Abstract
Optic atrophy (OA) and sensorineural hearing loss (SNHL) are key abnormalities in several syndromes, including the recessively inherited Wolfram syndrome, caused by mutations in WFS1. In contrast, the association of autosomal dominant OA and SNHL without other phenotypic abnormalities is rare, and almost exclusively attributed to mutations in the Optic Atrophy-1 gene (OPA1), most commonly the p.R445H mutation.
We present eight probands and their families from the US, Sweden, and UK with OA and SNHL, whom we analyzed for mutations in OPA1 and WFS1. Among these families, we found three heterozygous missense mutations in WFS1 segregating with OA and SNHL: p.A684V (six families), and two novel mutations, p.G780S and p.D797Y, all involving evolutionarily conserved amino acids and absent from 298 control chromosomes. Importantly, none of these families harbored the OPA1 p.R445H mutation. No mitochondrial DNA deletions were detected in muscle from one p.A684V patient analyzed. Finally, wolframin p.A684V mutant ectopically expressed in HEK cells showed reduced protein levels compared to wild-type wolframin, strongly indicating that the mutation is disease-causing.
Our data support OA and SNHL as a phenotype caused by dominant mutations in WFS1 in these additional eight families. Importantly, our data provide the first evidence that a single, recurrent mutation in WFS1, p.A684V, may be a common cause of ADOA and SNHL, similar to the role played by the p.R445H mutation in OPA1. Our findings suggest that patients who are heterozygous for WFS1 missense mutations should be carefully clinically examined for OA and other manifestations of Wolfram syndrome.
Keywords: autosomal dominant; hearing loss; optic atrophy; mutation; WFS1, Wolfram syndrome
INTRODUCTION
Optic atrophy (OA) and hearing impairment are features of several syndromes such as X-linked Mohr-Tranebjærg syndrome (OMIM 304700, deafness-dystonia-opticn neuronopathy syndrome), X-linked Charcot-Marie-Tooth disease-5 (OMIM 311070, OA, deafness, and polyneuropathy), and Gustavson syndrome (OMIM 309555, X-linked mental retardation with OA, deafness, and seizures). In addition, optic atrophy and deafness are features of the Wolfram syndrome type 1 (OMIM 222300) and 2 (OMIM 604928). In contrast, autosomal dominant OA (ADOA) and sensorineural hearing loss (SNHL) without any other phenotypic abnormality have been described in relatively few families, with the first report referring to a Swedish family that was clinically described in a Swedish Physician’s journal [Samuelson, 1940], and re-investigated as one of the families presented here.
Wolfram syndrome type 1 is a rare and severe autosomal recessive neurodegenerative disease, characterized by diabetes mellitus, optic atrophy, diabetes insipidus and deafness (DIDMOAD) and is caused by mutations in the WFS1 gene (reviewed in [Tranebjaerg et al., 2009]). Additional clinical features may include renal abnormalities, ataxia, dementia/mental retardation and diverse psychiatric illnesses. The minimal diagnostic criteria for Wolfram syndrome are OA and diabetes mellitus of juvenile onset. Hearing impairment in Wolfram syndrome is typically progressive and mainly affects the higher frequencies [Cryns et al., 2003], but a small fraction of affected individuals have congenital deafness [Barrett et al., 1995; Hansen et al., 2005]. Mutations in WFS1 are also a common cause of isolated autosomal dominant low-frequency nonsyndromic sensorineural hearing loss (LFSNHL) [Bespalova et al., 2001; Young et al., 2001]. In addition, a WFS1 p.E864K missense mutation has been reported in two families with dominantly inherited deafness with some members being affected by OA and impaired glucose regulation/diabetes [Eiberg et al., 2006; Valero et al., 2008], thereby mimicking Wolfram syndrome, but in an attenuated version. Finally, very recently, a p.K836N mutation in WFS1 has been found associated with autosomal dominant optic neuropathy and deafness in one family [Hogewind et al., 2010]. No plausible functional explanation has been found to explain the vast differences in clinical presentations and patterns of inheritance.
So far, more than 140 recessive mutations causing Wolfram syndrome have been identified. Most of these are truncating mutations, mainly located in exon 8 and unique to a particular individual or a few individuals/families. In the dominantly inherited disease, LFSNHL, the WFS1 mutations are mainly missense mutations in exon 8; 28 different missense mutations have been identified (http://www.khri.med.umich.edu/research/lesperance_lab/low_freq.php).
Until recently, only one gene, Optic Atrophy-1 (OPA1), had been found to underlie isolated optic atrophy and hearing loss. OPA1 encodes a dynamin-related GTPase involved in mitochondrial biogenesis (OMIM 605290) and is the gene most frequently underlying OA either isolated or in syndromic form [Amati-Bonneau et al., 2009; Yu-Wai-Man et al., 2010]. One recurrent OPA1 mutation, the heterozygous p.R445H missense mutation, underlies the majority of reported cases of isolated OA and hearing loss. This mutation was first identified in a Japanese and a French patient [Amati-Bonneau et al., 2003; Shimizu et al., 2003].
In this study, we demonstrate that heterozygous WFS1 missense mutations cause autosomal dominant isolated OA and hearing loss in eight families. Importantly, our data also suggest that a single, recurrent, non-founder WFS1 mutation, p.A684V, which was identified in six of the eight families studied, may be a common cause of isolated autosomal dominant OA and SNHL similar to the p.R445H mutation in OPA1.
PATIENTS AND METHODS
Clinical Data
Eight families of Caucasian origin from the US, Sweden and UK diagnosed with OA and SNHL were included in this study (Fig. 1). Initially, we analyzed 15 probands with OA and deafness in the study, but only identified mutations in WFS1 in 8 of these probands, and the families of these 8 probands are described here in detail. The clinical data were compiled from available medical records from different hospitals. Venous blood was drawn from available patients and relatives and DNA was extracted using standard methods. The study was approved by the Danish Research Ethical Committee (reference numbers KF 01-234/02 and KF 01-108/03) and the Institutional Review Board (IRBMED) of the University of Michigan Health System. When possible, patients underwent audiometric and ophthalmological evaluations. Normal hearing was defined as a pure tone average hearing level of less than or equal to 20 dB. Mild hearing loss was defined as 21–40 dB HL, moderate loss as 41–60 dB HL, severe loss as 61–90 dB HL, and profound loss was defined as exceeding 90 dB HL. OA was evaluated based on available ophthalmological examinations. Five of the families (NSDF916, NSDF1272, NSDF1865, NSDF1793, and NSDF2032) were ascertained through a study of genetic deafness in the alumni of Gallaudet University (a university for the education of the deaf and hard-of-hearing students, located in Washington, DC, USA) [Arnos et al., 2008] and the North American Repository of deaf individuals [Pandya et al., 2003].
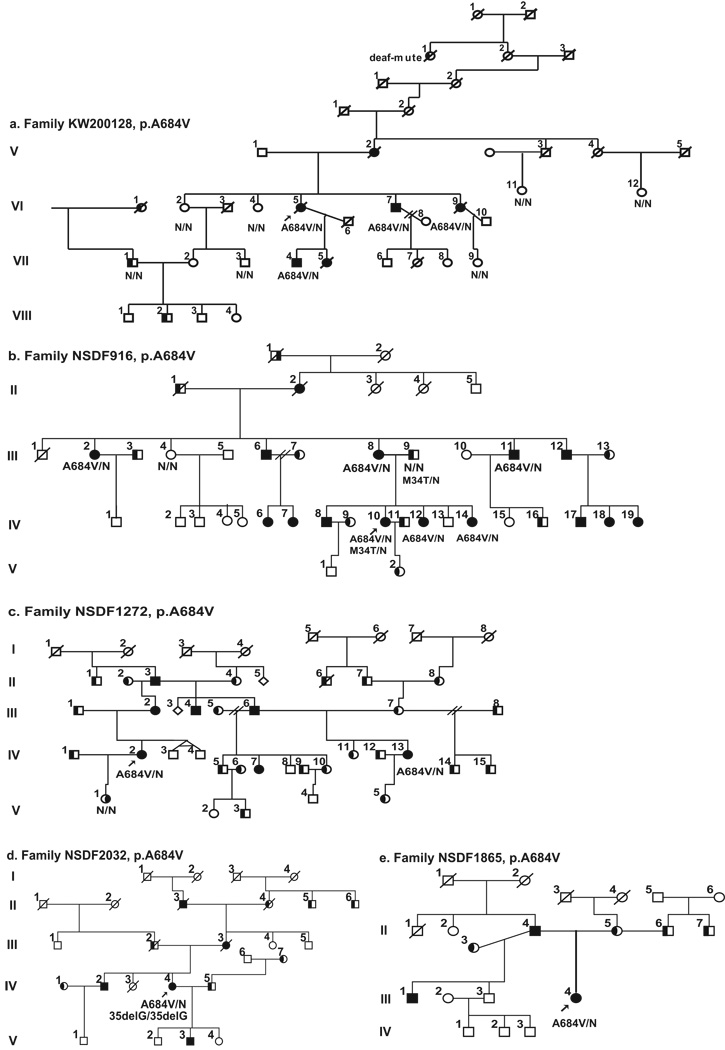
Figure 1.
