Abstract
A multi-step procedure was developed to register magnetic resonance imaging (MRI) and histological data from the same sample in the light microscopy image space, with the ultimate goal of allowing quantitative comparisons of the two datasets. The fixed brain of an owl monkey was used to develop and test the procedure. In addition to the MRI and histological data, photographic images of the brain tissue block acquired during sectioning were assembled into a blockface volume to provide an intermediate step for the overall registration process. The MR volume was first registered to the blockface volume using a combination of linear and nonlinear registration, and two dimensional (2D) blockface sections were registered to corresponding myelin stained sections using a combination of linear and nonlinear registration. Before this 2D registration, two major types of tissue distortions were corrected: tissue tearing and independent movement of different parts of the brain, both introduced during histological processing of the sections. The correction procedure utilized a 2D method to close tissue tears and a multiple iterative closest point (ICP) algorithm to reposition separate pieces of tissue in the image. The accuracy of the overall MR to micrograph registration procedure was assessed by measuring the distance between registered landmarks chosen in the MR image space and the corresponding landmarks chosen in the micrograph space. The average error distance of the MR data registered to micrograph data was 0.324 ± 0.277 mm, only 8% larger than the width of the MRI voxel (0.3 mm).
Introduction
Macroscopic anatomical structures and functional relationships of the central nervous system (CNS) have been extensively studied using non-invasive methods such as magnetic resonance imaging (MRI), computer tomography (CT), and positron emission tomography (PET). Recently, diffusion tensor imaging (DTI) has become another important tool in studying CNS structure and connectivity. On a microscopic scale, histological analysis provides information about the brain’s cytoarchitecture. Combining data across these modalities and distance scales provides new information: a better understanding of contrast mechanisms in the non-invasive images and the ability to infer microscopic tissue properties across the entire brain, in vivo.
The goal of this study was to develop a registration procedure that can successfully align MRI data with histological data within the histological image space. This would allow a direct comparison between MRI data and gold-standard information about the microscopic structural and chemical composition of brain tissue provided by histology. The result would be better characterized tools for understanding the CNS.
1. 1. Histology
Simply defined as the study of tissue, histology involves three main processes – fixation, sectioning, and visualization. Once a tissue sample is obtained through surgery, autopsy, or biopsy, it begins to undergo autolysis and the degenerative process starts almost immediately. The purpose of fixation is to preserve the structural and chemical composition of tissues by stopping this degenerative process. Use of formaldehyde as a fixative was first proposed by Blum [1], and 4% solution of formaldehyde is the most commonly used choice of fixative to this day. Formaldehyde has very low molecular weight (30 amu) and penetrates into tissues rapidly. However, the fixation process after its initial penetration is gradual and an incubation period of at least 24 hours at room temperature is recommended [2].
Tissue shrinkage is one of the biggest sources of concern during histology. Previous experiments have showed that the shrinkage during fixation itself is minimal [3,4], but the most severe forms of tissue distortions such as shrinkage, tearing, and folding, occur due to aggressive sectioning, staining, and mounting procedures following fixation [4]. Tissues are sectioned in thin slices that range from a few to a few hundred micrometers for microscopic examination, using a microtome. In this study, a freezing microtome is used to cut frozen brain tissues manually for light microscopy analysis.
Sectioned tissues are stained to increase the contrast of desired structures, increasing the quality of visualization. In this study, tissue sections are stained for myelin, which wraps around axons to provide insulation and facilitate the transmission of nerve impulses along the axons. The observation of stained myelin sheaths in histological sections provides valuable structural and directional information on fiber bundles. Many of the commonly used staining techniques for normal and degenerative myelin are listed in Bencroft et al [5], such as the Weigert-Pal method, Loyez method, and Luxol fast blue method. Gallyas silver staining [6] is another popular method for staining myelin that is used in this study. Once the desired contrast is achieved through staining, the tissue sections are available for analysis using light microscopy at high spatial resolution.
1. 2. Registration
In the field of medical imaging, the comparison of data between histological and MRI data is often required. The information provided by MRI is valuable by itself, but investigating it in conjunction with histological data can provide insight into the sources of contrast in the MRI images. However, image properties such as resolution, field of view, and contrast will likely be very different between the datasets and simply overlaying the data is usually insufficient for accurate alignment and comparison. Reliable comparison of multiple datasets requires transforming them to a common data space--the process of applying one or more transformations to an image to align it with a reference image is called registration. Two major classes of registration are linear and nonlinear registration.
Linear registration includes four simple operations - translation, rotation, scaling and shearing, which preserve vector addition and scalar multiplication relations. Rigid registration refers to a registration process that involves only rotation and translation operations while affine registration includes all four operations of translation, rotation, scaling, and shearing.
During the registration between histological and MRI images, we used linear registration first to address three dimensional (3D) global tissue deformations of the histological volumes. Volume shrinkage and shearing are caused by tissue fixation and embedding, as well as other mechanical effects. It has been shown in previous studies that photographs of the tissue block acquired during sectioning (‘blockface’ images) can be used to produce more robust registration results [7]. These images, assembled into a 3D volume dataset, provide a relatively undistorted intermediate reference space between the MRI and the histological data. Several studies have utilized this method to improve registration between histological volume and MRI data [8–10].
In addition to the global distortions described above, other tissue distortions are local and require complicated and often time consuming corrections. For this reason, many studies have either opted for a more qualitative data comparison without image registration [11,12] or utilized linear registration to correct for only the global distortions [13,14]. While these approaches may be time efficient, they are insufficient for studies that require more quantitative data analysis. In that case, a combination of both linear and nonlinear registration is usually required.
Nonlinear registration involves operations that do not preserve vector addition and scalar multiplication. For this reason, it is an appropriate method for correcting distortions that are more local. Before nonlinear registration is performed, linear registration is often used as a preliminary step to provide good initialization for the nonlinear algorithm. In many cases, linear registration provides sufficient initialization. However, in order to obtain good registration between MRI and histological data, one often has to address sources of deformations that are specific to histological data, such as tissue tearing and movement of separated tissue segments on the slide (e.g., different hemispheres or other smaller parts of the brain). Breen et al developed an interactive method for correcting spatial distortions in histology and used the method to register histology samples to corresponding MR images [15]. During their distortion correction procedure, significant tearing in the tissue section was corrected using a thin plate spline (TPS) warping method. Pitiot et al developed a piecewise affine registration method and took a more automated approach in addressing the issue of movement of gyri and other smaller parts of a brain [16]. More recently, Dauguet et al were able to successfully reconstruct a 3D histological volume and register it to the corresponding MRI volume by incorporating a hemi-rigid transformation. This approach was taken specifically to address the problem of movement of different hemispheres of the brain observed in histological sections [8].
