Abstract
Iron(III)-reducing bacteria have been demonstrated to rapidly catalyze the reduction and immobilization of uranium(VI) from contaminated subsurface sediments. Thus, these organisms may aid in the development of bioremediation strategies for uranium contamination, which is prevalent in acidic subsurface sediments at U.S. government facilities. Iron(III)-reducing enrichment cultures were initiated from pristine and contaminated (high in uranium, nitrate; low pH) subsurface sediments at pH 7 and pH 4 to 5. Enumeration of Fe(III)-reducing bacteria yielded cell counts of up to 240 cells ml−1 for the contaminated and background sediments at both pHs with a range of different carbon sources (glycerol, acetate, lactate, and glucose). In enrichments where nitrate contamination was removed from the sediment by washing, MPN counts of Fe(III)-reducing bacteria increased substantially. Sediments of lower pH typically yielded lower counts of Fe(III)-reducing bacteria in lactate- and acetate-amended enrichments, but higher counts were observed when glucose was used as an electron donor in acidic enrichments. Phylogenetic analysis of 16S rRNA gene sequences extracted from the highest positive MPN dilutions revealed that the predominant members of Fe(III)-reducing consortia from background sediments were closely related to members of the Geobacteraceae family, whereas a recently characterized Fe(III) reducer (Anaeromyxobacter sp.) and organisms not previously shown to reduce Fe(III) (Paenibacillus and Brevibacillus spp.) predominated in the Fe(III)-reducing consortia of contaminated sediments. Analysis of enrichment cultures by terminal restriction fragment length polymorphism (T-RFLP) strongly supported the cloning and sequencing results. Dominant members of the Fe(III)-reducing consortia were observed to be stable over several enrichment culture transfers by T-RFLP in conjunction with measurements of Fe(III) reduction activity and carbon substrate utilization. Enrichment cultures from contaminated sites were also shown to rapidly reduce millimolar amounts of U(VI) in comparison to killed controls. With DNA extracted directly from subsurface sediments, quantitative analysis of 16S rRNA gene sequences with MPN-PCR indicated that Geobacteraceae sequences were more abundant in pristine compared to contaminated environments,whereas Anaeromyxobacter sequences were more abundant in contaminated sediments. Thus, results from a combination of cultivation-based and cultivation-independent approaches indicate that the abundance/community composition of Fe(III)-reducing consortia in subsurface sediments is dependent upon geochemical parameters (pH, nitrate concentration) and that microorganisms capable of producing spores (gram positive) or spore-like bodies (Anaeromyxobacter) were representative of acidic subsurface environments.
As a by-product of nuclear weapons production during the Cold War era, large amounts of toxic and radioactive wastes have contaminated over 7,200 km2 of soils and groundwater surrounding many U.S. Department of Energy research and laboratory sites (9). The most predominant radionuclide contaminant found in these areas is uranium, which is chemically toxic and has half-lives ranging from 247,000 to 4.5 billion years, depending on the isotope (Natural and Accelerated Bioremediation Research [NABIR] website, January 1999, http://www.lbl.gov/NABIR/). Thus, there is increasing concern about the fate of uranium in contaminated areas.
The microbial catalysis of uranium(VI) reduction is a promising strategy for the potential remediation of uranium-contaminated groundwaters (14, 36). Upon reduction, the highly soluble and mobile U(VI) is converted to insoluble U(IV), which precipitates from groundwater. Therefore, stimulating microbial U(VI) reduction could potentially immobilize uranium contamination and prevent its migration through the subsurface.
Dissimilatory Fe(III)-reducing bacteria and sulfate-reducing bacteria are the two major groups of microorganisms capable of U(VI) reduction (36-38).. Bacterial U(VI) reduction may be catalyzed by both direct (enzymatic) and indirect (chemical) mechanisms. Both Fe(III)-reducing bacteria and sulfate-reducing bacteria utilize U(VI) as an electron acceptor, and a subset of these groups have been shown to conserve energy for growth via U(VI) reduction (39, 41, 42, 43, 65, 67). The products of microbial Fe(III) and sulfate reduction, Fe(II) and hydrogen sulfide, can also react abiotically to reduce U(VI) (34, 47). In the terrestrial subsurface, Fe(III)-reducing bacteria are likely to outcompete sulfate-reducing bacteria because Fe(III) is usually a much more abundant electron acceptor than sulfate in subsurface sediments (37, 38). Thus, Fe(III)-reducing bacteria are thought to have a high bioremediation potential in uranium-contaminated subsurface sediments.