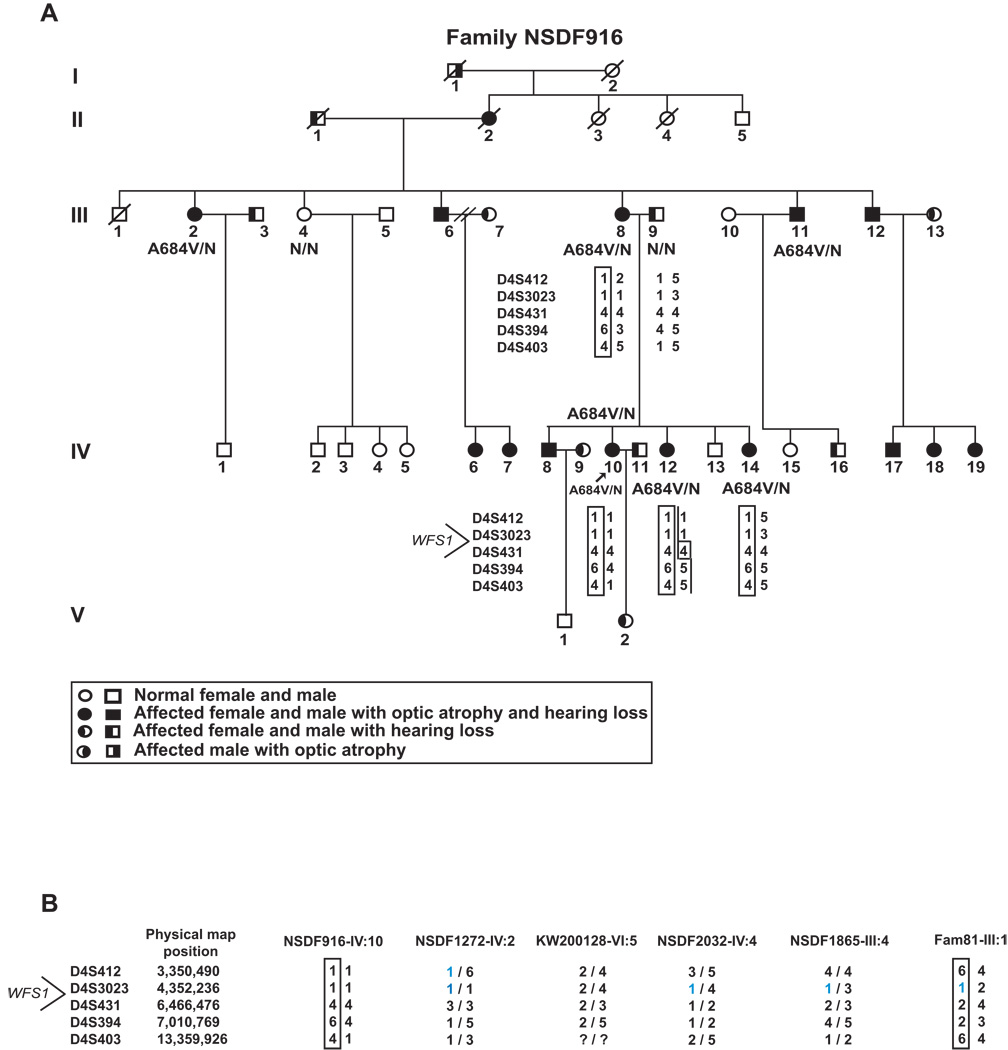
Pedigrees of the eight families investigated in this study. All the families show a segregation pattern compatible with the conclusion that the indicated WFS1 mutation causes autosomal dominant optic atrophy and hearing loss, except for family 81, where the father of two children with Wolfram syndrome is a sporadic case. Probands are indicated by arrows. Black symbols indicate optic atrophy with hearing loss patients. Black shading on the left part of symbol indicates individuals with isolated hearing loss and on the right part individuals with isolated optic atrophy. The WFS1 molecular result is indicated below individuals from whom DNA samples were available. N = normal allele. The presence of GJB2 mutation are indicated (c.35delG or p.M34T).
Sequence Analysis
Prior to WFS1 sequence analysis, GJB2 sequencing was performed in all probands and mutations were ruled out as the genetic cause of deafness in all but one proband, who had a complex family history of deafness. Primers were as described in Eiberg et al. [2006] or designed to PCR amplify exons and 20–50 bp of surrounding intronic regions of WFS1, OPA1, TIMM8A (RefSeq NM_006005.2, NM_015560.2 (OPA1 exons 1–28), NM_130837.2 (OPA1 exons 4B and 5B) and NM_004085.3, respectively). WFS1, OPA1 and TIMM8A were sequenced in DNA samples from all probands. WFS1 exon 8 contains several polymorphisms and to avoid using primers annealing to a region with polymorphisms, which could result in PCR amplification of only one allele, WFS1 exons 7 and 8 were amplified as one PCR product using the primers 5'-GCAGATCATGTTCGATGGAGCGGTTGGC-3' and 5'-CCTCATGGCAACATGCACTGGAAGCTCC-3', and AccuPrime™ Taq DNA Polymerase High Fidelity (Invitrogen) amplifying a fragment of 7930 bp. Furthermore, exon 8 sequencing primers were carefully positioned to avoid overlapping known polymorphisms. Primer sequences and PCR conditions are available upon request. PCR products were sequenced using BigDye Terminator chemistry (Applied Biosystems) and separated on an ABI 3130XL genetic analyzer (Applied Biosystems) according to the manufacturer’s instructions. Identified base pair changes were checked against databases of published polymorphisms and mutations (http://www.khri.med.umich.edu/research/lesperance_lab/low_freq.php; http://www.hgmd.cf.ac.uk/ac/index.php). In order to study the segregation of the mutations with the disease in each family, we used direct sequencing of PCR amplified genomic DNA. All detected mutations (Table I) were tested for their absence in 298 control chromosomes from 53 Danish and 96 UK ethnically matched Caucasian control individuals (from Sigma Aldrich).
TABLE I.
Clinical Manifestations in Eight Probands
| Family | KW200128a | NSDF916 | NSDF1272 | NSDF2032 | NSDF1865 | 81 | KW010862 | NSDF1793 |
|---|---|---|---|---|---|---|---|---|
| Proband ID | KW200128-VI:5 | 916-IV:10 | 1272-IV:2 | 2032-IV:4 | 1865-II:4 | Fam81-III:1 | KW01862-IV:1 | 1793-III:1 |
| Inheritance | ADOA + HI | ADOA + HI | ADOA + HI | ADOA + HI | ADOA + HI | Possibly sporadic | ADOA + HI | ADOA + HI |
| Age | Deceased age 63 | 32 | 38 | 54 | 46 | 55 | 14 | 49 |
| Nationality | Swedish | US/Caucasian | US/Caucasian | US/Caucasian | US/Caucasian | US/Caucasian | UK | US |
| Phenotype | OA + HI | OA + HI | OA + HI | OA + HI | OA + HI | OA + HI | OA + HI | OA + HI |
| WFS1 Mutation | p.A684V | p.A684V | p.A684V | p.A684V | p.A684V | p.A684V | p.G780S | p.D797Y |
| Optic atrophy | ||||||||
| Age of onset (years) | 9 | - Diagnosed at age 26) |
- Diagnosed at age 30) |
16 | 11 (Diagnosed at age 16) | - Diagnosed at age 41) |
12 | 20 |
| Visual acuity | 20/40 (R) 20/40 (L) |
20/40(R) 20/32 (L) |
- | - | 20/30 (R) 20/30 (L) |
20/60 (R) 20/60 (L) |
20/25 (R) 23/25 (L) |
20/80 (R) 20/70 (L) |
| Colour vision | - | Abnormal colour vision test (Ishihara plates) | - | - | Normal | - | Reduced | Not reported night and colour vision problems |
| Optic discs / optic nerves | Pale | Pale optic discs and cupped to 0.8 | - | - | Extreme pallor of optic discs with hypoplasia of the vessels and cupped to 0.7 | Extreme pallor of optic nerves | Bilateral pallor of optic nerves | Some pallor of the discs and narrowing of the vessels. Pale optic nerves bilaterally |
| Hearing impairment | ||||||||
| Age of onset (years) | Early childhood | Congenital | Early childhood | Congenital | 1.5y | 3y | Congenital | 3–4y |
| Severity | Severe, progressive | Severe to profound | Profound, progressive | ? | Severe to profound | Profound | Profound | Severe to profound, progressive |
| Diabetes | - | No | - | - | No | No | No | No |
| Psychiatric abnormalities | Depression, hallucinationsb | - | - | - | Depression | - | No | No |
| Other disorders / symptoms | - | - | - | - | Developed diabetes insipidus at age 20 following hypophysectomy for suspected pituitary tumor | - | No | No |
| Additional Information | Myopia in one family member | Humphrey visual fields with small enlargement of blind spot bilaterally. Retinal vessels, macula and retina periphery otherwise normal | Daughter with OA, normal hearing and bipolar illness does not have the mutation | GJB2 c.35delG homozygote | As part of evaluation for OA she had a CT scan. Mother is deaf from congenital rubella syndrome. Eye pressure 10 and 12 mm. Father is deaf, have OA and reported to have problems with colour vision | Has a son and a daughter with Wolfram syndrome and two WFS1 mutations: p.V415del and p.A684V c | Normal p-glucose and urine osmolality. Maternal great aunt with psychosis | OA was confirmed by CT/MRI scan. Except from deafness and OA physical examination normal. Eye pressure 10 and 12 mm. Normal macula and no evidence for retinal dystrophy. Also no diabetes in the mother of the proband who is deaf and has OA |
ADOA = autosomal dominant optic atrophy; HI = Hearing impairment; (−) No information; y = years;
the symptoms of the affected individuals have previously been described in Swedish (Samuelson, 1940);
some affected family members have anxiety and one affected family member (VII:5) committed suicide;
The p.V415del mutation in the children is inherited from their mother (the mutations are listed in the Kresge/WFS1 database);
mm = millimetres of mercury
The evolutionary conservation of wolframin amino acids among WFS1 orthologues was investigated using the ClustalW2 multiple sequence alignment program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and the Boxshade 3.21 program (hhtp://www.ch.embnet.org/software/BOX_form.html).