Methods
2. 1. Image Acquisition
2. 1. 1. Magnetic resonance imaging
All animal procedures were approved by the Vanderbilt Animal Care and Use Committee. A male owl monkey was given a lethal dose of barbiturate and perfused through the heart with buffered physiological saline. Fixation was performed by perfusing again with 4% paraformaldehyde in phosphate buffer, then by 4% paraformaldehyde in phosphate buffer with 10% sucrose. The fixed brain was removed from the skull and kept in 30% sucrose for approximately 24 hours. The brain was then transferred into a phosphate buffered saline (PBS) medium and scanned on a Varian 9.4 Tesla, 21 cm bore magnet using a multi-slice, pulse gradient spin echo sequence (b = 0 and 1309 s/mm2, 21 diffusion weighting directions, TE = 31.2 ms, TR = 17.1 s, 128 × 128 × 132 image volume matrix, 0.3 mm isotropic voxel resolution). After the image acquisition, one iteration of the anisotropic smoothing algorithm proposed by Ding et al [17] was performed to improve signal to noise ratio (SNR). Parameters used were ρ = 2SD and C = 3, where SD is the standard deviation of noise and C is a threshold parameter used to control the degree of smoothing. Non-diffusion weighted (T2-w) images were assembled into a 3D volume dataset, and the brain image was extracted from background for registration purposes [18].
2. 1. 2. Blockface
Twenty four hours after the MR imaging, the brain was embedded in dry ice and sectioned on a microtome at 50 micron thickness in the coronal plane, where the position of the brain stayed constant during cutting and only the blade of the microtome moved. Using a Cannon EOS20D digital camera with 70–300 mm zoom lens, the tissue block was digitally photographed prior to cutting every third section, resulting in a through-plane resolution of 150 microns. The initial in-plane resolution of the original blockface images was 16 µm isotropic. The original high resolution blockface images were downsampled to 256 × 256 with 0.15 mm isotropic voxel resolution for more efficient data processing.
Because the position of the brain stayed constant during sectioning, the 3D blockface volume could be constructed by simply stacking the two dimensional (2D) images of each section. The reconstructed volume was then corrected for the section-to-section intensity variation which was caused by the inconsistent light reflection from the frozen tissue block surface during photograph acquisition. This section-to-section intensity variation was corrected by adjusting the intensity of cortical gray matter of each section to be within a similar intensity range of the gray scale blockface images. The final blockface volume dataset was acquired by manually segmenting the brain from its dry ice background. Although time consuming, the manual segmentation of the brain was necessary because the contrast between the brain and dry ice was low and automatic segmentation methods failed to segment the edges of the brain successfully.
2. 1. 3. Light micrograph
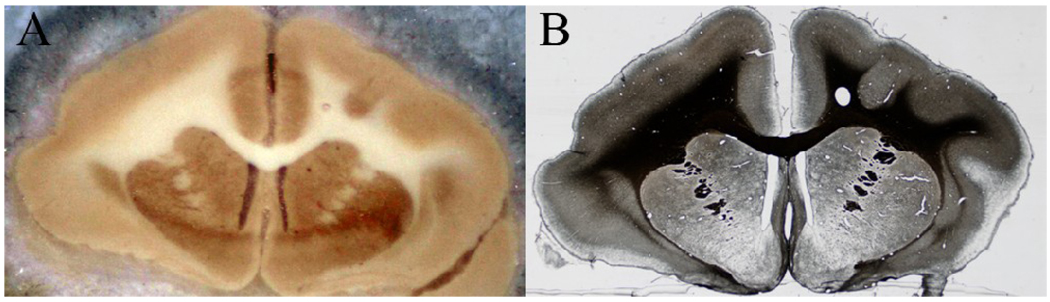
Sectioning of the brain block was followed by histochemical processing, where tissue sections were stained for myelin using Gallyas’ silver method [6]. Staining was performed on floating tissue sections, which were then mounted on glass slides manually for further investigation under a light microscope. A Nikon DXM1200F digital camera mounted on a Nikon E-800 microscope was used to take images of the stained sections at 0.5× magnification. The brain was segmented from the background before further data analysis. An example of a blockface image and the corresponding light micrograph (before brain segmentation) is shown in Fig 1.
Fig 1.
Blockface and histological images. (A) An example of a blockface image before the brain is segmented from its dry ice background (B) An example of a light micrograph image before the brain is segmented from its background. The section is the same as shown in (A), and is stained for myelin using the Gallyas silver staining method.
2. 2. Image Registration
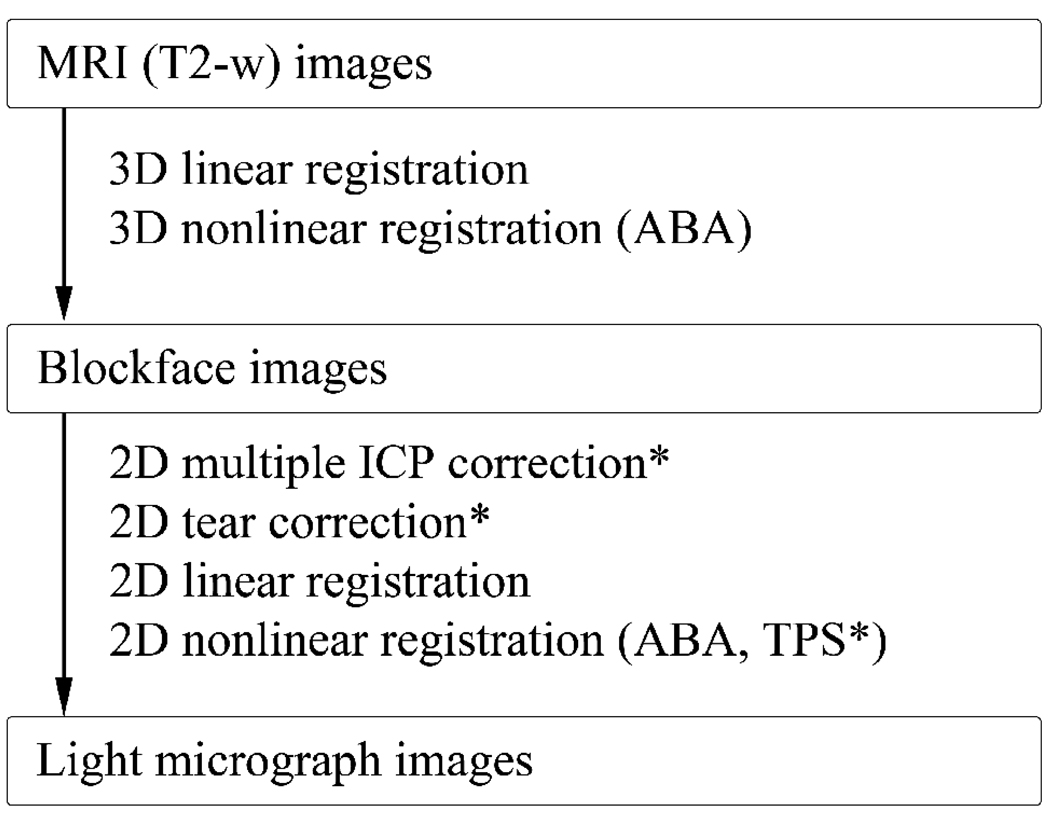
A multi-step registration scheme was developed in order to transfer MRI image data to the histological image space. First, volume datasets of the DTI and blockface images were constructed. Dimensions of the T2-wimage volume matrix were 128 × 128 × 132 with 0.3 mm isotropic voxel resolution. The original high resolution blockface and light micrograph volume dataset were down sampled to 256 × 256 × 222 with 0.15 mm isotropic voxel resolution for ease of data processing. The T2-w volume was then registered to the blockface volume using a combination of linear (i.e., rigid and anisotropic scaling) [19,20] and nonlinear registration with the Adaptive Bases Algorithm (ABA) [21]. Next, a section of interest was chosen and the corresponding blockface and light micrograph images were registered in two dimensions using both linear and nonlinear registration with ABA. Fig 2 summarizes the steps of this procedure. A more detailed description of the multi-step registration scheme is presented below.
