Abstract
A method for patterning narrow lines of biomolecules onto nitrocellulose membranes using laboratory syringe pumps is described. One syringe pump is used to drive the biomolecule solution through a needle, while another modified syringe pump acts as a one-dimensional translation stage, moving the needle across the membrane much like a pen. This method consumes very small volumes of reagents, and is a viable option for laboratory-scale fabrication and prototyping of point-of-care rapid diagnostic test strips.
Introduction
Paper-based biosensors such as lateral flow immunochromatographic test strips are of increasing interest to the lab-on-a-chip community1 because of advantages possessed by paper-based systems over conventional channel-based microfluidics.2 Paper-based systems do not require volumetric flow metering with precision pumps or pressure sources, as is required when dealing with conventional microfluidic systems. Instead, paper-based systems rely on capillary wicking of fluids through porous materials, most commonly made from nitrocellulose.3 Fluid flow characteristics are governed by physical properties of both the solution and substrate material such as viscosity, pore size, and membrane hydrophilicity.
Fabrication of conventional point-of-care lateral flow strips requires the patterning of capture lines of biomolecules (e.g., antibodies) onto porous membranes. These lines are typically ~1 mm in width and can be applied to the membranes using a variety of methods including aerosol deposition4, and contact striping.5 Although these approaches have had much success, currently available commercial instruments are expensive and costly to maintain. Others have reported conversion of more affordable office ink-jet printers for applications in patterning biomolecules6–9, but these approaches suffer from large minimum sample volume requirements. Conventional inkjet printers have minimum fluid volume requirements of several millilitres or more, leading to waste of the capture antibody reagent that may be expensive to produce and purify.
The requirements for our system to facilitate in-house assay development were that it be capable of patterning lines of variable width and spacing, that it consume as little of the antibody reagent as possible, and that it be moderatly high-throughput so as to produce dozens of tests for laboratory research purposes in a short period of time. The device and method we describe patterns lines of capture antibodies onto nitrocellulose membranes by taking advantage of the motion control capabilities of a commonly available piece of microfluidics lab equipment, the syringe pump.
We describe the design and setup of the apparatus, and characterize the linewidth as a function of flow rate and translation speed. We examine the uniformity of the linewidth as a function of distance patterned. Patterning of multiple lines of anti-streptavidin antibodies, and capture of a streptavidin-gold nanoparticle conjugate using single and multi-line flow strips is demonstrated. Using this method, forty strips can be patterned with a line of antibody in 3–5 minutes while consuming only 5–10 µL of capture molecule solution. This method will prove useful for laboratories currently possessing syringe pump infrastructure that are looking to produce lateral flow test strips in an inexpensive and rapid format.
Experimental
Materials
Syringe pumps one and two (SP1 and SP2) were from Kloehn, LTD., and Harvard Apparatus, respectively. SP1 was equipped with a 25 µL gas tight syringe (Kloehn). SP2 was modified by attaching a mechanical gripper arm to the pump’s movable backstop with a clamp. A manually adjustable z-translation stage was used from Optosigma. Flexible dispensing needles made of PEEK were purchased from Imagene Technology (part no. 7508), with an outer diameter of 508 µm, and an inner diameter of 250 µm. The needle was connected to SP1 using PEEK tubing, and fittings from The Lee Co. (part no. TMDA3212950z). A tee junction (Upchurch, part no. P-714) and a shut-off valve (Upchurch, part no. P-782) were included in the fluidic circuit to facilitate sample loading. Polyester-backed nitrocellulose membranes were from Millipore (HiFlow 180). Blue dye number 1 (blue food coloring) was mixed ~1:20 (v:v) with PBS buffer prior to use. Human plasma in disodium EDTA (HP; Valley Biomedical Inc., product No. HP1051) was centrifuged at 1000 × G for 30 minutes, and filtered through GDX graded syringe filters (Whatman) prior to use. Polyclonal rabbit anti-streptavidin IgG antibody was purchased from Abcam (part no. ab6676). Non-specific membrane blocking solution was from Invitrogen (part no. 00-0105). Gold nanoparticles (~20 nm in diameter) were synthesized using sodium citrate as the stabilizer and reducing agent.10 The gold nanoparticles were modified with a cationic diblock copolymer containing a terminal carboxyl group11, that was subsequently conjugated to streptavidin using carbodiimide chemistry. The streptavidin-gold reagent was purified using membrane ultrafiltration, and stored at 4 °C in PBS buffer for up to 3 months prior to use.