Most previous work on U(VI)-reducing bacteria has been conducted with pure cultures in the laboratory. A few studies of U(VI) reducers have been carried out with cultivation-independent techniques in subsurface environments, but these studies have largely been conducted in sediment slurries under controlled laboratory conditions or have been limited to studies of groundwater in the field. Among known Fe(III)-reducing bacteria, members of the Geobacteraceae family within the delta proteobacteria were most often detected in abundance from subsurface environments upon stimulation of concurrent U(VI) reduction and Fe(III) reduction through the addition of acetate as an electron donor (14, 23). Previous geochemical studies of microbial U(VI) reduction in neutrophilic subsurface sediments cocontaminated with nitrate have indicated that no net U(VI) reduction occurs until nitrate is reduced. Once nitrate is depleted, U(VI) and Fe(III) are reduced concurrently (14, 16). As for the sulfate-reducing bacteria, a study in a uranium-contaminated mill tailing site showed a correlation between high uranium concentrations and the occurrence of Desulfotomaculum-related sequences, suggestive of potential in situ uranium biotransformation by sulfate-reducing bacteria (7).
The study of anaerobic bacteria that carry out respiration in acidic environments is in its infancy. Though the impact of pH on uranium reduction has not been studied in detail, low pH is well known to hinder microbial activity through the inactivation of enzymes and the disruption of proton motive force (4, 33). Both sulfate-reducing bacteria and Fe(III)-reducing bacteria have been enriched from acidic sediments (17, 18, 26, 30-32), and a few pure cultures of Fe(III)-reducing bacteria which grow at low pH are available (26, 30). However, to date no sulfate-reducing bacteria that grow below pH 5 have been isolated.
Here we used a combination of cultivation-based and cultivation-independent approaches to characterize Fe(III)-reducing bacterial consortia from acidic subsurface sediments. Through elucidation of metal-reducing bacterial communities and the sediment properties controlling their activity, our observations will aid in the development of bioremediation strategies for uranium-contaminated subsurface sediments.
MATERIALS AND METHODS
Site description and sampling procedures.
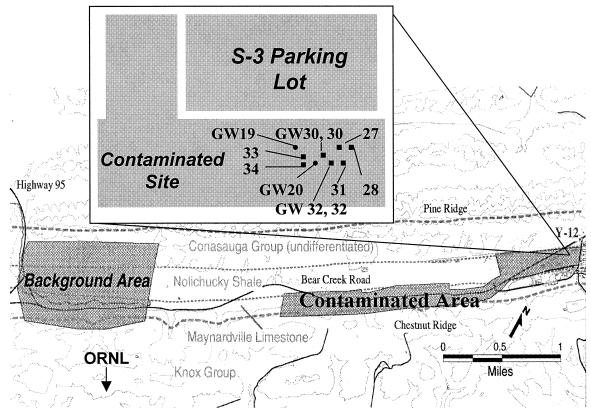
This study focused on contaminated subsurface sediments collected from the Field Research Center designated by the NABIR program of the U.S. Department of Energy. The Field Research Center is located at the Y-12 complex within the Oak Ridge National Laboratory (ORNL) reservation at Oak Ridge, Tennessee. The contaminated plot lies adjacent to a parking lot which caps three former waste ponds (S-3 ponds) containing uranium and nitric acid waste generated during weapons production (coring and well locations given in Fig. 1). Contaminated subsurface sediments were sampled from the saturated zone of shallow residuum overlying Nolichucky shale, where elevated concentrations of uranium and nitrate have been observed (NABIR website, January 1999, http://www.lbl.gov/NABIR/). The background area is a pristine site ∼163 ha in size, located in West Bear Creek Valley, approximately 2 km away from the S-3 ponds.
FIG. 1.
Map of sample sites at the U.S. Department of Energy-NABIR Field Research Center, Oak Ridge, Tennessee. The contaminated plot is 7 by 25 m just south of the S-3 ponds, and contamination extends to a depth of approximately 22.8 m below the surface.
Sediment cores were sampled on 16 February 2001, 31 May 2001, and 30 August 2001 with an Acker Drill Co. (TBD-II) Holegator track drill equipped with polyurethane sleeves lining the corer. Cores (3.75 cm in diameter, 182 cm in length) were immediately transferred to a Coy anaerobic chamber adjacent to the field sites, where they were subsampled under aseptic and strictly anoxic conditions with alcohol-rinsed equipment. Samples were sealed under argon and shipped to Florida State University on blue ice via FedEx priority overnight. Groundwater samples of approximately 1-liter volume were withdrawn from an injection well approximately 6.4 to 6.7 m deep and 3.0 cm in diameter into airtight bottles for shipment as described above (NABIR website, January 1999, http://www.lbl.gov/NABIR/).
Groundwater chemistry.
Samples were tested for pH with a calibrated digital pH meter. Nitrate analysis was conducted after reduction with V(III) to NOx with a chemiluminescence detector (5). A kinetic phosphorescence analyzer, which uses a pulsed nitrogen dye laser in conjunction with a complexing solution, was used to measure hexavalent uranium(6).
Solid-phase geochemistry.