Fig 2.
Multi-step registration workflow summary. Three major datasets – MRI, blockface, and light microscopy datasets – were acquired. The datasets were registered to each other using a combination of linear and nonlinear registration. For selected histological sections with tissue tearing or relative displacement of different parts of brain, 2D rigid tear correction and/or 2D multiple ICP correction was performed as a preprocessing step. In some cases, an additional step using TPS was necessary after nonlinear registration of the blockface images.
2. 2. 1. T2-w → Blockface
Linear registration was performed using a mutual information (MI) based method similar to that of Maes [19]. Partial volume (PV) interpolation was used for intensity interpolation of the transformed reference image [19]. Powell’s multidimensional direction set method was used to maximize the MI registration criterion, using Brent’s optimization algorithm for line minimization [22]. Powell’s criterion was set to 10−5, Brent’s to 10−3, and the maximum number of iterations was set to 600. The number of bins for joint histogram calculation was set to 64 × 64 and three resolution levels were used. Transformation in the MRI to blockface step was performed by optimizing first the in-plane parameters, then the through-plane parameters.
In addition to the linear transformations, nonlinear registration was performed using ABA [21]. Fifteen control point levels and two resolution levels were used to determine the transformation scale and spatial resolution. Sixty four bins were used for joint histogram calculation and a Jacobian threshold of 0.05 was used as an optimization constraint. Optimization of a basis function was halted when the cost function's improvement was below 0.0005.
2. 2. 2. Blockface → Light micrograph
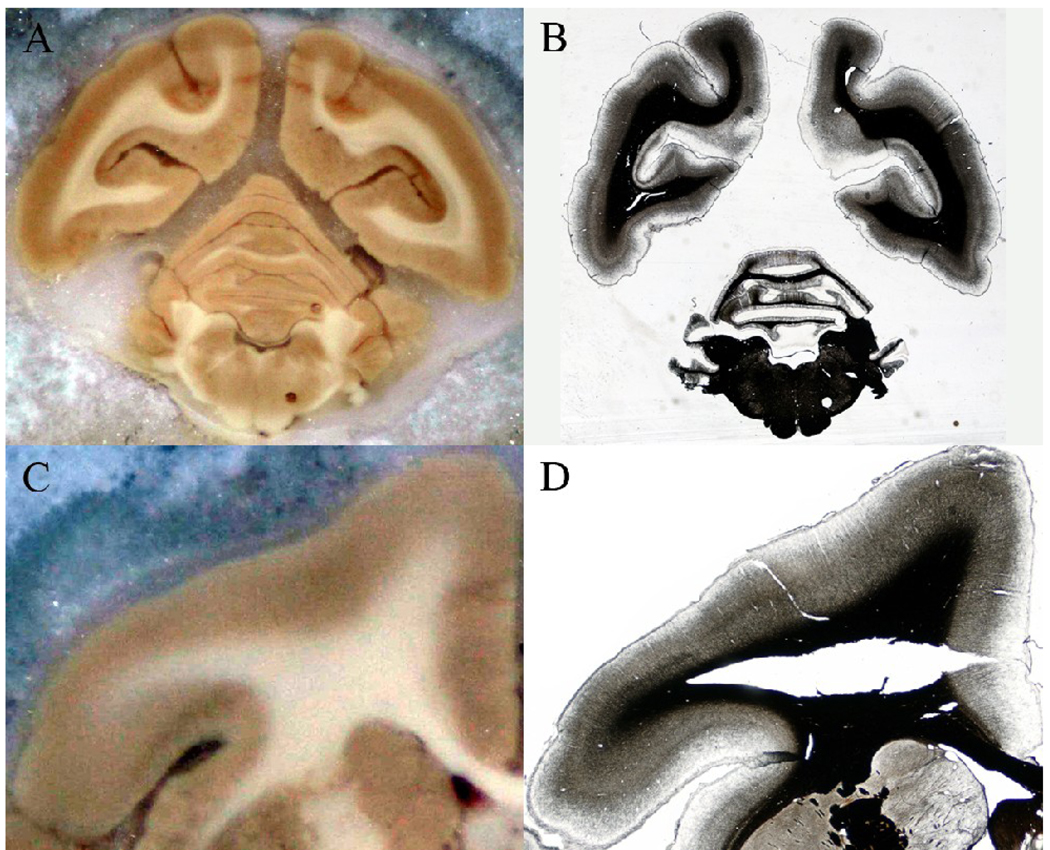
Due to the extensive artifacts in some micrographs [see Fig 3], additional preprocessing was necessary for the affected sections to ensure robust registration results. The preprocessing procedure was developed to address two major types of artifacts: tissue tearing and relative displacement of different pieces of tissue on the slide.
Fig 3.
Image artifacts introduced during histological processing. (A), (C) Undistorted blockface images. (B) Example of relative displacement of the hemispheres and the cerebellum. (D) Example of tissue tearing.