Striping Procedure
A solution of rabbit polyclonal anti-streptavidin IgG (1 mg/mL in 0.01 M PBS, pH 7.3) was loaded into the PEEK tubing using a micropipette as follows. The shut-off valve was opened to atmospheric pressure, and 10 µL of the antibody solution was injected into the PEEK tubing at the needle end. The valve was closed, and the pipette tip was removed. The dispensing needle was then attached to the PEEK tubing, and primed by slowly pumping with SP1 to expel the air from the needle.
The nitrocellulose membrane was affixed to the z-translation stage at each end using laboratory tape. The stage was raised so that the membrane was a few millimeters below the needle tip. SP2 was manually positioned so as to align the needle tip’s range of motion with the desired start and end points on the membrane. After alignment, the stage was raised to bring the needle tip into contact with the laboratory tape at one end of the membrane.
Once the needle and tape were brought into contact, SP1 was triggered, and a small amount of fluid was allowed to accumulate at the tip. After a few seconds, the translation motion of SP2 was triggerred, and the needle was translated off of the tape and onto the membrane. Both syringe pumps continued to run concurrently, translating the needle tip across the membrane while the biomolecule solution was simultaneously pumped through the needle orifice. The end point of the line occurred when the needle tip came into contact with the tape at the distal end of the membrane, which lifted the needle tip off the membrane, at which point dispensing was stopped. Multiple lines were patterned serially onto the same membrane.
Effects of Flow Rate and Translation Speed on Linewidths
Blue food coloring in PBS buffer was striped onto HiFlow nitrocellulose membranes at variable flow rates and translation speeds. The flow rate of SP1 was varied from 1.6 to 22 µL/min. The translation speed of SP2 was set to 1.8, 2.4, or 3.0 mm/sec. Care was taken to maintain a constant angle between the needle and substrate. This was achived by setting all samples at the same z-position during striping. After striping and drying of the substrates for 30 minutes at room temperature, the lines of dye were photographed using a stereomicroscope. The linewidth measured 3 cm from the starting point of the stripe was plotted as a function of flow rate. Uniformity was evaluated by measuring the linewidth at 0.5 mm intervals along the entire length of the line.
Lateral Flow Immunossay of Gold-Labeled Streptavidin
Lateral flow immunochromatographic test strips were fabricated by writing single or multiple lines of rabbit polyclonal anti-streptavidin IgG at 1 mg/mL on a rectangular piece of nitrocellulose (1.6 × 7 cm). The flow rate of SP1 was set to 9.38 µL/min, and the needle translation speed was 3.0 mm/sec. After striping of the capture antibody, the nitrocellulose membrane was treated with non-specific blocking solution for 30 minutes at room temperature. The membrane was then dried overnight in a vacuum dessicator. Finally the membrane was cut perpendicular to the patterned line at 3 mm intervals using a CO2 laser cutting system12 (Universal Laser). If users do not have access to a laser cutter, a common rotorary cutter and ruler can be used. The strips were fixed onto adhesive-coated Mylar in contact with an absorbent pad. Devices were stored for up to two weeks at room temperature in a dessicator.
A 5 µL droplet of streptavidin-coated gold nanoparticles (~20 nm in diameter, 4 nM) in 50% human plasma was deposited onto a petri dish. The lateral flow strip was placed in contact with the droplet, which completely wicked into the flow strip within 5 minutes. Next the lateral flow strip was moved onto a wet absorbent pad saturated with PBS rinse buffer. The buffer rinse was allowed to proceed for 5 minutes. The devices were then air-dried and imaged using a flatbed scanner.