Haplotype Analysis
For haplotype analysis, five polymorphic dinucleotide repeat DNA markers from the 4p16.2-p15.33 WFS1 genomic region (Tel- D4S412, D4S3023, D4S431, D4S394, D4S403-Cen) were used according to standard protocols (Table IS online). Information about primer sequences and physical map positions were obtained from UCSC Genome Browser Human Mar. 2006 (hg18) assembly web site (http://genome.ucsc.edu/index.html?org=Human). An 18-bp extension sequence (5’-TGACCGGCAGCAAAATTG-3’) was added to the 5’ end of the forward primer to allow amplification of a fluorescently labelled third primer for visualization on an ABI 3130XL genetic analyzer [Mellersh et al., 2006]. Genotypes were scored semi-automatically using Genemapper (Applied Biosystems) and the haplotypes were constructed manually.
mtDNA Deletion Analysis
Total DNA was extracted from skeletal muscle using QIAamp® DNA Mini Kit (Qiagen). Aliquots of 0.1 µg DNA were digested with the restriction enzyme PvuII (New England Biolabs) and fractionated by electrophoresis in 0.5% agarose gels. The DNA was then transferred to Hybond-C nitrocellulose filter (GE Healthcare) by capillary blotting under standard procedures. The filter was hybridized with an equimolar mix of radiolabelled mtDNA probes corresponding to nucleotides 1–12640 and 14956–16569, as described previously [Larsson et al., 1990]. Hyperfilm MP (GE Healthcare) was exposed to the filter for 4 hours before development.
Protein Expression Analysis of Wolframin Mutants
Expression vector for wild-type, myc epitope-tagged wolframin was kindly provided by Dr. Timothy G. Barrett, UK. Mutations were introduced in wolframin using the QuickChange procedure (Stratagene) and confirmed by sequencing. Each mutant was generated 3 independent times and subjected to protein expression analysis. HEK293 cells were cultured and transfected as described [Doehn et al., 2009]. Briefly, 1.6 × 105 cells were seeded per 3.1 cm2 dish and co-transfected the following day, using 0.5 µg of wolframin and enhanced green-fluorescent protein (EGFP) expression plasmid, respectively, complexed with 3 µl FuGENE 6 reagent (Roche), according to the manufacturer’s instructions. After 16 hours, the cells were lysed with SDS-PAGE sample buffer (2% sodium dodecyl sulphate, 62 mM Tris-HCL (pH 6.8), 10% glycerol, 50 mM dithiothreitol, 0.12% bromophenol blue). Aliquots of the cell lysates were subjected to SDS-PAGE and immunoblotting analysis with antibody against the myc-tag on wolframin or EGFP, using standard immunoblotting procedures.
RESULTS
Families
The pedigrees of the eight families studied here and that all present autosomal dominant OA and SNHL are shown in Figure 1. Clinical and genetic findings are summarized in Tables I, II, and III. None of the probands contained the OPA1 mutations p.R445H or p.G439V nor any other mutations in the entire OPA1 or TIMM8A genes that might cause disease (one a single silent OPA1 variant, p.R393R was identified in one proband). Below, we describe the WFS1 mutations that were identified in each family and absent from 298 ethnically matched control chromosomes, as well as the clinical manifestations. Identified polymorphisms are listed in Table IIS online. GJB2 results were negative, except in a few cases that are indicated in Figure 1. Representative audiograms are provided in Figure 3.
TABLE II.
Clinical Manifestations in Additional Family Members
| Family | NSDF916 | KW010862 | KW010862 | KW010862 |
|---|---|---|---|---|
| Patient id | III:2 | III:2 | III:5 | IV:2 |
| Inheritance | ADOA + HI | ADOA + HI | ADOA + HI | ADOA + HI |
| Age | 69 | 48 | 42 | 9 |
| Nationality | US/Caucasian | UK | UK | UK |
| Phenotype | OA + HI | OA + HI | OA + HI | OA + HI |
| WFS1 Mutation | p.A684V | p.G780S | Not tested | p.G780S |
| Optic atrophy | ||||
| Age of onset (years) | 26 | Symptoms from age 20 | Diagnosed at age 42 (as a consequence of diagnosis in IV:1) | Diagnosed at age 9 (as a consequence of diagnosis in IV:1) |
| Visual acuity | 20/70 (R) 20/25 (L) |
20/40+3 (R) 20/40+2 (L) |
20/30 (R) 20/30 (L) |
10/10 (R) 10/10 (L) |
| Colour vision | - | - | Reduced | - |
| Optic discs | Pallor and shallow cup | Bilateral pallor of optic discs, particularly temporal | Bilateral pallor | Bilateral pallor |
| Hearing impairment | ||||
| Age of onset (years) | Congenital | Congenital | Congenital | Congenital |
| Severity | - | Profound | Profound | Profound |
| Diabetes | - | - | No | No |
| Psychiatric abnormalities | - | Schizophrenia. Treated for psychosis | No | Autistic |
| Neurological abn.. | - | No | No | No |
| Mental retardation | - | No | No | No |
| Other disorders / symptoms | Late onset bilateral cataract | No | No | No |
| Additional Information | Eye pressure 13 mm, blurred vision | Chlorpromazine cataracts | Normal plasma glucose | Normal urine osmolality |
ADOA = autosomal dominant optic atrophy; HI = Hearing impairment; (−) No information; mm = millimetres of mercury
TABLE III.
Ophthalmological Examination Results in Family KW200128
| Patient | Year of birth |
Age at diagnosis of optic nerve atrophy |
BCVA 0–20 y |
BCVA 21–40 y |
BCVA 41–50 y |
BCVA 51–60 y |
BCVA 61–70 y |
BCVA 71 – 80 y |
BCVA 81–100 y |
Papillae (by ophthalmoscopy) |
Age at diagnosis of glaucoma |
Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V:2 | 1895 | 42 | - | - | R 0.5 L 0.3 |
- | R 0.1 L 2/60 |
R 2/60 L 1/60 |
R 1/120 L <1/120 |
Bilateral pale especially temporally | 68 | - |
| VII:5 | 1951 | 8 | R 0.9 L 0.9 |
R 0.9 L 0.9 |
R<0.1 L 0.2 |
- | - | - | - | Bilateral pale optic nerves (from age 8) | 43 | - |
| VII:4 | 1955 | 18 | R 1.0 L 1.0 |
R 1.0 L 1.0 |
R 0.5 L 0.6 |
- | - | - | - | Bilateral pale optic nerves | No glaucoma | Myopia |
| VI:5 (Proband) | 1928 | 22 | R 0.5 L 0.5 |
R 0.5 L 0.5 |
- | R 0.1 L 0.1 |
R A L 1/120 |
- | - | Bilateral pale optic nerves | R 56 L 65 + CVO |
- |
| VI:9 | 1925 | 12 | - | R 0.4 L 0.5 |
- | R 0.1 L 0.1 |
R 0.1 L 0.1 |
- | - | Bilateral pale optic nerves | No glaucoma | Visual field reduced to 20 degrees at age 70 |
| VI:7 | 1931 | 25 | - | - | - | R 0.1 L 0.1 |
R 0.1 L 1/60 |
R 0.1 L 1/60 |
- | Bilateral pale optic nerves | 48 | - |
y = age in years; BCVA = best corrected visual acuity by Snellen test; R = Right eye; L = Left eye; (−) no information; A = Amaurosis; CVO = Central venous occlusion.
Additional ophthalmological description: In family KW200128 there was a characteristic course of the progressive visual impairment due to optic nerve atrophy. The visual acuity is affected moderately in childhood, slowly decreasing in middle age and causing daily problems from 50 – 60 years of age. Four out of eight individuals affected by optic nerve atrophy, in addition, developed glaucoma, which leads to further visual decrease and one individual developed bilateral venous trombosis leading to very low vision.
The optic neuropathy showed the typical picture of inherited optic nerve atrophy, i.e. early thinning of the nerve fibre layer especially in the papillomacular bundle. Defects in this part of the nerve fibre layer early influence the visual acuity and all affected family members have very low visual acuity after age 60. The optic nerve appears pale and flat, influencing the quality of vision i.e. colour vision, contrast vision and mobility-orientation. Visual fields show generally increasing low sensitivity and are concentrically narrowed until about 40–50 degrees in the late stages. Individuals with glaucoma, moreover, were affected by typical Bjerrum scotoma which further narrowed the visual fields.