Correction for severe tissue tearing artifacts
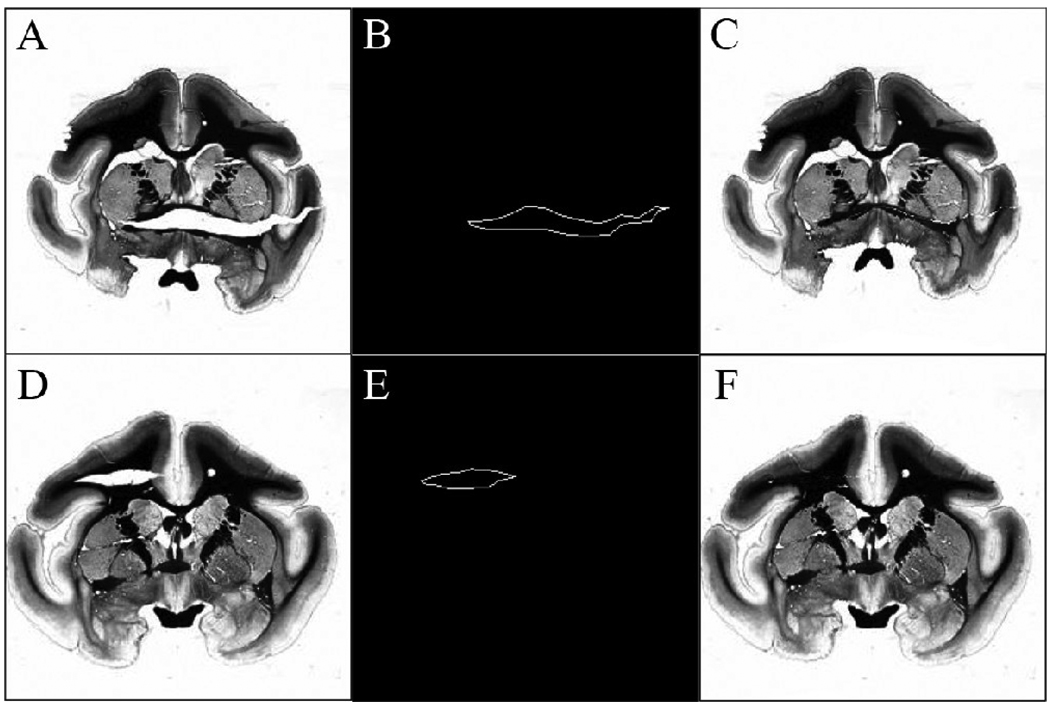
Fig 4A and D shows light micrographs of myelin stained tissue sections with severe tearing of tissue. In order to correct a large tearing artifact, the contour of the torn region was first selected by a user. Examples of the outlines are shown in Fig 4B and E. Because of the procedure used to manipulate the tissue sections, tissue tears were nearly horizontal (anatomical right-left direction) in the coronal sections. According to the location of the tear, the user can choose to fix the tear using one of three options: 1) translate the image data below the tear upward to meet the top edge of the tear, 2) translate the image data above the tear downward to meet the lower edge of the tear, or 3) translate image data both above and below the tear to meet the center line of the torn region. If the user chooses the third scheme, the center line is computed automatically based on the contour of the torn region and the distance between each pixel on the contour and the centerline is also calculated. Pixels in image columns passing through the tear are translated towards the center line, according to the calculated distances. Fig 4C shows the image after translating the tissue up while Fig 4F shows the image after moving the tissues toward the center line. Generally, the option that minimized the mean pixel displacement was chosen.
Fig 4.
Contour selection for 2Dtear correction. (A), (D) Original light micrographs with severe tissue tearing artifacts. (B), (E) User selected contours of tissue tear edges. (C), (F) Result of tear correction method.
Correction for relative displacement of different pieces of tissue
Each of the myelin stained sections was mounted on a glass slide manually. In some sections, different pieces of the tissue, such as left and right hemispheres, are not physically connected and so must be mounted and oriented on a slide separately. Fig 3B shows an example of a stained section in which the two hemispheres and the cerebellum have all moved away from each other. This relative displacement of different parts of the tissue section is more obvious when compared to the corresponding undistorted blockface section, as shown in Fig 3A.
Once the sections that need to be corrected for excessive relative displacements were identified, regions of interest (ROIs) containing the same piece of tissue in the blockface image and micrograph were selected manually. The iterative closest point (ICP) algorithm [23] was then applied to the selected ROIs.
The ICP algorithm is a technique that has been applied widely to surface-based registration in medical images. It calculates the transformation and distance between two point sets iteratively. Once the distance is found to converge to a user-selected threshold, the algorithm is terminated. In this study, we applied the ICP algorithm to multiple tissue components. For each pair of corresponding ROIs, suppose X = {xi, i = 1,2, …N} is a point set in the micrograph and Y the point set in the blockface image. The algorithm then proceeds as follows (also refer to [24] for more information):
∀ xi ε X, find the closest point yi in the point set Y;
- Compute the rotation R and translation t through optimizing the root mean square disparity function D:
(1) Apply the R and t to the point set X to obtain the new X′ = RX+ t;
Compute the new distance D′ between X′ and Y. If the absolute difference between D and D′ is less than 1e-5 (selected empirically), terminate the procedure. Otherwise, let X = X′, and repeat the procedure from step 1.
The ICP algorithm was applied to each pair of ROIs to generate the corresponding rigid body transformations. The transformations are applied to the different tissue pieces to deform the micrograph data to the (undistorted) blockface image space.
Following the preprocessing steps described above, a combination of 2D linear and nonlinear registration was performed on each of the corresponding block and light micrograph sections, using the same registration parameters as used during T2-w→ blockface registration. Two different deformation fields were generated after performing both registration steps. The first deformation field described the transformation of the 3D T2-w image volume into the 3D blockface image volume space, and the second deformation field described the transformation of a 2D blockface image into the 2D light micrograph image space. Each of the deformation fields was applied to the original T2-w volume data to generate a registered T2-w image that could be aligned with the corresponding light micrograph data in the histological data space.
2. 2. 3. Accuracy measurement
Alignment of structures after each registration step (T2-w → blockface, blockface → micrograph) was evaluated both qualitatively and quantitatively. Qualitative assessment was performed by superimposing the transformed target images (MRI data) onto reference images (light micrographs) for visual inspection. After the initial visual inspection, landmarks were manually selected throughout the MRI data volume. Corresponding landmarks in the blockface and light micrograph volumes were selected, and the distance between corresponding landmarks in the registered target and reference volumes provided a measure of registration accuracy.