Results and Discussion
Striping System Design
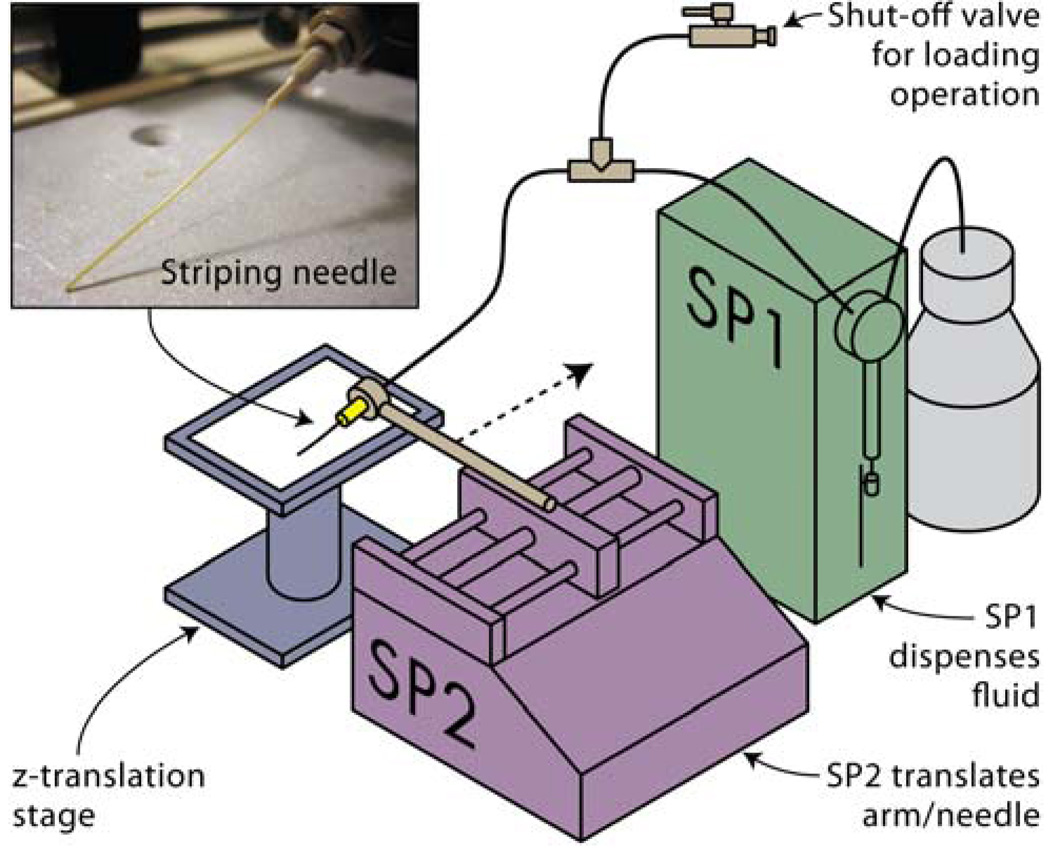
The key components of the protein striping system, shown in Figure 1, are two syringe pumps (SP1 and SP2), a needle with a compliant tip, and a z-translation stage. SP1 drives the fluid flow to the needle tip, effectively metering the rate at which the biological solution is deposited. A tee junction and stop valve were included between SP1 and the needle to facilitate sample loading. The pumping speed of SP1 was controlled through a PC serial port. SP2 is a free-standing benchtop syringe pump with a movable backstop that translates along a threaded rotating rod. The translation speed of SP2 was controlled by changing the flow rate setting in the display panel. An arm with a mechanical claw was attached to the backstop of SP2 to hold the needle. Lines of biomolecules were written when the flexible tip of the needle was translated across the substrate by the movement of SP2, while fluid was simulteneously pumped through the needle orifice by SP1. The nitrocellulose substrate was secured below the needle tip on a z-translation stage that was used to bring the substrate and needle into and out of contact during alignment and striping.
Figure 1.
Experimental setup of the protein striping system showing system components.