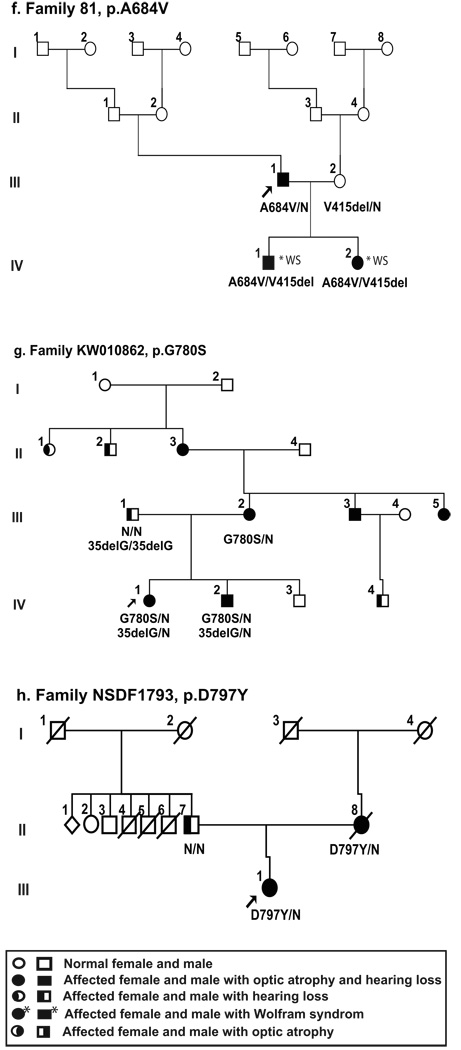
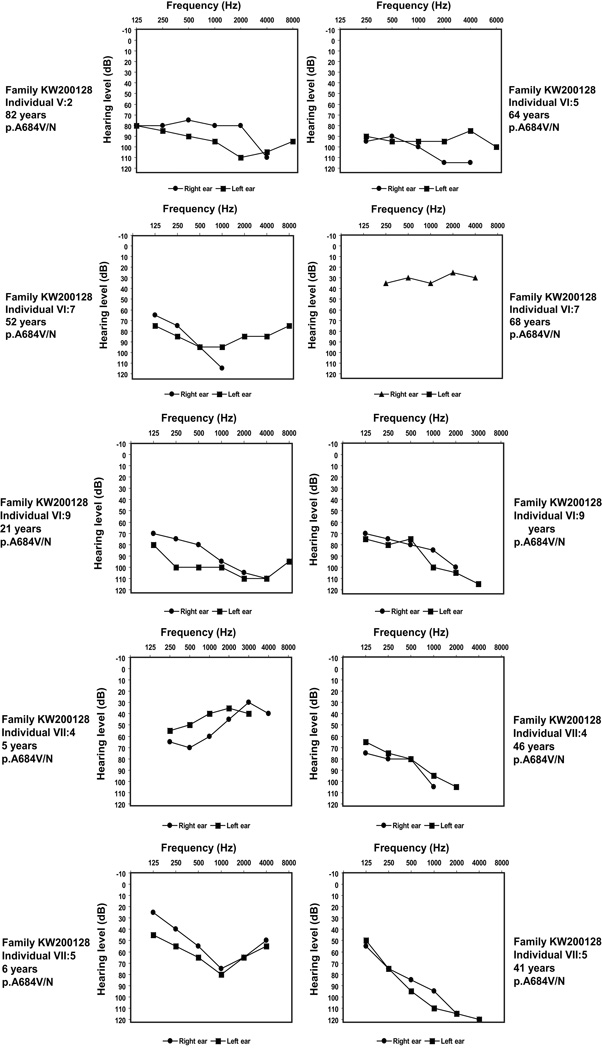
Figure 3.
Pure-tone audiograms of left and right ear of representative, affected family members heterozygous for WFS1 mutation from six of the families studies (KW2001128 (audiograms from 6 individuals), NSDF916, NSDF1865, NSDF1272, and NSDF1793). For some individuals two audiograms are shown to illustrate the hearing loss over time. Individual VI:7 (family KW200128) had cochlear implant at age 57 years, resulting in an improved hearing between age 52 and age 68.
Family KW200128
This is a large eight-generation family (Fig. 1a) from Sweden with six affected individuals and is the family described in the first report of isolated ADOA and SNHL [Samuelson, 1940]. Four members of the family, V:2, VI:5, VI:7 and VI:9, suffered from the same complaints, but the severity of vision loss and hearing impairment varied among the family members [Samuelson, 1940]. Samuelson [1940] suggested that the cause of the reduced vision and hearing was atrophy of the optic nerve, and the cochlear nerve, respectively.
In our follow-up study, the proband, VI:5, now deceased, had OA from age 9 and severe bilateral SNHL from early childhood as well as psychiatric problems (depression and hallucinations). Furthermore, one affected family member (VII:5) committed suicide, and four affected family members have regular psychiatric treatment in order to cope with their anxiety. In one branch of the family a different and unknown dominant cause of SNHL segregated, since individuals VI:1, VII:1 and VIII:2 all had moderate sensorineural hearing loss, but no visual problems and none had the WFS1 mutation described below. GJB2 sequencing in DNA from VII:1 was normal. Cochlear implantation was performed at age 49, 57 and 77 years, respectively, in individuals VII:4, VI:7, and VI:9 with variable degree of benefit. Individual VI:7 experienced considerably improved hearing (Fig. 3), but the outcome was questionable for the other two individuals due to old age, general medical frailty and inability to perform relevant training efforts after CI treatment.
The OA in the family presented with temporal paleness of the optic nerves similar to the optic atrophy due to OPA1 mutations. The visual impairment progressed to low visual acuity after the age of 60, and a subsequent need for special visual adjustments (Table III). In four of the six individuals, glaucoma developed and responded well to standard treatment. The glaucoma treatment may have caused a relative improvement of visual capacity due to the optic atrophy, until late stages of the course where restriction in visual field occurred. These family members were followed in many instances several years ago, and details of visual acuity were not recorded in accordance with current standards.
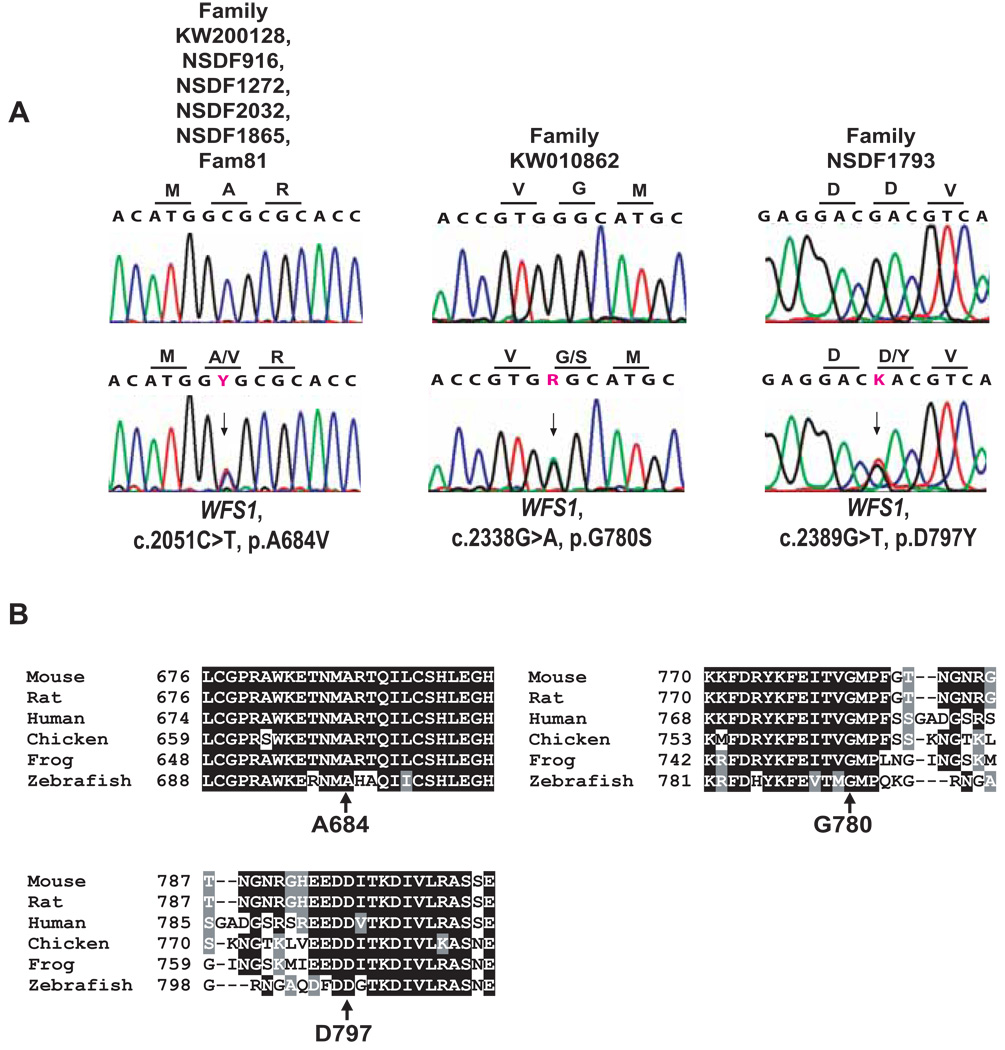
To establish a molecular diagnosis, DNA was collected from nine individuals, four of whom were affected with OA and SNHL (VI:5, VI:7, VI:9, and VII:4, Fig. 1). Since the clinical features suggested that the disease was caused by OPA1 mutation, we first sequenced this gene. However, no OPA1 mutation was identified. Next the gene for Mohr-Tranebjærg syndrome, TIMM8A, that also present with similar features was also sequenced, but again no mutation was identified. Since Wolfram syndrome presents with OA and deafness and sometimes with psychiatric problems, as in the proband, we speculated that mutation of WFS1 might be the cause of disease in the family. Indeed, sequencing of WFS1 in the proband identified a heterozygous sequence change c.2051C>T in exon 8, leading to a substitution of alanine for valine at position 684 of wolframin (p.A684V). A684 is located in the hydrophilic C-terminus of wolframin and is conserved in evolutionarily distant species such as mouse, rat, chicken, frog and zebrafish (Fig. 2B). The mutation was absent in 289 control chromosomes. These facts along with the co-segregation of the p.A684V mutation with the disease strongly indicate that this mutation underlies optic atrophy and hearing loss in the family. The mutation appears to be de novo, first manifested in affected individual V:2 who had unaffected sibs and parents, who died in old age with preserved hearing and vision.
