulose, Avicel PH-101. rXyn5(120k) or rXyn5(90k) was incubated with various amounts of Avicel PH-101 at 0°C for 30 min, and the mixture was centrifuged at 13,000 × g for 10 min to precipitate Avicel PH-101 particles, followed by Western immunoblotting for rXyn5 in the supernatants and precipitates obtained. As shown in Fig. 4A, the amount of unbound rXyn5(120k) in the supernatants decreased with increasing amounts of Avicel PH-101, and the amount of coprecipitated rXyn5(120k) increased concomitantly. In contrast, we observed no significant differences in the amounts of rXyn5(120k) in the precipitates and supernatants irrespective of the amount of Avicel PH-101 used (Fig. 4B). These results indicate that rXyn5(120k) bound to Avicel PH-101 particles, while rXyn5(90k) did not bind significantly to the crystalline cellulose, suggesting that Xyn5 has a cellulose-binding activity that is attributable to the CBM9 of the enzyme. In addition, binding of rXyn5(120k) to Avicel PH-101 was not significantly affected by the addition of soluble oat spelt xylan (0 to 20 mg in 300 μl) to the reaction mixtures for the cellulose-binding assay (results not shown), suggesting that CBM9 has a cellulose-specific binding ability. Because xylan-binding activity was anticipated for family 22 CBMs, we assayed the binding of rXyn5(120k) to insoluble oat spelt xylan as for cellulose binding. However, rXyn5(120k) did not bind significantly to insoluble oat spelt xylan (results not shown). Furthermore, the mobility of rXyn5(120k) in polyacrylamide gel electrophoresis was not decreased in the presence of 0, 0.2, 0.5, or 1.0% (wt/vol) soluble oat spelt xylan (results not shown). In affinity polyacrylamide gel electrophoresis, the binding of soluble xylan to the CBMs of xylanases can be detected as retarded mobility of the protein band (8, 18, 25, 40). These results suggested that family 22 CBMs of Xyn5 have no substantial xylan-binding activity.
FIG. 4.
Adsorption of rXyn5(120k) (A) and rXyn5(90k) (B) to a crystalline cellulose, Avicel PH-101. Ten micrograms of rXyn5(120k) or rXyn5(90k) was incubated with Avicel PH-101 (10 to 40 mg) at 4°C for 30 min in 300 μl of 50 mM potassium phosphate buffer (pH 7.0). After centrifugation, the supernatants (S) obtained were stored at 0°C and the precipitates (P) were washed and suspended in the same buffer. Portions of the S and P fractions were analyzed by Western immunoblotting. M, molecular markers. The solid and open arrowheads indicate the positions of rXyn5(120k) and rXyn5(90k), respectively.
Cell surface localization of Xyn5.
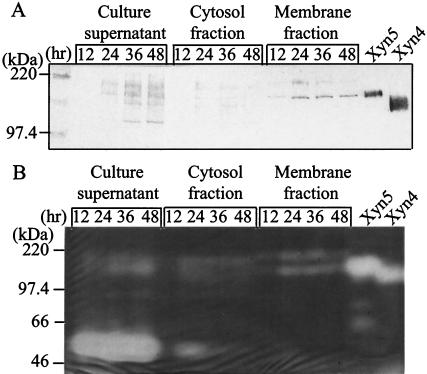
Culture supernatants, membrane fractions, and cytosol fractions of strain W-61 were analyzed for Xyn5 using Western immunoblotting and zymography. When strain W-61 was cultivated for 12 h, an immunoreactive and xylanolytic band corresponding to 140 kDa (i.e., Xyn5) was detected in the membrane fraction (but not in the culture supernatant and cytosol fractions), and the amount of Xyn5 in the membrane fraction increased and reached a plateau in the following 12 h (Fig. 5). After cultivation for 24 h or longer, the band corresponding to Xyn5 appeared in the culture supernatants as well, and another immunoreactive and xylanolytic band corresponding to 120 kDa (i.e., Xyn4, a C-terminally truncated Xyn5) was detected in the culture supernatants (Fig. 5A). Because we used 10-fold more culture supernatants than cytosol and membrane fractions in the assay, the results showed that newly synthesized Xyn5 was exclusively cell bound, and a small portion of the cell-bound Xyn5 was thereafter released into the medium. The release of Xyn5 into the medium is consistent with previous observations that strain W-61 excreted Xyn5 and Xyn4 into the medium after cultivation for 36 h or longer (28). Based on its molecular mass, the truncated C-terminal 20-kDa fragment of Xyn5 would correspond to the SLH domains, suggesting that the membrane-bound Xyn5 was released into the medium by limited proteolysis of the anchoring modules. It should be noted that another immunoreactive and xylanolytic band corresponding to 200 kDa was visible in the membrane fraction after cultivation for 24 h or longer (Fig. 5), suggesting that Xyn5 forms an SDS-resistant complex on the cell surface. Incidentally, a xylanolytic band corresponding to 58 kDa (i.e., Xyn3) was detected in the culture supernatants (but not in the membrane fractions) after cultivation for 12 to 48 h (Fig. 5B), indicating that Xyn3 was secreted into the medium.