Characterization of Linewidths
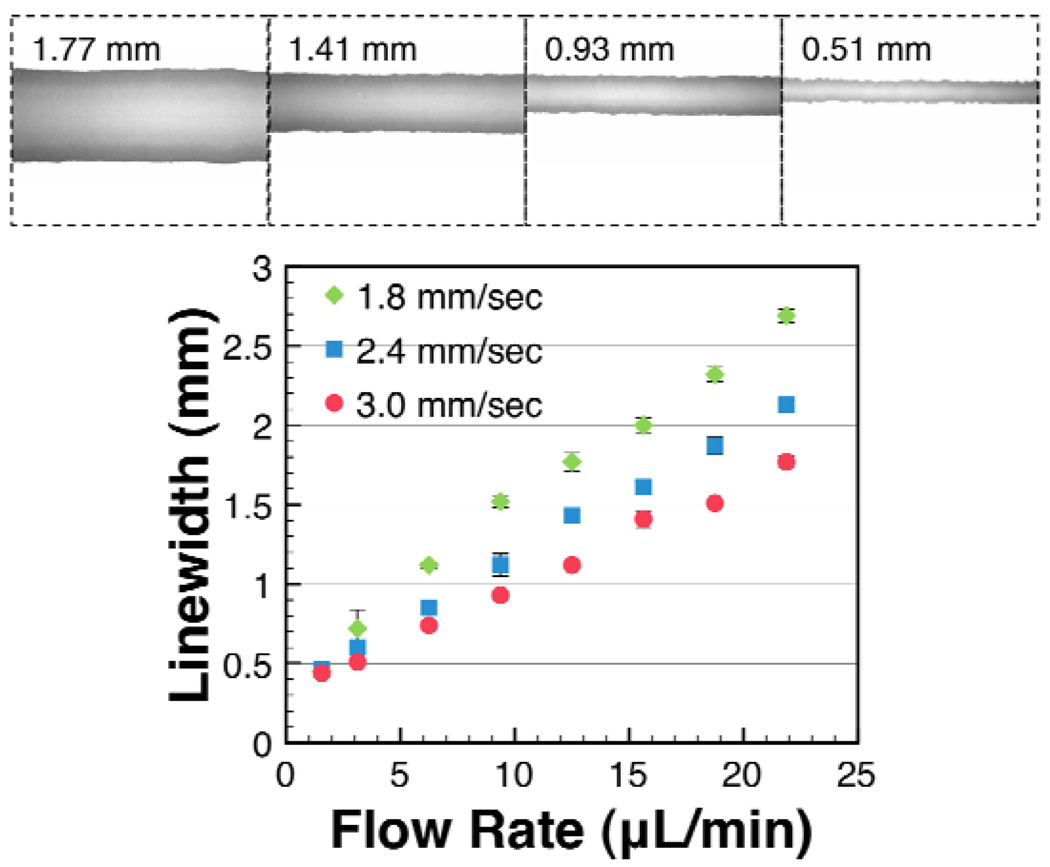
A solution of blue dye in PBS buffer was striped at variable flow rates of SP1, and variable translation speeds of SP2, in order to evaluate the effects of these parameters on linewidths and uniformity. As shown in Figure 2 (bottom), linewidths were found to be linearly correlated with the flow rate of SP1. The narrowest line that could be patterned was 440 µm, which was achieved at an SP1 flow rate of 1.6 µL/min, and an SP2 translation speed of 3.0 mm/sec. Lowering the flow rate below 1.6 µL/min resulted in discontinuous lines for all three of the translation speeds tested. Minimum linewidths achieved with this system are below the optimal linewidth for lateral flow immunoassay devices, reported to be 0.8–1 mm.13 It should be noted that the achievable linewidths are dependent not only on flow rate and translation speed, but also on membrane properties such as pore size and wettability, and fluid properties such as viscosity, and vapor pressure. Furthermore, when striping solutions of capture antibodies, we observed the functionalized region to be smaller than the region wet by buffer. Biomolecules in striping solutions may rapidly bind to the bare membrane, resulting in capture lines that are narrower than the spread of liquid.
Figure 2.
Characterization of dye deposition (top) and linewidths (bottom). Blue food coloring in PBS buffer was striped onto nitrocellulose membranes. The top panel shows line images and linewidths for dye deposited at an SP2 translation speed of 3 mm/sec, and SP1 flow rates of 21.9, 15.6, 9.4, and 3.1 µL/min, respectively (top, left to right). Microscopy images (top) showed the dye was more concentrated at the lines’ edges than in the middle. The distribution of the dye may not resemble the distribution of proteins, which often bind more rapidly to the membrane than do small dye molecules. The linewidth measured 3 cm from the starting point of the line was found to scale linearly with the flow rate (bottom). For a given flow rate, faster translation speeds resulted in narrower lines.