Figure 2.
(A) Representative sequence chromatograms for each of the WFS1 missense mutations compared to a normal control. The arrows indicate the nucleotide changes of the heterozygous missense mutations. Nomenclature of mutations refers to the WFS1 RefSeq NM_006005.2, with nucleotide number +1 being A of the start codon ATG. Each mutation is heterozygous in affected individuals. (B) Alignment in different species shows strong evolutionary conservation of the relevant amino acid mutated in the patients.
We then speculated whether the p.A684V WFS1 mutation could lead to multiple mithochondrial DNA (mtDNA) deletions since some early studies identified mtDNA deletions in Wolfram syndrome patients [Gomez-Zaera et al., 2001] and since some OPA1 mutations lead to multiple mtDNA deletions in skeletal muscle [Amati-Bonneau et al., 2008]. Therefore, skeletal muscle biopsy specimens from one p.A684V mutation carrier (VI:7) was analyzed for deletions of mtDNA by Southern blotting. However, no mtDNA deletions could be detected (data not shown).
In conclusion, these data suggested that ADOA and SNHL in the family were caused by the p.A684V mutation in WFS1.
Family NSDF916
This is a five-generation, large Caucasian American family of English origin with 15 affected individuals (Fig. 1b). DNA was collected from eight individuals, six of whom were affected (III:2, III:8, III:11, IV:10, IV:12, and IV:14, Fig. 1). The 32-year-old proband (IV:10) has had optic atrophy since childhood and has bilateral, congenital severe to profound hearing impairment. Sequencing of WFS1 in the proband revealed the same heterozygous c.2051C>T mutation in exon 8 leading to the p.A684V mutation identified in the above-mentioned Swedish family. The mutation consistently co-segregated with optic atrophy and hearing loss in the family. The proband was heterozygous for the p.M34T sequence variant in GJB2, as is her father, who is deaf, but does not have OA.
Family NSDF1272
This is a five-generation, Caucasian American family of Polish/Russian and mixed European ancestry with seven affected family members (Fig. 1c). DNA was available from 3 individuals, of whom two were affected with optic atrophy and hearing loss. The proband (IV:2) has optic atrophy which was diagnosed at approximately age 30 and bilateral profound hearing loss, which was progressive and started in early childhood.
The p.A684V mutation in WFS1 was identified in the proband and in a remote family member (individual IV:13), who also has bilateral OA and SNHL. In contrast, the daughter (V:1) of the proband, has hydrocephalus, a complex medical history, bipolar illness and optic atrophy, but has normal hearing and does not have the mutation. Thus, the isolated case of optic atrophy in V:1 cannot be explained by results of WFS1 or OPA1 sequencing, suggesting that her optic atrophy is due to another, genetic or environmental, cause.
Family NSDF2032
This is a five-generation Caucasian American family of Dutch ancestry with five affected individuals (Fig. 1d). Individual IV:4 had OA diagnosed at the age of 16 years and congenital deafness, and was found to have the p.A864V mutation in WFS1. In addition, this individual is also homozygous for the c.35delG mutation in GJB2. It is quite conceivable that she has two forms of hereditary deafness given her complex family history of deaf parents and deaf maternal grandparents.
Family NSDF1865
The proband (III:4) of this Caucasian American family (Fig. 1e) has optic atrophy, diagnosed at age 16, and bilateral, severe to profound sensorineural hearing loss (Fig. 3). She has also been treated for depression since the age of 38. Sequencing of WFS1 again revealed the p.A684V mutation.
Family 81
Four members, two parents and their children, of this Caucasian American family were analyzed (Fig. 1f). The father (III:1) is of Dutch and Italian descent, while the mother (III:2) is of Dutch-Polish, and German-Irish descent. III:1 is the only relative with optic atrophy and deafness without other manifestations of Wolfram syndrome. He had bilateral, profound SNHL diagnosed at age 3 years and OA diagnosed at age 41 by the ophthalmologist (M.B.M) evaluating his children. Both IV:1 and IV:2 were diagnosed with Wolfram syndrome based on juvenile onset insulin-dependent diabetes mellitus, optic atrophy, bilateral profound sensorineural hearing loss, and congenital cataracts. IV:1 had hearing loss diagnosed at birth and IV:2 had hearing loss diagnosed at age 3 months and premature birth at 30 weeks of gestation.
III:1 is heterozygous for p.A684V, and III:2 is apparently asymptomatic and heterozygous for p.V415del mutation. Their children, IV:1 and IV:2, are compound heterozygotes for the WFS1 mutations p.A684V and p.V415del (c.1243_1245delGTC). Unfortunately, no medical data nor DNA were available from the parents of the father, III:1, and therefore it can not be determined whether the p.A684V mutation is a de novo event or transmitted from one of his parents. In any case, the phenotype of the children suggest that the p.V415del mutation aggravates the effect of the p.A684V mutation, resulting in Wolfram syndrome.
Family KW010862
This is a Caucasian UK family with six affected individuals (Fig. 1g). DNA was available from 4 individuals (three affected and one unaffected). The proband (IV:1) is reported to have bilateral, prelingual, profound hearing loss in addition to optic atrophy. The proband has normal plasma glucose and urine osmolality.
Sequencing of WFS1 revealed a novel heterozygous mutation, c.2338G>A, in exon 8, which results in the substitution of a glycine codon for a serine codon at position 780 (p.G780S). p.G780S segregates with disease in the available family members. Furthermore, the mutation was absent from 298 control chromosomes. Amino acid p.G780 is located in the hydrophilic C-terminus of wolframin and is conserved in wolframin from species such as mouse, rat, chicken, frog and zebrafish (Fig. 2B). Upon testing for the c.35delG mutation in GJB2, the deaf father (Fig. 1g, III:1) of the proband was homozygous, and individuals IV:1 and IV:2 were heterozygous.
Family NSDF1793
This is a small Caucasian American family with 2 affected individuals (Fig. 1h). The father of the proband is of Polish descent and her mother of Scottish descent. The proband (III:1) had hearing loss first suspected at the age of 3–4 years, which progressed to a severe to profound, sensorineural hearing loss. Optic atrophy was recognized at approximately age 20 years. The proband had normal mental and intellectual status.
Sequencing of WFS1 in the proband revealed a novel heterozygous mutation c.2389G>T in exon 8. The mutation results in substitution of an aspartic acid codon for a tyrosine codon at position 797 (p.D797Y). The affected mother of the proband is also heterozygous for this mutation. The mutation was absent from 298 control chromosomes. Amino acid p.D797 is located in the hydrophilic C-terminus of wolframin and is conserved in wolframin from species such as mouse, rat, chicken, frog and zebrafish (Fig. 2B).
Haplotype Analysis
Haplotype analysis was performed in family NSDF916, with the recurrent p.A684V mutation, by using five polymorphic markers (D4S412, D4S3023, D4S431, D4S394, and D4S403) flanking WFS1. A common haplotype for all affected individuals was found (Fig. 4A). Marker analysis with the same five markers in probands from the other five p.A684V families showed that one of the probands (NSDF1272-IV:2) had allele sizes for two markers (D4S412 and D4S3032) in common with family NSDF916, and three probands (NSDF-2032:IV:4, NSDF1865-III:4, and Fam81-III:1) had allele size for one marker (D4S3023) in common with family NSDF916 (Fig. 4B). However, there is no indication of one ancient shared haplotype between these families that originate from very different parts of the world, and it is most likely that the mutation has arisen independently several times.
Figure 4.
Haplotype analysis. (A) Haplotype analysis in Family NSDF916, harboring the p.A684V mutation, using five polymorphic markers flanking WFS1. (B) Marker analysis with the same five markers in probands from the other five p.A684V families. A bar indicates the haplotype carrying the mutation in those families where this could be deduced.