FIG. 5.
Subcellular localization of Xyn5. After cultivation of strain W-61 at 23°C for 12, 24, 36, or 48 h, cells were collected and disrupted as described in Materials and Methods. Small portions of culture supernatants, cytosol fractions, and membrane fractions (equivalent to 0.1, 0.01, and 0.01 ml of the culture, respectively) were analyzed by Western immunoblotting (A) and zymography (B).
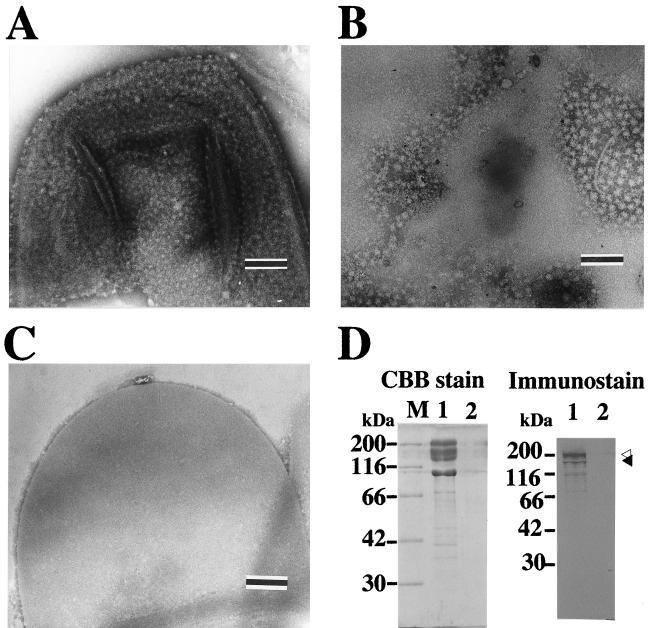
To further study the localization of Xyn5, the membrane fraction was fractionated by discontinuous sucrose gradient centrifugation. After the centrifugation, a white and a reddish-brown band were visible in the positions corresponding to 2.02 M sucrose and the upper part of 1.44 M sucrose, respectively. Assay of absorbance at 280 nm for the fractions obtained revealed two major peaks, which corresponded to the white and the reddish-brown bands (results not shown). The peak fractions corresponding to the white and the reddish-brown bands were designated H (heavy) and L (light) fractions, respectively. Western immunoblotting and zymography showed that Xyn5 was exclusively distributed in the H fraction (results not shown). Succinate dehydrogenase was preferentially detected in the L fraction, and a number of protein bands were visible on SDS-PAGE for the L fraction (results not shown), indicating that the L fraction had characteristics of the cytoplasmic membrane. In contrast, the H fraction revealed a major protein band corresponding to 100 kDa and a small number of minor bands on SDS-PAGE, and the 100-kDa protein was localized in the H fraction (results not shown). The N-terminal amino acid sequence of the 100-kDa protein was determined to be Asp-Thr-Ala-Thr-Ser-Pro-Gln-Gln-Gln-Phe-Asp-Ala-Leu-Lys-Ala-Lys-Gly-Ile-Phe-Asn-Gly-Tyr-Pro-Asp-. The N-terminal amino acid sequence of the 100-kDa protein showed similarity to the SLH domain on a search using the National Center for Biotechnology Information Conserved Domain Database(http://www.ncbi.nih.gov/Structure/cdd/wrpsb.cgi). Electron microscopy for negatively stained specimens of the H fraction revealed sheet-like structures on which particles with diameters of ∼25 nm were arrayed, and side views of the particles were also visible on the edges of the sheets (Fig. 6A). Treatment of the H fraction with 5 M guanidinium HCl detached the particles from the sheet-like structures (Fig. 6B), and the remaining sheets with smooth surfaces were digested by egg white lysozyme (results not shown). The soluble fraction obtained by the guanidinium HCl treatment contained particles with the same dimensions (Fig. 6C), and the soluble fraction contained the majority of Xyn5 and the 100-kDa protein (Fig. 6D). These results indicate that the sheet-like structures in the H fraction consisted of a peptidoglycan-containing layer and an associated S layer and that Xyn5 was associated with the S layer. Incidentally, each particle of the S layer may comprise several molecules of the 100-kDa proteins, based on the size.