As shown in Figure 2 (bottom), faster SP2 translation speeds (3.0 mm/sec vs. 1.8 mm/sec) were found to decrease the linewidth for a given flow rate. This was expected because as the needle is translated more rapidly at a given flow rate, the same volume of fluid must be distributed over a longer distance, causing the lines to narrow.
As shown in Figure 2 (top), the patterned lines were highly uniform in width, but the distribution of blue dye was not uniform across the linewidth. We observed that more dye was deposited at the line edges. This effect is likely explained by evaporation-induced capillary flow from the center to the edges at the time of deposition.14
The striping protocol involved effectively “inking” the microfluidic needle by allowing a small volume of fluid to pool at the tip of the needle on a piece of laboratory tape before SP2 translation was actuated. Once translation of SP2 was actuated, some of this extra liquid was transferred onto the nitrocellulose membrane, causing the lines to taper immediately following the starting point.
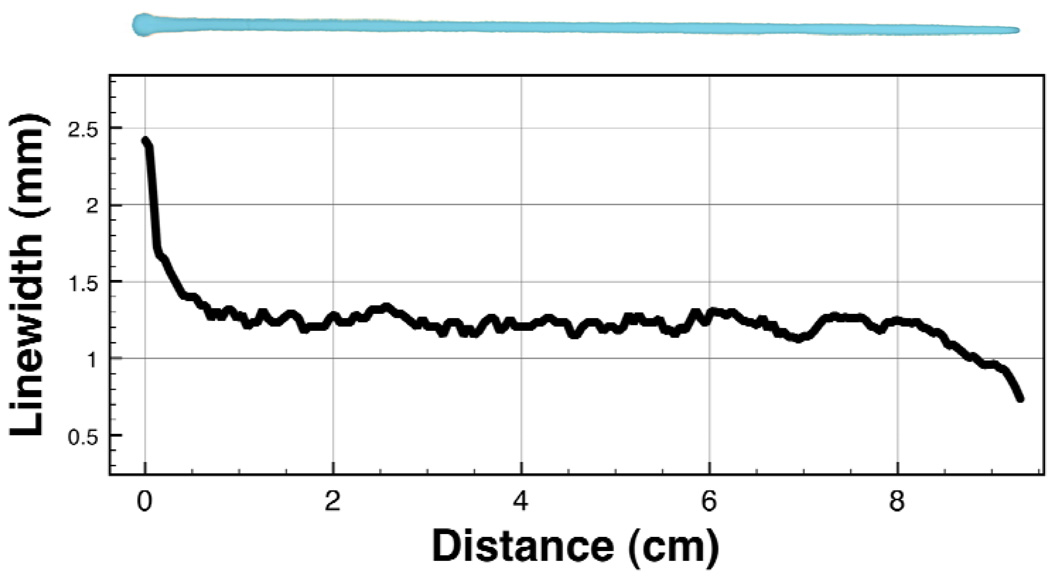
A representative plot of the linewidth vs. distance is shown in Figure 3 (bottom), with the corresponding line image shown in Figure 3 (top). From zero to one centimeter, the linewidth decreased dramatically as the needle shed the excess fluid that had built up at the tip due to pumping of SP1 before SP2 translation was actuated. After about one centimeter of translation, the deposititon process reached a steady state until 8 cm when the line began to narrow near the end.
Figure 3.
Characterization of line uniformity and tapering. A mixture of bovine serum albumin and blue dye in PBS buffer was deposited onto the nitrocellulose membrane (15 µL/min, 3 mm/sec). The width of the observable blue dye was measured at 0.5 mm intervals. Tapering at the start is due to excess fluid pooled at the needle tip, and at the end is due to a loss of steady state deposition as the needle translated off of the membrane and onto the tape used to fix the membrane to the stage.