Results
3. 1. T2-w → Blockface
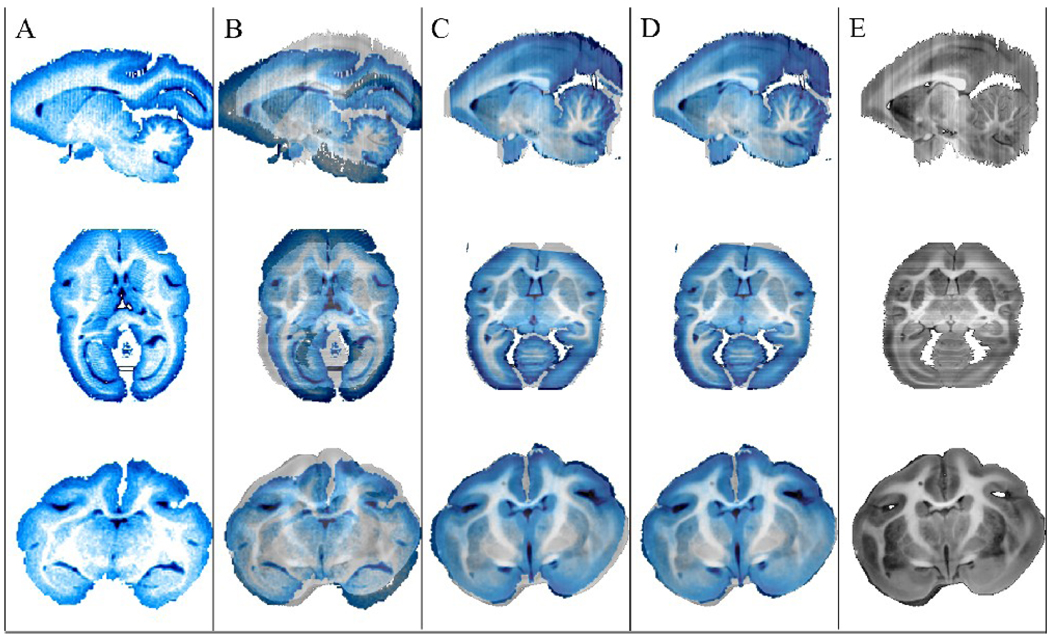
Construction of the blockface volume involved stacking the original 2D images without image registration because each of the blockface photographs was acquired from a stationary brain sample, cut with a moving blade. For this reason, the alignment of the T2-w and blockface volume datasets was good in many regions after only linear registration. The result of the 3D registration of the MRI (more specifically, T2-w) volume to blockface volume was qualitatively assessed by aligning the volumes in the original blockface volume data space, as shown in Fig 5. Column A of Fig 5 shows orthogonal views of the T2-w volume (displayed in blue for better contrast with the blockface images). Column B shows the same T2-w data overlaid on the original blockface volume. Column C shows the T2-w volume after it was linearly registered to the blockface volume, overlaid on the original blockface volume. Notice the significant decrease of misalignment when compared to the superimposed images of column B. Any remaining misalignment of structures after linear registration, such as in some cortical and cerebellar areas, was addressed through nonlinear registration using ABA. Column D shows the T2-w volume after both linear and nonlinear registration to the blockface volume, overlaid on the blockface volume. Good overall alignment of structures was observed after registration.
Fig 5.
3D registration of MRI (T2-w) to blockface volume. (A) Orthogonal views of the original non-diffusion weighted (T2-w) image volume. (B) Original T2-w images superimposed on blockface images, (C) T2-w images after linear registration, and (D) T2-w images after linear and nonlinear registration are overlaid on original blockface images, reproduced in (E).
3. 2. Blockface → Light micrograph
Visual inspection of each of the myelin stained sections was performed in order to identify those with severe tissue tearing and/or relative displacement of separated pieces of the tissue. Once identified, those sections were preprocessed using tear correction and multiple ICP methods to prepare the sections better for the linear and nonlinear registration steps to follow. Comparison of results between affected sections that had not and those that had been preprocessed demonstrated that the preprocessing step provided more robust registration, as shown in Fig 6 and Fig 7.
Fig 6.
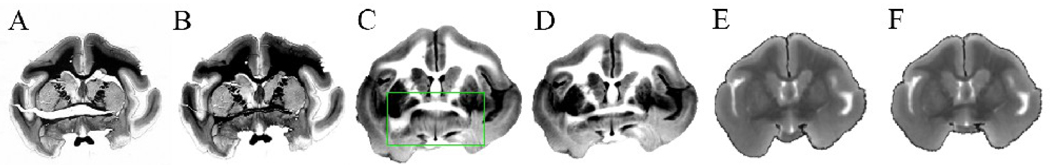
Example of application of the tear correction method. (A) An original light micrograph, (B) the micrograph after closing the tear, (C) & (D) the corresponding blockface image deformed to match (A) and (B), respectively. (E) & (F) MR images registered to (A) & (B), respectively. Note the green region in (C) was locally stretched by the nonlinear registration algorithm in order to match the hole in (A), causing a distortion.
Fig 7.
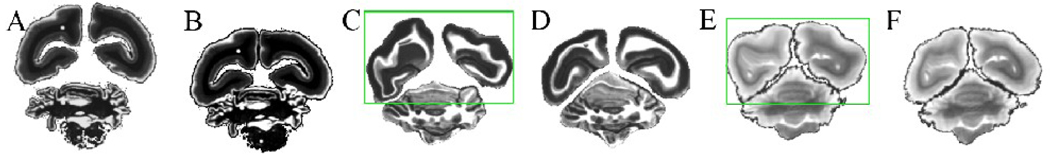
Example of application of the multiple component ICP method. (A) The original light micrograph of mounted tissue, (B) the corrected micrograph using the ICP algorithm, (C) & (D) the deformed blockface images and (E) & (F) MR images registered to (A) & (B), respectively. The green region shows the large distortion when the ICP algorithm was not applied to the light micrograph.
Fig 6 demonstrates an example of tear correction performed on a myelin stained section and its effect on image registration. Fig 6A shows a section with severe tissue tearing artifacts caused by a vertical tensile force along the anterior commissure as well as another tear between the corpus callosum and internal capsule. Fig 6B shows the result of tear correction on the torn section. Fig 6C and E show the result of overall registration (linear and nonlinear registration) of the blockface and MR images, respectively, when they were registered to the myelin stained section that was not preprocessed. Fig 6D and F show the result of the overall registration of the blockface and MR images, respectively, when they were registered to the myelin stained section that was preprocessed. The results shown in Fig 6D and F demonstrate that the tear correction produces more accurate registration (compare to Fig 6C and E).