FIG. 6.
Electron microscopy of the H fraction (A) and the guanidinium HCl-soluble (B) and -insoluble (C) fractions obtained from the H fraction, as well as Western immunoblot analysis for distribution of Xyn5 in the fractions (D). (A to C) The H fraction was treated with 5 M guanidinium HCl at room temperature for 1 h and centrifuged at 105,000 × g and 4°C for 1 h. The H fraction and the guanidinium HCl-soluble and -insoluble fractions (i.e., the supernatant and the precipitate obtained, respectively) were subjected to electron microscopy after dialysis against 10 mM phosphate buffer (pH 7.2). The scale bars represent 200 nm. (D) Portions of the guanidinium HCl-soluble and -insoluble fractions were analyzed by SDS-PAGE (left), followed by Western immunoblotting using specific antiserum against rXyn5(120k) (right). Lanes 1 and 2, guanidinium HCl-soluble and -insoluble fractions, respectively. The solid and open arrowheads indicate the bands corresponding to 140 and 200 kDa, respectively. CBB, Coomassie brilliant blue R-250; M, molecular markers.
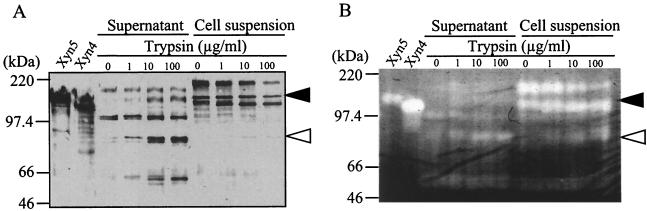
To elucidate whether Xyn5 is exposed on the cell surface, we assayed the susceptibility of the cell-bound Xyn5 to trypsin digestion and analyzed liberated Xyn5 fragments in the supernatants. As shown in Fig. 7, treatment of strain W-61 with trypsin produced immunoreactive and xylanolytic bands corresponding to ∼80 kDa in the supernatants, and the intensities of the bands increased with increasing concentrations of trypsin. Concomitantly, the intensity of the immunoreactive band corresponding to 140 kDa in the cell suspensions was decreased by trypsin treatment (Fig. 7A). Furthermore, the intensity of the immunoreactive and xylanolytic band corresponding to 200 kDa was also decreased by trypsin treatment (Fig. 7). Thus, Xyn5 is localized on the cell surface and is present, at least in part, as a 200-kDa complex. In addition, immunoreactive bands corresponding to 140 and 60 kDa were detected in the supernatants (Fig. 7A). The band corresponding to 140 kDa would be Xyn5 and/or a truncated form of Xyn5 that has lost a small part of the protein but retained xylanolytic activity. In contrast, the bands corresponding to ∼60 kDa were not xylanolytic (Fig. 7B), suggesting that the Xyn5 fragments lost xylanase activity because of the truncation of part of the catalytic domain.