The average width (Figure 3) measured every 0.5 mm along the line’s steady state from 1–8 cm was found to be 1.23 ± 0.05 mm (mean ± SD), a 4% coefficient of variation. The length of membrane patterned by a single stripe was limited by the maximum translation distance of the syringe pump, which in this case was 12.5 cm. Given a strip width of 3 mm, this corresponds to a maximum of ~40 strips patterned per striping cycle.
Lateral Flow Immunoassay of Gold-Labeled Streptavidin
For the striping system to be useful, the deposited capture reagents (e.g., antibodies) must remain bioactive after deposition, non-specific blocking, drying, and storage. Additionally, patterning of multiple lines is necessary because tests require a minimum of two lines (test and control), or more for multiplexed detection.
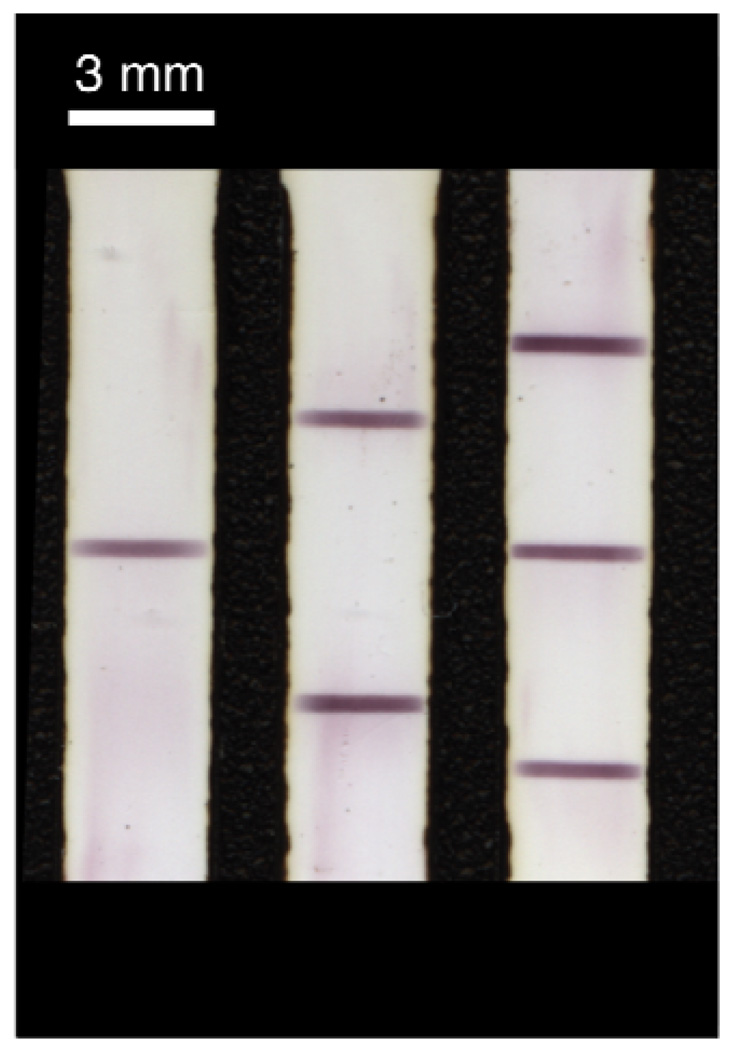
Bioactivity of the deposited antibodies was evaluated by performing a lateral flow immunoassay on 5 µL of a sample containing streptavidin-coated gold nanoparticles (~20 nm in diameter, 4 nM) spiked into a 1:1 (v/v) human plasma:PBS mixture. Figure 4 shows the reflectivity scan obtained after drying of the strips patterned with one (left), two (middle), or three (right) lines of anti-streptavidin capture antibody. Lateral flow strips stored in a dessicator for up to two weeks gave comparable results to those shown in Figure 4.
Figure 4.
Demonstration of multiline patterning. Anti-streptavidin IgG was patterned in one (left), two (middle), or three (right) lines onto the nitrocellulose membrane. The gold-labeled streptavidin reagent in 50% human plasma was wicked through the capture zones and rinsed with PBS buffer. Imaging was performed using a flatbed scanner.