Protein Expression Analysis of Identified Wolframin Mutations
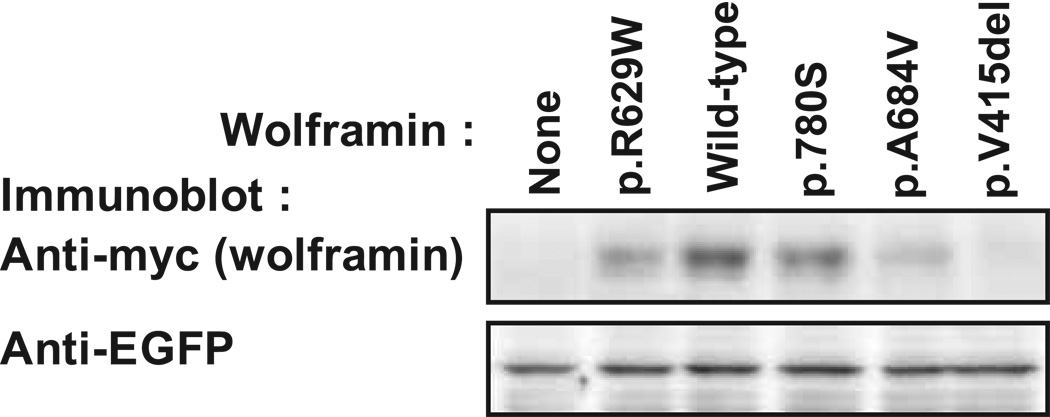
We next performed functional expression analysis of certain of the indentified mutations to further validate that they are pathogenic. Thus, we transiently transfected HEK cells with plasmid expressing myc epitope-tagged wild-type wolframin or wolframin harboring selected mutations and analyzed the cells for the expression level of the exogenous wolframin by immunoblotting for the myc tag. This analysis revealed that the p.A684V and the p.V415del mutants showed greatly and the p.G780S mutant mildly decreased protein expression compared to wild-type wolframin (Fig. 5, upper panel). The p.R629W mutation previously shown to confer instability to wolframin protein, resulting in decreased protein expression and Wolfram syndrome, was included as a positive control [Hofmann et al., 2003]. To control for equal transfection efficiencies across the cell populations, we immunoblotted for enhanced green-fluorescent protein (EGFP) that we had co-transfected together with wolframin. As shown in Figure 5 lower panel, all samples showed equal expression of EGFP, confirming equal transfection across all the cell populations analyzed.
Figure 5.
Functional protein expression analysis of WFS1 mutations. HEK293 cells were co-transfected with plasmid expressing myc-tagged wild-type or mutant wolframin together with plasmid expressing EGFP. The cells were lysed 16 hours post-transfection and the cell extracts were subjected to SDS-PAGE and immunoblotting for the myc tag and EGFP. The experiment was repeated 3 times with independently generated mutants with similar results.
DISCUSSION
We have identified heterozygous missense mutations in WFS1 as the cause of ADOA and SNHL in eight families. Previously, isolated ADOA and SNHL were reported only in association with OPA1 mutations, but our present report of multiple families, supported by the very recent report of one family [Hogewind et al., 2010], shows that WFS1 mutations can also underlie isolated ADOA and SNHL. Furthermore, we have discovered that a p.A684V WFS1 mutation underlies this dual sensory impairment in the original Swedish family [Samuelson 1940] and in 5 of 7 additional families suggesting that the p.A684V WFS1 mutation may in fact be a frequent (maybe even primary) cause of isolated ADOA and SNHL. Finally, we demonstrate that ectopic expression in HEK cells of wolframin with the p.A684V, p.V415del and p.G.780S WFS1 mutations, respectively, results in reduced levels of mutant wolframin as compared to wild-type wolframin, supporting the conclusion that these mutations are disease-causing.
Wolfram syndrome is an autosomal recessive condition with minimal diagnostic criteria of juvenile OA and juvenile onset diabetes mellitus. Importantly, these clinical criteria were developed prior to identification of the responsible gene, WFS1. Several studies on families with Wolfram syndrome have been performed. In these studies, Wolfram syndrome is in agreement with an autosomal recessive mode of inheritance and mutations have been identified on both alleles in Wolfram syndrome patients and carriers of one mutation are healthy. Thus, isolated optic atrophy and hearing loss has not to our knowledge been reported as a feature in heterozygous carriers of these WFS1 mutations. However interestingly, heterozygous carriers of WFS1 mutations may have an increased incidence of psychiatric disorders including suicide attempts, endogenous depression, short-term memory loss, and anxiety (reviewed in [Cryns et al., 2003]).
Our study supports recent data that a dominant form of partial Wolfram syndrome exists with the key features of optic atrophy and sensorineural hearing loss, not necessarily of juvenile onset. Furthermore, psychiatric illness was present in family KW200128 and family NSDF1865.. A WFS1 p.E864K missense mutation has been reported in two families with dominantly inherited deafness and some members affected by OA and impaired glucose regulation/diabetes [Eiberg et al., 2006; Valero et al., 2008]. In two Japanese families, however, the p.E864K mutation only caused isolated LFSNHL [Fukuoka et al., 2007]. Finally, most recently, a p.K836N mutation in WFS1 has been found associated with autosomal dominant optic neuropathy and deafness in a family with three affected members [Hogewind et al., 2010]. Interestingly, simultaneously with the family described by Hogewind et al. [2010] Fujikawa et al. [2010] published a family with 14 affected members segregating SNHL associated with mutation of the same WFS1 amino acid, K836, but here mutated to threonine instead of an asparagine. However, in this family, the deafness-affected individuals had no OA, nor any other WFS1-associated abnormalities, such as diabetes insipidus or mellitus (impaired glucose tolerance was reported in an individual with no deafness and severe depression in another with no deafness in the family, but genetic studies could not be performed in these individuals). These findings suggest that there may be a spectrum of phenotypes associated with WFS1 based on differences in functional implications of the mutations, as suggested in the review by Tranebjærg [2008].
We found three different wolframin missense mutations (p.A684V, p.G780S, and p.D797Y) to be associated with a dominant phenotype of OA and SNHL in eight families. The causative changes were all identified in exon 8 of WFS1, similar to the many known WFS1 mutations causing Wolfram syndrome or LFSNHL. Several lines of evidence support the conclusion that they are pathogenic, rather than rare polymorphisms. Firstly, the three mutations were heterozygous and in perfect dominant co-segregation with optic atrophy and hearing loss. Secondly, they were absent from 298 ethnically matched control chromosomes and from the NCBI SNP database. Thirdly, all three mutations affect residues evolutionary conserved in human, rat, mouse, chicken, frog and zebrafish wolframin (Fig. 2B), suggesting that altering these residues have deleterious consequences for the wolframin protein. Finally, we obtained experimental evidence for this later hypothesis by showing that the p.A684V, and to a smaller extent the p.G780S mutation, decreased wolframin protein expression levels. It may be noted that although the expression analysis provide strong evidence for a deleterious effect of the mutation, it does not provide a straight forward relationship between wolframin expression levels and severity of disease, since the pV415del mutation caused the greatest decrease in wolframin expression, yet was associated with a phenotype only in compound heterozygosity with the p.A684V mutation. Furthermore, it may be noted that although the expression data suggest that the mutations cause disease, they do not provide evidence that they can cause dominant disease (as opposed to recessive disease). Rather, combined with the absence of mutations in any other known OA genes in any of our probands, these data collectively suggest that the identified mutations act dominantly to cause OA and SNHL. It can be speculated that the mutations cause a misfolded wolframin protein. Part of the misfolded wolframin pool is degraded, but the remaining pool act as dominant negative mutants in the wolframin pathway to cause dominant disease. In conclusion, our findings support the involvement of missense mutations in WFS1 in an isolated autosomal dominant monogenic form of hearing loss and optic atrophy.
Six of the eight families with ADOA and SNHL reported here harbored the same heterozygous WFS1 mutation, p.A684V (c.2051C>T). Haplotype analysis in p.A684V individuals suggested that the c.2051C>T mutation arose independently in the families studied, and may represent a mutational hotspot. Importantly, these data open the possibility that a single, recurrent mutation in WFS1 i.e. p.A684V, may be a common cause of isolated ADOA and SNHL, similar to the role played by the OPA1 p.R445H mutation in autosomal dominant optic atrophy.