Fig 7 demonstrates an example of the multiple ICP algorithm applied to a myelin stained section and its effect on the image registration. Fig 7A shows a myelin stained section where three pieces of brain tissue were displaced from their original locations during the mounting procedure. Fig 7C and E show the blockface image and T2-w image, respectively, registered to the myelin stained section that was not preprocessed. It can be seen that the registration algorithm is not able to account for large displacements of structures and produces an incorrectly deformed result. Fig 7D and F, on the other hand, show the blockface image and T2-w image, respectively, when they were registered to the myelin stained section that was preprocessed using the multiple ICP algorithm. The improvement compared to the results shown in Fig 7C and E is evident. Hence, the multiple ICP algorithm provides a better initialization for the overall registration process, leading to a more accurate result. It should be noted that although cerebellar sections that had moved during mounting were also preprocessed, the smaller size and complex structure of the cerebellum sometimes resulted in tearing and movement, as well as missing pieces of the tissue that rendered it almost impossible to register with its corresponding blockface sections, even after the preprocessing step. For this reason, and because it was not the focus of this study, the cerebellum was excluded from any further analysis.
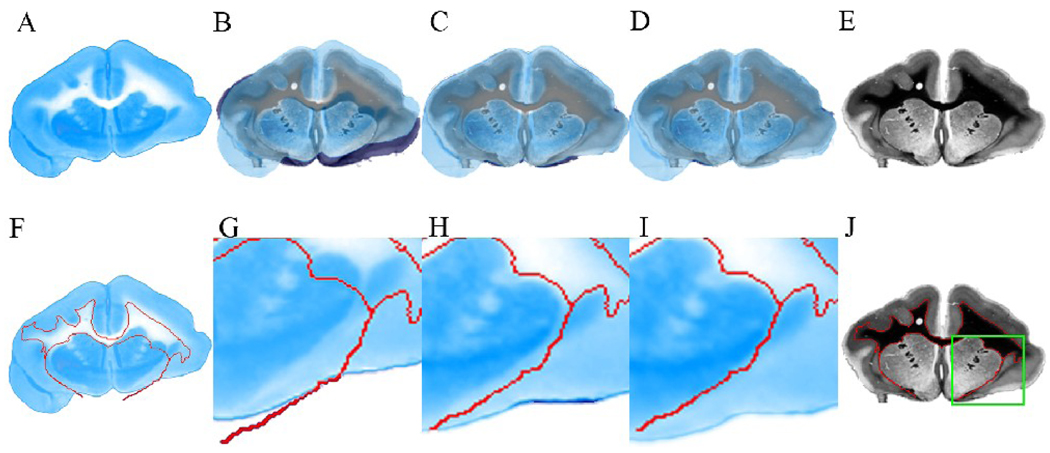
An example of the 2D registration of a blockface section and the corresponding light micrograph is shown in Fig 8. The top row of Fig 8 allows for visual inspection of the registration over the whole section. Fig 8A and E are the images of the original blockface and myelin stained section. Fig 8B–D show the original blockface section, the blockface section after linear registration, and after linear and nonlinear registration, respectively, superimposed on the myelin stained section. It can be seen that linear registration improves the alignment of the sections substantially, and some remaining tissue artifacts around the cortical area are corrected with further, nonlinear registration. The bottom row of Fig 8 shows the registration result in a more local region around the left external capsule, highlighted within the green box shown in Fig 8J. Fig 8F shows the original blockface section with the outline of the myelin stained section’s white matter (WM) overlaid in red. Fig 8G–I provide a zoomed-in view of the change in the external capsule after each registration step. Notice how, in Fig 8I, the outlines of the external capsule of blockface and myelin stained sections are aligned well.
Fig 8.
2D registration of a blockface section to a corresponding myelin stained section. (A) Original blockface section. (B) Original blockface section, (C) blockface section after linear registration, and (D) blockface section after linear and nonlinear registration overlaid on the myelin stained section, respectively. (E) Original myelin stained section. (F) Original blockface section. The red line represents the white matter (WM) outline of the corresponding myelin stained section, also outlined in (J). (G) A region of interest (ROI), outlined in green in (J), is selected from the original blockface section. (H) ROI in the blockface section after linear registration. (I) ROI in the blockface section after linear and nonlinear registration. In (G–I), the WM outline (in red) of the corresponding myelin stained section is also overlaid for comparison.
3. 3. T2-w → Blockface → Light micrograph
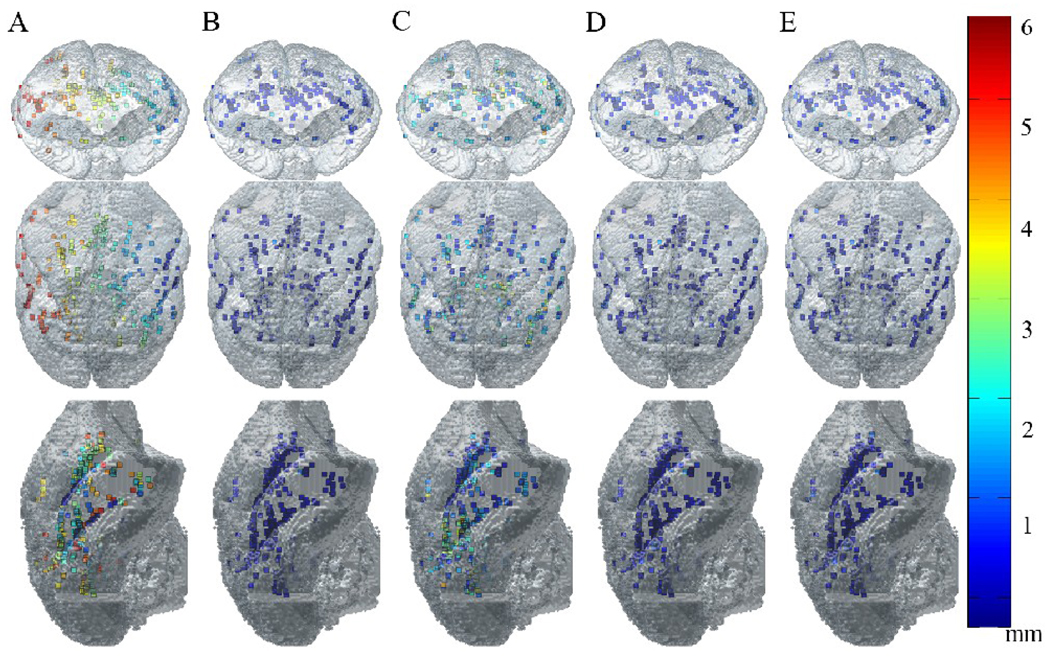
A total of 291 landmarks were selected over the whole brain volume within the original T2-w volume. Corresponding points were then identified manually in the blockface and light micrograph volumes. The corresponding points provided the ‘true’ locations of the landmarks within their respective image spaces. The landmarks selected in the T2-w dataset were then transferred into the blockface and micrograph image spaces to provide a measure of error for different stages of the overall registration process (i.e., T2-w → blockface linear, T2-w → blockface linear and nonlinear, blockface → light micrograph linear, blockface→ light micrograph linear and nonlinear, and T2-w → light micrograph linear and nonlinear). The measure of registration error for each stage was the distance between the corresponding points transformed to the same image space. In Fig 9, the distribution of the landmarks used is visualized within the surface rendered image of the original MRI volume. Each of the landmarks is color coded according to the corresponding error measurement, which is summarized in Table 1.