FIG. 7.
Susceptibility of cell-bound Xyn5 to tryptic digestion. Washed cells were treated with trypsin (final concentration, 0, 1, 10, or 100 μg/ml) at 37°C for 30 min. After the addition of 0.1 mM N-tosyl-l-lysyl chloromethyl ketone, the cell suspensions were centrifuged at 8,000 × g for 10 min. Portions of the trypsin-treated cell suspensions and the supernatants (1.6 and 16 μl, respectively) were withdrawn and analyzed using Western immunoblotting (A) and zymography (B). Note that the amounts of the supernatants used were 10-fold more than those of the cell suspensions. The solid and open arrowheads indicate the bands corresponding to 140 and 80 kDa, respectively.
DISCUSSION
In this study, we showed that Xyn5 of Paenibacillus sp. strain W-61 is a cell surface-anchored family 10 xylanase possessing a functional cellulose-binding module and SLH domains. The N-terminal part of Xyn5 has the same modular structure as those of C. josui Xyn10A and several other family 10 xylanases, whereas the C-terminal part of Xyn5 has the same domain organization as C. thermocellum SdbA. SLH domains have been shown to anchor S-layer proteins and extracellular hydrolases acting on carbohydrate (i.e., pullulanase and xylanases) to the cell surface (7, 33). Therefore, the SLH domains would be involved in anchoring Xyn5 to the cell surface. In this study, we also showed that Xyn5 is associated with the cell envelope, consisting of a peptidoglycan-containing layer and an S layer (Fig. 5 and 6). Furthermore, after treatment of the H fraction with guanidinium HCl, Xyn5 was distributed exclusively in the S-layer fraction (Fig. 6). Taken together with the susceptibility of Xyn5 to trypsin digestion (Fig. 7), this indicates that Xyn5 could be enmeshed in the outermost S layer rather than being linked to the underlying peptidoglycan-containing layer through the SLH domains. In this context, it should be noted that a significant portion of Xyn5 was detected as an immunoreactive and xylanolytic band corresponding to 200 kDa, and this band was susceptible to trypsin digestion (Fig. 5 and 7). Thus, Xyn5 may be present, at least in part, as an SDS-resistant complex(s) with another protein(s) on the cell surface. A possible counterpart of Xyn5 in the complex is the 100-kDa S-layer protein, because the SLH domain is thought to mediate association between SLH domain-bearing polypeptides and because complexes of S-layer proteins from T. thermosulfurigenes were resistant to dissociation treatment with SDS (6, 30). However, previous studies suggested that a secondary cell wall polymer consisting mainly of N-acetylglucosamine and N-acetylmannosamine is involved in the association of the S-layer protein of Bacillus stearothermophilus PV72/p2 with the cell surface (27) and that common attachment sites for S-layer proteins and modular xylanase from T. thermosulfurigenes EM1 may be secondary cell wall polymers (6, 30). It was also shown that purified proteins of C. stercorarium Xyn10B and T. thermosulfurigenes EM1 XynA (i.e., family 10 xylanases with SLH domains) (Fig. 2) bound to cell wall fractions of the bacteria in vitro through SLH domains (6, 12). Therefore, the secondary cell wall polymer of strain W-61 is another molecule possibly responsible for the anchoring of Xyn5. Further study is needed to elucidate what molecule(s) is involved in anchoring Xyn5 to the cell surface.