Cost Analysis
We evaluated our system on a cost basis by calculating the cost per data point of the primary capture antibody reagent required to perform a typical microplate ELISA. We estimate that a typical ELISA protocol uses a 100 µL volume of plated primary capture antibody at a concentration of 20 µg/mL, which at $320.00/mg of antibody would cost $0.64 per data point. A similar test performed with lateral flow strips patterned using our approach at an SP1 flow rate of 2 µL/min, an SP2 translation speed of 3 mm/sec, and an antibody concentration of 1 mg/mL would have a cost per data point of only $0.02. This cost includes the small volume (~1 µL) of reagent wasted by fluid pooling during initiation of striping. Our approach therefore represents a ~32-fold reduction in cost of the primary capture antibody as compared with a typical ELISA.
Capture antibody loadings of 1–3 µg/cm, or 10–30 µg/cm2, are typically used in commercial striping systems. The non-optimized capture antibody loading used here represents approximately 1/10 of the loading found in a typical commercial system. Instrumentation costs for our system, including the two syringe pumps and the z-translation stage, add up to approximately $3,000–4000. This cost is significantly lower than a commercial striping system for depositing lines of capture molecules, which costs approximately $18,000, depending on options selected. It should be noted that SP1 is not absolutely necessary, and a cheaper system could be designed where hydrostatic pressure is used to drive the biomolecule solution through the needle instead of pumping with SP1. In practice, however, it was found that controlling the flow rate with SP1 gave more consistent and uniform lines as compared with hydrostatic pressure driven flow.
Conclusion
We have demonstrated how ubiquitous laboratory syringe pumps can be engineered into a protein striping system for writing lines of biomolecules onto porous membranes for use in immunochromatographic biosensors. Our approach is inexpensive, and given that many lab-on-a-chip practioners may already own the necessary infrastructure, this method should prove useful to many readers.
Acknowledgements
Michael A. Nash gratefully acknowledges funding from the National Science Foundation Graduate Research Fellowship Program. This work was supported by the National Institutes of Health (Grant EB000252) and by the Bill and Melinda Gates Grand Challenges Program.
Notes & References
- 1.Pelton R. Trac-Trends Anal. Chem. 2009;28:925–942. doi: 10.1016/j.trac.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Analytical Chemistry. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 3.Posthuma-Trumpie GA, Korf J, van Amerongen A. Analytical and Bioanalytical Chemistry. 2009;393:569–582. doi: 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- 4.Lyashchenko KP, Singh M, Colangeli R, Gennaro ML. J. Immunol. Methods. 2000;242:91–100. doi: 10.1016/s0022-1759(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 5.Handali S, Klarman M, Gaspard AN, Noh J, Lee YM, Rodriguez S, Gonzalez AE, Garcia HH, Gilman RH, Tsang VCW, Wilkins PP. Clin. Vaccine Immunol. 17:68–72. doi: 10.1128/CVI.00339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe K, Suzuki K, Citterio D. Analytical Chemistry. 2008;80:6928–6934. doi: 10.1021/ac800604v. [DOI] [PubMed] [Google Scholar]
- 7.Cai KY, Dong HD, Chen C, Yang L, Jandt KD, Deng LH. Colloids and Surfaces B-Biointerfaces. 2009;72:230–235. doi: 10.1016/j.colsurfb.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Derby B. Journal of Materials Chemistry. 2008;18:5717–5721. [Google Scholar]
- 9.Hossain SMZ, Luckham RE, Smith AM, Lebert JM, Davies LM, Pelton RH, Filipe CDM, Brennan JD. Analytical Chemistry. 2009;81:5474–5483. doi: 10.1021/ac900660p. [DOI] [PubMed] [Google Scholar]
- 10.Frens G. Nature-Physical Science. 1973;241:20–22. [Google Scholar]
- 11.Nash MA, Lai JJ, Hoffman AS, Yager P, Stayton PS. Nano Letters. 2010;10:85–91. doi: 10.1021/nl902865v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spicar-Mihalic P, Stevens DY, Yager P. MicroTAS. Paris: 2007. pp. 667–670. [Google Scholar]
- 13.Jones KD. IVD Technology. 1999 [Google Scholar]
- 14.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Nature. 1997;389:827–829. doi: 10.1103/physreve.62.756. [DOI] [PubMed] [Google Scholar]