Fig 9.
Distribution of landmarks used for registration accuracy measurements. Chosen landmarks are visualized within the surface rendered image of the original MRI volume. Each of the landmarks is color coded according to the registration error between landmarks in (A) original MRI image space and original blockface image space, (B) registered (linear & nonlinear) MRI image space and original blockface image space, (C) original blockface image space and original light micrograph space, (D) registered (linear & nonlinear) blockface image space and original light micrograph space, (E) registered (linear & nonlinear) MRI image space and original light micrograph space. The size of the voxels is 0.3 mm3.
Table 1.
Registration accuracy as measured by the distance between corresponding landmarks chosen from T2-w, blockface, and light micrograph volume data.
| T2-w → blockface | T2-w (in blockface image space) → light micrograph |
T2-w → light micrograph |
|
|---|---|---|---|
| Initial error (mm) | 3.84 ± 1.39 | 1.87 ± 1.07 | 4.55 ± 1.80 |
| After linear registration (mm) | 0.326 ± 0.18* (t-stat: 44.02, p < 0.01) | 1.08 ± 1.16* (t-stat: 11.62, p < 0.01) | n/a |
| After linear and nonlinear registration (mm) | 0.261 ± 0.18* (t-stat: 8.46, p < 0.01) | 0.319 ± 0.28* (t-stat: 11.25, p < 0.01) | 0.324 ± 0.28* (t-stat: 30.40, p < 0.01) |
Statistically significant (p < 0.05) reduction in error relative to the previous step.
The original T2-w volume was resized from 128 × 128 × 132 to 256 × 256 × 222 through interpolation only for the purpose of calculating the initial error between the MR volume and blockface and light micrograph volumes before registration. Note that the initial error measurements include rotation as well, resulting in large error measurements. The measured initial error between MR and blockface volumes was 3.84 ± 1.39 mm. Error between the blockface data and the T2-w data that was linearly registered was 0.326 ± 0.18 mm, which is slightly over the size of the original MR voxel (0.3 mm). This confirmed the previous visual observation that the T2-w → blockface registration was good, even after performing only a linear transformation. Error between the blockface and T2-w data that was linearly and nonlinearly registered to the blockface was 0.261 ± 0.18 mm, which shows improvement over simple linear registration (0.326 ± 0.18 mm). It should also be noted that the error after both linear and nonlinear registration is approximately the size of the MR voxel.
The large initial error (1.87 ± 1.07 mm, ~ 6 MR voxels) between the light micrograph points and the T2-w data in the blockface space is due to large tissue distortions that were introduced during histological processing and mounting. Even after linear registration, the error is relatively high at 1.08 ± 1.16 mm. Combination of the preprocessing step, linear and nonlinear registration improves the error between the micrograph points and the T2-w data registered to the micrograph to 0.319 ± 0.28 mm. Finally, the overall registration accuracy measurement between T2-w data and light micrograph data was measured using the landmarks selected in the original T2-w data space and light micrograph space. The initial error was 4.55 ± 1.80 mm and the overall error was 0.324 ± 0.28 mm. Note that the large initial error measurement results from including rotation error between the two data sets. The change in error after each of the registration steps was statistically significant (p < 0.05).
Discussion
In this study, a registration workflow that transfers MR data into histological image space was developed and its accuracy was measured. The acquisition of blockface data was essential because it provided an undistorted three dimensional image of the brain before sectioning and served as an intermediate step in registering MR data to distorted light micrographs. The post processing of the image data proved to be very time consuming, however, due to the lack of an automated means to segment the brain from the surrounding dry ice. Other studies have encountered similar problems [8] and reported progress using new dyes for dry ice.
One of the biggest challenges for the study was the severe tissue distortion that was introduced into light micrographs during histological processing and mounting. This distortion was difficult to control because the whole process – sectioning, staining, and mounting - was performed manually. There are several possible solutions to this problem, for example the use of an automated cryomicrotome stage and acetate tape transfer system that would allow one to preserve the histological structures. Other studies have used this method successfully and also reported improved registration results due to decreased tissue distortions [25,26]. The use of acetate film tape, however, leaves a residue on sectioned tissues and may not be ideal for studies that require staining of floating sections. Another possible solution is to use a celloidin medium that would stay with the sectioned tissues throughout the staining and mounting procedure [27,28].
Despite these limitations, we were able to successfully register MRI data to the distorted light micrographs by preprocessing to correct severe local distortions then applying linear and nonlinear registration steps. The use of robust nonlinear registration (using ABA), in particular, was critical because it provided spatially adaptive and topologically consistent deformation fields [21]. The procedure has allowed us to register light micrograph sections with mean error less than the size of an MR voxel.
The result of our study suggests that through the use of a carefully designed registration scheme, it is possible to register MR to histological data, even in the presence of severe tissue distortions, such as tearing and displacement of different pieces of tissue. This procedure can be useful for those who would like to study and compare histological microstructures with other imaging methods such as MRI.
Conclusion
Despite the rapid growth of different neuroimaging modalities, histological analysis of CNS still provides the gold standard for information about the brain’s cytoarchitecture. Quantitative comparison of neuroimaging and histological data is facilitated by effective tools for cross-modality registration. In this study, a multi-step registration procedure is presented that enables an effective overlay of MRI and histological data in the histological image space. A blockface volume was reconstructed to provide an intermediate step for the overall registration process, which allowed for a more robust registration result. Two major types of tissue distortions– tissue tearing and movement of separated pieces of tissue- were corrected using a preprocessing procedure that implemented 2D tearing correction and the 2D multiple ICP algorithm. The accuracy of the overall (linear and nonlinear) registration workflow was assessed by measuring the discrepancy between the position of landmarks chosen in the MR image space, then transformed to the micrograph space, and the position of the corresponding points chosen in the micrograph space. In this study, it was shown that the registration procedure provides an effective means to quantitatively compare MRI and histological data with the average error comparable to the size of the original MR voxel (0.3mm).