The cellulose-binding activity of Xyn5 is attributable to CBM9, because a truncation of the N-terminal half of the module abolished the activity (Fig. 4). It has been shown that family 10 xylanases, such as T. maritima Xyn10A and C. stercorarium Xyn10B, have cellulose-binding activity, and the cellulose-binding activity is attributed to CBM9 (2, 38). Furthermore, eight amino acid residues of T. maritima Xyn10A CBM9 have been shown to be pivotal for its cellulose-binding activity (4, 22). Seven of the pivotal eight amino acid residues are conserved in the Xyn5 CBM9 (i.e., Trp781, Glu787, Gln806, Arg808, Gln861, Gly868, and Asn881), and one residue (Trp881 of T. maritima Xyn10A) is replaced with an aromatic amino acid, Tyr884, in Xyn5. Such a replacement of Trp with Tyr occurs in another family 10 xylanase, C. josui Xyn10A, which has cellulose-binding activity (12). Taken together with the fact that CBM9 of Xyn5 has 46% amino acid sequence identity with T. maritima Xyn10A, this indicates that the eight amino acid residues would be important for the cellulose-binding activity of CBM9. In addition to the cellulose-binding activity, Xyn5 would have xylan-binding activity because of the presence of the family 22 CBMs. It has been shown that family 22 CBMs of T. maritima Xyn10A and C. thermocellum Xyn10B have xylan-binding and thermostabilizing functions (3, 8, 10, 15, 18, 35). Four amino acid residues (Trp53, Tyr103, Tyr136, and Glu138) of the family 22-2 CBM of C. thermocellum Xyn10B have been shown to be pivotal for xylan-binding activity (8, 40), and they are conserved in other family 10 xylanases. Although CBM22-1 of Xyn5 has 26.4% amino acid sequence identity with CBM22-2 of C. thermocellum Xyn10B, it retains only one (Tyr103) of four important amino acid residues required for ligand binding (i.e., Glu138 and Trp53 are replaced with Lys and Tyr, respectively, and Tyr136 is missing). The replacement of Glu138 with Lys and the lack of Tyr136 in the family 22-1 CBM of Xyn5 would markedly decrease or abolish xylan-binding activity of the module. In contrast to CBM22-1, three (i.e., Tyr103, Tyr136, and Glu138) of four amino acid residues pivotal for substrate binding are conserved in CBM22-2 of Xyn5, and Trp53 is replaced with another aromatic amino acid, Tyr. In addition, CBM22-2 of Xyn5 has 28.2% amino acid sequence identity with C. thermocellum Xyn10B. Therefore, it seems likely that CBM22-2 of Xyn5 is intrinsically functional but inaccessible to xylans, possibly due to steric hindrance by the other domain(s) of the enzyme. Furthermore, the family 22 CBMs of Xyn5 have no substantial heat-stabilizing function, because the optimal temperature for Xyn5 is 40°C and it lost 70% of its xylanolytic activity upon incubation at 60°C for 10 min (28).
Finally, as far as the biological significance of the production of multiple xylanases by single species of microorganisms is concerned, the modular structure of Xyn5 would suggest a cooperative action of the multiple xylanases produced by Paenibacillus sp. strain W-61. The SLH domains of Xyn5 would anchor Xyn5 to the cell surface of the bacterium, and the cellulose-binding CBM9 of Xyn5 could combine the bacterial cells with cellulose microfibrils of the plant cell wall, leading to efficient hydrolysis of neighboring β-1,4-xylans by Xyn5 and the other xylanases secreted by the bacterium.
Acknowledgments
This work was supported in part by the Pioneering Research Project in Biotechnology of the Ministry of Agriculture, Forestry, and Fisheries of Japan. N. Roy was the recipient of a fellowship from the Ministry of Education, Science, Sports, and Culture of Japan. N. Sugawara-Tomita is the recipient of a postdoctoral fellowship from the Japan Society for Promotion of Science.