Prior to our study, the p.A684V mutation had not been reported in association with an isolated OA and hearing loss phenotype. However, this mutation had been identified in a patient from Italy with Wolfram syndrome [Tessa et al., 2001] (Family 3 at the WFS1 Gene Mutation and Polymorphism Database at http://www.khri.med.umich.edu/research/lesperance_lab/wfs_delete.php). The Italian patient had diabetes mellitus, optic atrophy, hearing loss, diabetes insipidus, ataxic gait and psychiatric abnormalities. In addition to the p.A684V mutation, the patient also harbored a c.1387delCTCT mutation. The p.A684V mutation was absent in 100 Italian control chromosomes. Unfortunately, it is not known whether one of the parents had the p.A684V mutation in trans and associated with impaired vision and hearing (personal feed back from the corresponding author A. Tessa, who reported that the parents were unavailable for follow-up). By contrast, in the present study, DNA was available from the parents of two children (Family 81) with Wolfram syndrome, who are compound heterozygous for p.A684V and p.V415del in WFS1. The children both have juvenile onset of insulin-dependent diabetes mellitus, OA, bilateral profound sensorineural hearing loss and congenital cataracts [Mets et al., 2010]. Interestingly, we identified the p.A684V mutation in heterozygous state in the father, who has OA and hearing loss only, while the mother, heterozygous for the p.V415del mutation, had no symptoms. These data support that heterozygosity for p.A684V alone causes isolated OA and hearing loss. Since the mother had no obvious disease symptoms, we went on to demonstrate that the p.V415del mutation greatly decreased wolframin protein expression. This demonstrates that the p.V415del alteration is indeed the responsible mutation that aggravates the effect of the p.A684V mutation, causing Wolfram syndrome in the children. We believe that a similar scenario also underlies the Wolfram syndrome phenotype of the Italian patient reported by Tessa et al., [2001], although his parents would have to be mutation analyzed to substantiate this conclusion.
The current lack of knowledge about the physiological and cellular and biochemical functions of wolframin makes it difficult to explain the disease-causing mechanism of the mutations. To this end, we addressed whether WFS1 mutations could lead to multiple mitochondrial DNA deletions, by analyzing one p.A684V patient for deletions of mtDNA in skeletal muscle. However, no deletions were detected suggesting that the p.A684V mutation does not cause ADOA and hearing loss by increasing mtDNA instability. Unfortunately, muscle DNA was not available from the other patients presented here.
To date, only mutations in OPA1 have been repeatedly linked to the rare phenotype featuring isolated OA with sensorineural deafness. One recurrent OPA-1 mutation, the heterozygous p.R445H missense mutation, underlies the large majority of reported cases of isolated OA and hearing loss. This mutation was first identified in a Japanese and a French patient [Amati-Bonneau et al., 2003; Shimizu et al., 2003] and subsequently in families from the US, and Belgium [Payne et al. 2004] and finally in five patients/families from France, Spain, and of Caucasian origin [Amati-Bonneau et al., 2005; Li et al., 2005]. In a few of these families, some patients also developed ptosis, ophthalmoplegia, ataxia and/or nonspecific myopathy at middle age [Payne et al., 2004; Yu-Wai-Man et al., 2010]. Only two other OPA1 mutations (c.970delCGTTCTCCA and p.G401D) have so far, to our knowledge been associated with isolated OA and hearing loss [Puomila et al., 2005; Ke et al., 2006; Amati-Bonneau et al., 2009]. Phenotypic characteristics of most of these patients include onset of hearing loss at the age of 6–30 years [Amati-Bonneau et al., 2005]. By contrast, all probands with WFS1 mutation ascertained in the present study were congenitally deaf or had hearing loss onset in early childhood (Table I). Further patients need to be studied to evaluate whether WFS1 mutation, and in particular the p.A684V mutation, is a predominant cause of dominant OA and SNHL, and whether there is a correlation between early onset of hearing loss and WFS1 mutation versus OPA1 mutation, to assist in molecular diagnosis of this disease.
The fact that carriers of WFS1 mutations associated with Wolfram syndrome do not have hearing impairment and usually not significant if present, and the previous reports of heterozygosity for other WFS1 mutations resulting in nonsyndromic autosomal dominant LFSNHL emphasize the difficulties in predicting the consequences of the different types of WFS1 mutations [Eiberg et al., 2006; Tranebjærg, 2008]. Our report supports this notion, since our study is the first study of several families with OA and hearing loss without other major clinical features (two families did have psychiatric abnormalities when carefully followed) found to be associated with mutations in WFS1. The first family segregating optic atrophy and hearing loss in a dominant pattern with a WFS1 mutation (p.E864K) was described by Eiberg et al. [2006]. However, in this family, careful metabolic evaluations of three out of four mutation carriers, revealed that they had undiagnosed diabetes, impaired glucose tolerance and/or impaired insulinogenic index indicating that the p.E864K mutation may affect pancreatic beta-cell function as well [Eiberg et al., 2006]. It is entirely possible that additional abnormalities are actually present in a higher fraction of the families with WFS1 related disorder, as illustrated by psychiatric disease in the two families in the present report (KW200128 and NSDF1865) and reported by Eiberg et al [2006] and Valero et al. [2008]. A similar modification of clinical presentation became evident when OPA1 patients in larger numbers were carefully clinically examined, showing that about 20% actually turned out to have neurological abnormalities [Yu-Wan-Man et al., 2010].
In several of our families there are several spouses with SNHL, who may introduce one or more additional SNHL genes. It can therefore be speculated that these genes may contribute to the phenotype or are acting digenically with the identified WFS1 mutation in the respective families. In all cases, where we have both phenotypic and molecular genetic results and found a heterozygous missense mutation in WFS1, the OA and SNHL phenotype was present, pointing to that the respective WFS1 mutations alone are capable of causing the disease phenotype. However, in one family (NSDF2032) the proband is congenital deaf, and in addition to being heterozygous for the p.A864V mutation in WFS1, this individual is homozygous for the c.35delG mutation in GJB2. Thus, in this case, it cannot be solved which of the two genes is causing her deafness.
In conclusion, we fully support the recent suggestion by Valéro et al. [2008] of systematic sequencing analysis of WFS1 in patients with diabetes and deafness (+/− optic atrophy), in particular when mtDNA mutations have been excluded, and a broad clinical examination in such patients. We also recommend WFS1 sequencing in patients with optic atrophy and early onset hearing loss, especially when OPA1 mutations have been excluded [Tranebjærg, 2008]. Hopefully, future WFS1 screening of larger patient cohorts with hearing loss and optic atrophy will determine the extent to which WFS1 mutations underlies this phenotype. Finally, supplementary efforts are essential to determine the precise functions of wolframin in the inner ear and eye and the role of genetic or environmental modifying factors. Studies focusing on the functional consequences of WFS1 mutations identified in patients are needed and may help to resolve why different WFS1 mutations may cause either recessive Wolfram syndrome, dominant LFSNHL or dominant optic atrophy with deafness, respectively, which would have major implications for the involved families in terms of appropriate genetic counseling and relevant clinical long-term follow-up.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the families who participated in this study. We acknowledge valuable technical assistance from Elvira Chapka at Department of Cellular and Molecular Medicine, University of Copenhagen. Bernd Wissinger, Tübingen, Germany, is acknowledged for OPA1 sequencing in family KW200128. The Lundbeck Foundation (grant no. 32011) and Widex AS are acknowledged for financial support to the Audiogenetic Research Group at ICMM. This work was also supported by the US National Institutes of Health grant 0R01DC006707 awarded to KSA. The project was hosted by Wilhelm Johannsen Centre for Functional Genome Research, established by the Danish National Research Foundation.
Footnotes
Additional supporting information may be found in the online version of this article.