Acknowledgements
The authors acknowledge support from NIH grants 1R01NS58639, 1RO1EB002777, 1S10RR17799, and 5R01DA028303-02.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blum F. Notiz uber die anwendung des formaldehyds (formol) als harrungs-und koservierungsmittel. Anat Anz. 1894;9:229. [Google Scholar]
- 2.Underhill BML. The rate of penetration of fixatives. J R Microsc Soc. 1932;52:113. [Google Scholar]
- 3.Durgun-Yucel B, Hopwood D, Yucel AH. The effects of mercaptoethanol-formaldehyde on tissue fixation and protein retention. Histochem J. 1996;28(5):375–383. doi: 10.1007/BF02331400. [DOI] [PubMed] [Google Scholar]
- 4.Fox CH, Johnson FB, Whiting J, Roller PP. Formaldehyde fixation. J Histochem Cytochem. 1985;33(8):845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 5.Bancroft JD, Cook HC. Manual of Histological Techniques and their Diagnositic Application. Edinburgh: New York Churchill Livingstone; 1994. pp. 349–356. [Google Scholar]
- 6.Gallyas F. Silver staining of myelin by means of physical development. Neurol Res. 1979;1(2):203–209. doi: 10.1080/01616412.1979.11739553. [DOI] [PubMed] [Google Scholar]
- 7.Toga AW, Ambach KL, Schluender S. High-resolution anatomy from in situ human brain. NeuroImage. 1994;1(4):334–344. doi: 10.1006/nimg.1994.1018. [DOI] [PubMed] [Google Scholar]
- 8.Dauguet J, Delzescaux T, Conde F, Mangin JF, Ayache N, Hantraye P, Frouin V. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. Journal of neuroscience methods. 2007;164(1):191–204. doi: 10.1016/j.jneumeth.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Palm C, Axer M, Grassel D, Dammers J, Lindemeyer J, Zilles K, Pietrzyk U, Amunts K. Towards Ultra-High Resolution Fibre Tract Mapping of the Human Brain - Registration of Polarised Light Images and Reorientation of Fibre Vectors. Frontiers in human neuroscience. 2010;4:9. doi: 10.3389/neuro.09.009.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarty MM, Bertrand G, Hodge CP, Sadikot AF, Collins DL. The creation of a brain atlas for image guided neurosurgery using serial histological data. NeuroImage. 2006;30(2):359–376. doi: 10.1016/j.neuroimage.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 11.Scholtes F, Phan-Ba R, Theunissen E, Adriaensens P, Brook G, Franzen R, Bouhy D, Gelan J, Martin D, Schoenen J. Rapid, postmortem 9.4 T MRI of spinal cord injury: correlation with histology and survival times. Journal of neuroscience methods. 2008;174(2):157–167. doi: 10.1016/j.jneumeth.2008.06.038. [DOI] [PubMed] [Google Scholar]
- 12.Engelhorn T, Eyupoglu IY, Schwarz MA, Karolczak M, Bruenner H, Struffert T, Kalender W, Doerfler A. In vivo micro-CT imaging of rat brain glioma: a comparison with 3T MRI and histology. Neuroscience letters. 2009;458(1):28–31. doi: 10.1016/j.neulet.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Grate LL, Golden JA, Hoopes PJ, Hunter JV, Duhaime AC. Traumatic brain injury in piglets of different ages: techniques for lesion analysis using histology and magnetic resonance imaging. Journal of neuroscience methods. 2003;123(2):201–206. doi: 10.1016/s0165-0270(02)00361-8. [DOI] [PubMed] [Google Scholar]
- 14.Ma B, Lin Z, Winkelbach S, Lindenmaier W, Dittmar KE. Automatic registration of serial sections of mouse lymph node by using Image-Reg. Micron. 2008;39(4):387–396. doi: 10.1016/j.micron.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Breen MS, Lancaster TL, Wilson DL. Correcting spatial distortion in histological images. Comput Med Imaging Graph. 2005;29(6):405–417. doi: 10.1016/j.compmedimag.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Pitiot A, Bardinet E, Thompson PM, Malandain G. Piecewise affine registration of biological images for volume reconstruction. Medical image analysis. 2006;10(3):465–483. doi: 10.1016/j.media.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Ding Z, Gore JC, Anderson AW. Reduction of noise in diffusion tensor images using anisotropic smoothing. Magn Reson Med. 2005;53(2):485–490. doi: 10.1002/mrm.20339. [DOI] [PubMed] [Google Scholar]
- 18.Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16(2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 20.Li R. Automatic Placement of regions of interest in medical images using image registration [Master's Thesis] Nashville: Vanderbilt University; 2001. [Google Scholar]
- 21.Rohde GK, Aldroubi A, Dawant BM. The adaptive bases algorithm for intensity-based nonrigid image registration. IEEE Trans Med Imaging. 2003;22(11):1470–1479. doi: 10.1109/TMI.2003.819299. [DOI] [PubMed] [Google Scholar]
- 22.Press WH. Numerical Recipes in C. Cambridge, U.K.: Cambridge Univ. Press; 1992. pp. 412–419. [Google Scholar]
- 23.Besl PJ, McKay HD. A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell. 1992;14(2):239–256. [Google Scholar]
- 24.Fitzpatrick JM, Hill DL, Maurer CR. Handbook of Medical Imaging. In: Fitzpatrick JM, Sonka M, editors. Medical Image Processing and Analysis. Volume 2. Bellingham, Washington: SPIE International Society for Optical Engineering; 2000. pp. 447–513. [Google Scholar]
- 25.Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, Dawant BM, Caprioli RM, Gore JC. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nature methods. 2008;5(1):57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Analytical chemistry. 2006;78(18):6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Yankeelov TE, Rosen GD, Gore JC, Dawant BM. Enhancement of histological volumes through averaging and their use for the analysis of magnetic resonance images. Magn Reson Imaging. 2009;27(3):401–416. doi: 10.1016/j.mri.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosen GD. [cited 2010 July 30];MBL Training Manual - Detailed Protocol. 2003 Jan 27; Available from: http://www.mbl.org/tutorials/MBLTrainingManual/index.html.