REFERENCES
- 1.Alexander, B., and F. G. Priest. 1989. Bacillus glucanolyticus, a new species that degrades a variety of β-glucans. Int. J. Syst. Bacteriol. 39:112-115. [Google Scholar]
- 2.Ali, M. K., H. Hayashi, S. Karita, M. Goto, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci. Biotechnol. Biochem. 65:41-47. [DOI] [PubMed] [Google Scholar]
- 3.Black, G. W., G. P. Hazlewood, S. J. Millward-Sadler, J. I. Laurie, and H. J. Gilbert. 1995. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem. J. 307:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boraston, A. B., A. L. Creagh, M. Alam, J. M. Kormos, P. Tomme, C. A. Haynes, R. A. J. Warren, and D. G. Kilburn. 2001. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40:6240-6247. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brechtel, E., and H. Bahl. 1999. In Thermoanaerobacterium thermosulfurigenes EM1 S-layer homology domains do not attach to peptidoglycan. J. Bacteriol. 181:5017-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brechtel, E., M. Matuschek, A. Hellberg, E. M. Egelseer, R. Schmid, and H. Bahl. 1999. Cell wall of Thermoanaerobacterium thermosulfurigenes EM1: isolation of its components and attachment of xylanase XynA. Arch. Microbiol. 171:159-165. [DOI] [PubMed] [Google Scholar]
- 8.Charnock, S. J., D. N. Bolam, J. P. Turkenburg, H. J. Gilbert, L. M. Ferreira, G. J. Davies, and C. M. Fontes. 2000. The X6 thermostabilizing domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry 39:5013-5021. [DOI] [PubMed] [Google Scholar]
- 9.Claus, D., and R. C. W. Berkeley. 1986. Genus Bacillus Cohn 1872, p. 1105-1140. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams and Wilkins Co., Baltimore, Md.
- 10.Dupont, C., M. Roberge, F. Shareck, R. Morosoli, and D. Kluepfel. 1998. Substrate-binding domains of glycanases from Streptomyces lividans: characterization of a new family of xylan-binding domains. Biochem. J. 15:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, U., T. Rogall, H. Bloecker, M. Emde, and E. Boetter. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, J. X., S. Karita, E. Fujino, T. Fujino, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Cloning, sequencing, and expression of the gene encoding a cell-bound multi-domain xylanase from Clostridium josui, and characterization of the translated product. Biosci. Biotechnol. Biochem. 64:2614-2624. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, H. J., and G. P. Hazlewood. 1993. Bacterial cellulases and xylanases. J. Gen. Microbiol. 139:187-194. [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lee, Y. E., S. E. Lowe, B. Henrissat, and J. G. Zeikus. 1993. Characterization of the active site and thermostability regions of endoxylanase from Thermoanaerobacterium saccharolyticum B6A-RI. J. Bacteriol. 175:5890-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leibovitz, E., and P. Beguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leibovitz, E., H. Ohayon, P. Gounon, and P. Beguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner, K., D. Wassenberg, and W. Liebl. 2000. The thermostabilizing domain of the modular xylanase Xyn10A of Thermotoga maritima represents a novel type of binding domain with affinity for soluble xylan and mixed-linkage β-1,3/β-1,4-glucan. Mol. Microbiol. 36:898-912. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura, L. K. 1984. Bacillus amylolyticus sp. nov., nom. rev.,Bacillus lautus sp. nov., nom. rev., Bacillus pabuli sp. nov., nom. rev., and Bacillus validus sp. nov., nom. rev. Int. J. Syst. Bacteriol. 34:224-226. [Google Scholar]
- 20.Nguyen, V. D., Y. Kamio, N. Abe, J. Kaneko, and K. Izaki. 1991. Purification and properties of β-1,4-xylanase from Aeromonas caviae W-61. Appl. Environ. Microbiol. 57:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen, V. D., Y. Kamio, N. Abe, J. Kaneko, and K. Izaki. 1993. Purification and properties of β-1,4-xylanase 2 and 3 from Aeromonas caviae W-61. Biosci. Biotechnol. Biochem. 56:1708-1712. [Google Scholar]