REFERENCES
- Amati-Bonneau P, Guichet A, Olichon A, Chevrollier A, Viala F, Miot S, Ayuso C, Odent S, Arrouet C, Verny C, Calmels MN, Simard G, Belenguer P, Wang J, Puel JL, Hamel C, Malthiery Y, Bonneau D, Lenaers G, Reynier P. OPA1 R445H mutation in optic atrophy associated with sensorineural deafness. Ann Neurol. 2005;58:958–963. doi: 10.1002/ana.20681. [DOI] [PubMed] [Google Scholar]
- Amati-Bonneau P, Milea D, Bonneau D, Chevrollier A, Ferre M, Guillet V, Gueguen N, Loiseau D, de Crescenzo MA, Verny C, Procaccio V, Lenaers G, Reynier P. OPA1-associated disorders: phenotypes and pathophysiology. Int J Biochem Cell Biol. 2009;41:1855–1865. doi: 10.1016/j.biocel.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Amati-Bonneau P, Odent S, Derrien C, Pasquier L, Malthiery Y, Reynier P, Bonneau D. The association of autosomal dominant optic atrophy and moderate deafness may be due to the R445H mutation in the OPA1 gene. Am J Ophthalmol. 2003;136:1170–1171. doi: 10.1016/s0002-9394(03)00665-2. [DOI] [PubMed] [Google Scholar]
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, Iommarini L, Labauge P, Figarella-Branger D, Marcorelles P, Furby A, Beauvais K, Letournel F, Liguori R, La MC, Montagna P, Liguori M, Zanna C, Rugolo M, Cossarizza A, Wissinger B, Verny C, Schwarzenbacher R, Martin MA, Arenas J, Ayuso C, Garesse R, Lenaers G, Bonneau D, Carelli V. OPA1 mutations induce mitochondrial DNA instability and optic atrophy 'plus' phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- Arnos KS, Welch KO, Tekin M, Norris VW, Blanton SH, Pandya A, Nance WE. A comparative analysis of the genetic epidemiology of deafness in the United States in two sets of pedigrees collected more than a century apart. Am J Hum Genet. 2008;83:200–207. doi: 10.1016/j.ajhg.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TG, Bundey SE, Macleod AF. Neurodegeneration and diabetes: UK nationwide study of Wolfram (DIDMOAD) syndrome. Lancet. 1995;346:1458–1463. doi: 10.1016/s0140-6736(95)92473-6. [DOI] [PubMed] [Google Scholar]
- Bespalova IN, Van CG, Bom SJ, Brown DJ, Cryns K, DeWan AT, Erson AE, Flothmann K, Kunst HP, Kurnool P, Sivakumaran TA, Cremers CW, Leal SM, Burmeister M, Lesperance MM. Mutations in the Wolfram syndrome 1 gene (WFS1) are a common cause of low frequency sensorineural hearing loss. Hum Mol Genet. 2001;10:2501–2508. doi: 10.1093/hmg/10.22.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryns K, Sivakumaran TA, Van den Ouweland JM, Pennings RJ, Cremers CW, Flothmann K, Young TL, Smith RJ, Lesperance MM, Van CG. Mutational spectrum of the WFS1 gene in Wolfram syndrome, nonsyndromic hearing impairment, diabetes mellitus, and psychiatric disease. Hum Mutat. 2003;22:275–287. doi: 10.1002/humu.10258. [DOI] [PubMed] [Google Scholar]
- Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, Cohen MS, Johansen JV, Winther BR, Lund LR, Winther O, Taunton J, Hansen SH, Frodin M. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol. Cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberg H, Hansen L, Kjer B, Hansen T, Pedersen O, Bille M, Rosenberg T, Tranebjaerg L. Autosomal dominant optic atrophy associated with hearing impairment and impaired glucose regulation caused by a missense mutation in the WFS1 gene. J Med Genet. 2006;43:435–440. doi: 10.1136/jmg.2005.034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa T, Noguchi Y, Ito T, Takahashi M, Kitamura K. Additional heterozygous 2507A>C mutation of WFS1 in progressive hearing loss at lower frequencies. Laryngoscope. 2010;120:166–171. doi: 10.1002/lary.20691. [DOI] [PubMed] [Google Scholar]
- Fukuoka H, Kanda Y, Ohta S, Usami S. Mutations in the WFS1 gene are a frequent cause of autosomal dominant nonsyndromic low-frequency hearing loss in Japanese. J Hum Genet. 2007;52:510–515. doi: 10.1007/s10038-007-0144-3. [DOI] [PubMed] [Google Scholar]
- Gomez-Zaera M, Strom TM, Rodriguez B, Estivill X, Meitinger T, Nunes V. Presence of a major WFS1 mutation in Spanish Wolfram syndrome pedigrees. Mol Genet Metab. 2001;72:72–81. doi: 10.1006/mgme.2000.3107. [DOI] [PubMed] [Google Scholar]
- Hansen L, Eiberg H, Barrett T, Bek T, Kjaersgaard P, Tranebjaerg L, Rosenberg T. Mutation analysis of the WFS1 gene in seven Danish Wolfram syndrome families; four new mutations identified. Eur J Hum Genet. 2005;13:1275–1284. doi: 10.1038/sj.ejhg.5201491. [DOI] [PubMed] [Google Scholar]
- Hofmann S, Philbrook C, Gerbitz KD, Bauer MF. Wolfram syndrome: structural and functional analyses of mutant and wild-type wolframin, the WFS1 gene product. Hum Mol Genet. 2003;12:2003–2012. doi: 10.1093/hmg/ddg214. [DOI] [PubMed] [Google Scholar]
- Hogewind BF, Pennings RJ, Hol FA, Kunst HP, Hoefsloot EH, Cruysberg JR, Cremers CW. Autosomal dominant optic neuropathy and sensorineual hearing loss associated with a novel mutation of WFS1. Mol Vis. 2010;16:26–35. [PMC free article] [PubMed] [Google Scholar]
- Ke T, Nie SW, Yang QB, Liu JP, Zhou LN, Ren X, Liu JY, Wang Q, Liu MG. The G401D mutation of OPA1 causes autosomal dominant optic atrophy and hearing loss in a Chinese family. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:481–485. [PubMed] [Google Scholar]
- Larsson NG, Holme E, Kristiansson B, Oldfors A, Tulinius M. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr Res. 1990;28:131–136. doi: 10.1203/00006450-199008000-00011. [DOI] [PubMed] [Google Scholar]
- Li C, Kosmorsky G, Zhang K, Katz BJ, Ge J, Traboulsi EI. Optic atrophy and sensorineural hearing loss in a family caused by an R445H OPA1 mutation. Am J Med Genet A. 2005;138A:208–211. doi: 10.1002/ajmg.a.30794. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Pettitt L, Forman OP, Vaudin M, Barnett KC. Identification of mutations in HSF4 in dogs of three different breeds with hereditary cataracts. Vet Ophthalmol. 2006;9:369–378. doi: 10.1111/j.1463-5224.2006.00496.x. [DOI] [PubMed] [Google Scholar]
- Mets R, Emery S, Lesperance M, Mets M. Congenital Cataracts in Two Siblings with Wolfram Syndrome: a Newly Associated Finding. Ophthalmic Genetics. 2010 doi: 10.3109/13816810.2010.516056. In revision. [DOI] [PubMed] [Google Scholar]
- Pandya A, Arnos KS, Xia XJ, Welch KO, Blanton SH, Friedman TB, Garcia SG, Liu MD, X, Morell R, Nance WE. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet Med. 2003;5:295–303. doi: 10.1097/01.GIM.0000078026.01140.68. [DOI] [PubMed] [Google Scholar]
- Payne M, Yang Z, Katz BJ, Warner JE, Weight CJ, Zhao Y, Pearson ED, Treft RL, Hillman T, Kennedy RJ, Meire FM, Zhang K. Dominant optic atrophy, sensorineural hearing loss, ptosis, and ophthalmoplegia: a syndrome caused by a missense mutation in OPA1. Am J Ophthalmol. 2004;138:749–755. doi: 10.1016/j.ajo.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Puomila A, Huoponen K, Mantyjarvi M, Hamalainen P, Paananen R, Sankila EM, Savontaus ML, Somer M, Nikoskelainen E. Dominant optic atrophy: correlation between clinical and molecular genetic studies. Acta Ophthalmol Scand. 2005;83:337–346. doi: 10.1111/j.1600-0420.2005.00448.x. [DOI] [PubMed] [Google Scholar]
- Samuelson A. Familjärt uppträdande synnervsatrofi och dövhet. Nordisk Medicine (Hygiea) 1940;6:769–772. [Google Scholar]
- Shimizu S, Mori N, Kishi M, Sugata H, Tsuda A, Kubota N. A novel mutation in the OPA1 gene in a Japanese patient with optic atrophy. Am J Ophthalmol. 2003;135:256–257. doi: 10.1016/s0002-9394(02)01929-3. [DOI] [PubMed] [Google Scholar]
- Tessa A, Carbone I, Matteoli MC, Bruno C, Patrono C, Patera IP, De LF, Lorini R, Santorelli FM. Identification of novel WFS1 mutations in Italian children with Wolfram syndrome. Hum Mutat. 2001;17:348–349. doi: 10.1002/humu.32. [DOI] [PubMed] [Google Scholar]
- Tranebjærg L. Wolframin 1-related disease and hearing. In: Kóks S, Vasar E, editors. Emerging link between the emotional brain and endocrine pancreas. 2008. pp. 107–124. [Google Scholar]
- Tranebjaerg L, Barrett T, Rendtorff ND. WFS1-Related Disorders. GeneReviews at GeneTests: Medical Genetics Information Resource. Seattle: University of Washington; 2009. pp. 1997–2008. [database online]. [Google Scholar]
- Valero R, Bannwarth S, Roman S, Paquis-Flucklinger V, Vialettes B. Autosomal dominant transmission of diabetes and congenital hearing impairment secondary to a missense mutation in the WFS1 gene. Diabet Med. 2008;25:657–661. doi: 10.1111/j.1464-5491.2008.02448.x. [DOI] [PubMed] [Google Scholar]
- Young TL, Ives E, Lynch E, Person R, Snook S, MacLaren L, Cater T, Griffin A, Fernandez B, Lee MK, King MC. Non-syndromic progressive hearing loss DFNA38 is caused by heterozygous missense mutation in the Wolfram syndrome gene WFS1. Hum Mol Genet. 2001;10:2509–2514. doi: 10.1093/hmg/10.22.2509. [DOI] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Gorman GS, Lourenco CM, Wright AF, Auer-Grumbach M, Toscano A, Musumeci O, Valentino ML, Caporali L, Lamperti C, Tallaksen CM, Duffey P, Miller J, Whittaker RG, Baker MR, Jackson MJ, Clarke MP, Dhillon B, Czermin B, Stewart JD, Hudson G, Reynier P, Bonneau D, Marques W, Jr, Lenaers G, McFarland R, Taylor RW, Turnbull DM, Votruba M, Zeviani M, Carelli V, Bindoff LA, Horvath R, Amati-Bonneau P, Chinnery PF. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010;133:771–786. doi: 10.1093/brain/awq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatyka M, Ricketts C, da SX, Minton J, Fenton S, Hofmann-Thiel S, Rutter GA, Barrett TG. Sodium-potassium ATPase 1 subunit is a molecular partner of Wolframin, an endoplasmic reticulum protein involved in ER stress. Hum Mol Genet. 2008;17:190–200. doi: 10.1093/hmg/ddm296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.