- 22.Notenboom, V., A. B. Boraston, D. G. Kilburn, and D. R. Rose. 2001. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry 40:6248-6256. [DOI] [PubMed] [Google Scholar]
- 23.Okai, N., M. Fukasaku, J. Kaneko, T. Tomita, K. Muramoto, and Y. Kamio. 1998. Molecular properties and activity of a carboxyl-terminal truncated form of xylanase 3 from Aeromonas caviae W-61. Biosci. Biotechnol. Biochem. 62:1560-1567. [DOI] [PubMed] [Google Scholar]
- 24.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 25.Peter, T., B. Alisdair, M. Jeffery, R. Kormos, J. Antony, and G. Douglas. 2000. Affinity electrophoresis for the identification and characterization of soluble sugar binding by carbohydrate-binding modules. Enzyme Microb. Technol. 27:453-458. [DOI] [PubMed] [Google Scholar]
- 26.Prade, R. A. 1995. Xylanases: from biology to biotechnology. Biotechnol. Genet. Eng. Rev. 13:101-129. [DOI] [PubMed] [Google Scholar]
- 27.Ries, W., C. Hotzy, I. Schocher, U. B. Sleytr, and M. Sara. 1997. Evidence that a secondary cell wall polymer recognizes the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2. J. Bacteriol. 179:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy, N., N. Okai, T. Tomita, K. Muramoto, and Y. Kamio. 2000. Purification and some properties of high-molecular-weight xylanases, the xylanases 4 and 5 of Aeromonas caviae W-61. Biosci. Biotechnol. Biochem. 64:408-413. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sara, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shida, O., H. Takagi, K. Kadowaki, L. K. Nakamura, and K. Komagata. 1997. Emended description of Paenibacillus amylolyticus and description of Paenibacillus illinoisensis sp. nov. and Paenibacillus chibensis sp. nov. Int. J. Syst. Bacteriol. 47:299-306. [DOI] [PubMed] [Google Scholar]
- 32.Smit, J., Y. Kamio, and H. Nikaido. 1975. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J. Bacteriol. 124:942-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Specka, U., A. Spreinat, G. Antranikian, and E. Mayer. 1991. Immunocytochemical identification and localization of active and inactive α-amylase and pullulanase in cells of Clostridium thermosulfurigenes EM1. Appl. Environ. Microbiol. 57:1062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunna, A., and G. Antranikian. 1997. Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol. 17:39-67. [DOI] [PubMed] [Google Scholar]
- 35.Sunna, A., M. D. Gibbs, and P. L. Bergquist. 2000. A novel thermostable multidomain β-1,4-xylanase from Caldibacillus cellulovorans and effect of its xylan-binding domain on enzyme activity. Microbiology 146:2947-2955. [DOI] [PubMed] [Google Scholar]
- 36.Viikari, V., A. Kantelinen, J. Sundquist, and M. Linko. 1994. Xylanases in bleaching: from an idea to the industry. FEMS Microbiol. Rev. 13:335-350. [Google Scholar]
- 37.Whistler, R. L., and E. L. Richard. 1970. Hemicellulose in the carbohydrates, p. 447-469. In W. Pigman and D. Horton (ed.), The carbohydrates: chemistry and biochemistry, 2nd ed. Academic Press, New York, N.Y.
- 38.Winterhalter, C., P. Heinrich, A. Candussio, G. Wich, and W. Liebl. 1995. Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 15:431-444. [DOI] [PubMed] [Google Scholar]
- 39.Wong, K. Y., L. U. L. Tan, and J. N. Saddler. 1988. Multiplicity of β-1,4-xylanase in microorganisms: functions and application. Microbiol. Rev. 52:305-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie, H., H. J. Gilbert, S. J. Charnock, G. J. Davies, M. P. Williamson, P. J. Simpson, S. Raghothama, C. M. Fontes, F. M. Dias, L. M. Ferreira, and D. N. Bolam. 2001. Clostridium thermocellum Xyn10B carbohydrate-binding module 22-2: the role of conserved amino acids in ligand binding. Biochemistry 40:9167-9176. [DOI] [PubMed] [Google Scholar]