Density, dry/wet ratio, porosity, and percent carbon loss on combustion were measured for each sediment core obtained from the background and contaminated sites. The sediment pH was determined by diluting 2 ml of sediment with 2 ml of deionized water. The 1:1 dilution was shaken for 1 h and centrifuged, and then the pH of the supernatant was measured with a calibrated digital pH meter (46).
Nitrate was extracted from the solid phase with a 1-h extraction with a 1:1 dilution of deionized water followed by centrifugation. The supernatant was then analyzed for nitrate as described above. Poorly crystalline Fe(III) oxide minerals were quantified with a 1-h 0.5 M HCl extraction (28). Crystalline forms of Fe(III) oxide minerals were determined with a 1-h dithionite-citrate-bicarbonate extraction followed by colorimetric determination of total Fe (28).
Bacterial enrichment, enumeration, and screening.
Iron(III)-reducing bacterial populations were enumerated by the three-tube most-probable-number (MPN) assay with serial dilutions of sediment in growth medium. A minimal freshwater medium was prepared and dispensed according to Widdel and Bak (68) into Hungate tubes. Amorphous Fe(III) oxyhydroxide (FeOOH) was prepared as described previously (40). FeOOH was added as the electron acceptor to 50 mM final concentration, and FeCl2 was added as a mild reductant (1). Carbon substrates (lactate, acetate, glycerol, and glucose) were added from anoxic sterile stock solutions to a final concentration of 10 mM. MPN tubes were purged with either a 90%-10% mixture (pH 7 tubes) or 80%-20% mixture (pH 4 to 5 tubes) of N2-CO2 gas before being sealed with a butyl rubber stopper.
Selected sediment samples were washed three times with a 10-fold dilution of distilled water to eliminate excess nitrate. Samples were centrifuged between washing steps to separate sediment and to retain cells from wash water. Medium prepared at pH 7 was buffered with carbonate, and initial medium prepared at pH 5 was buffered with 20 mM acetate. Later, medium for pH 4 to 5 experiments contained no buffer and 10 mM glucose was added as the carbon substrate. Tubes were incubated at 30°C in the dark for approximately 10 months. Iron(III) reduction activity was scored by visual screening (accumulation of magnetite) and colorimetric quantification of accumulated Fe(II) in HCl extracts as described above. Organic acid utilization was quantified by ion exclusion chromatography (Dionex 600X system). The MPN index was determined to 95% certainty from statistical tables published by the American Public Health Association (2). Direct cell counts were carried out on sediments with acridine orange and epifluorescence microscopy as previously described (22) and modified for sediments (52).
To screen enrichment cultures for U(VI) reduction, inocula were transferred to fresh medium (prepared as described above) with 50 mM ferric citrate as the electron acceptor and lactate as the electron donor (10 mM final concentration). After 1 week, the cells were centrifuged and washed three times in an anaerobic 30 mM bicarbonate solution (20). Cells were then transferred to anoxic minimal medium containing 1 to 10 mM uranium as the electron acceptor and a 10 mM concentration of the electron donor. Heat-killed controls contained cells that had been autoclaved prior to introduction into the media. The cultures were incubated in the dark over a period of 1 to 2 weeks. At specific time points, samples were filtered, diluted anaerobically 1:100 with deionized water, and measured for the decrease of uranium(VI) with the kinetic phosphorescence analyzer (20). Samples were not acidified prior to U(VI) measurements.
DNA extraction and 16S rRNA gene analysis in enrichment cultures.
Microbial community DNA for use in cloning and terminal restriction fragment length polymorphism (T-RFLP) assays was extracted from enrichment cultures with the Ultra Clean Soil DNA kit (Mo Bio Laboratories, Solana Beach, Calif.). A variety of MPN dilutions (101, 102, and 103) and transfers (initial, second, fifth, and seventh) were used for both cloning and T-RFLP analyses to ensure data validity and consistency. The contents of each MPN tube were centrifuged, and the resulting pellet was used in the extraction; 250 μl of sterile water was extracted in parallel as a control. Immediately after extraction, aliquots (1 μl) of DNA were added to PCRs for both cloning and T-RFLP. Bacterial 16S ribosomal RNA genes were amplified from community DNA with primers 8F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1392R (5′-ACG GGC GGT GTG TRC-3′) as previously described (11).
The PCR products were cloned with a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) and screened by digestion with restriction enzyme HaeIII (New England Biolabs, Beverly, Mass.) as previously described (13). Cloned inserts with unique restriction patterns were amplified with M13 primers from whole cells, purified for sequencing with QIAquick PCR purification columns (Qiagen, Valencia, Calif.), and sequenced with primers G and H (35). Sequences were aligned against close relatives from the Ribosomal Database Project with the ARB software package (61) and rRNA secondary structure diagrams. Dendrograms were constructed with ARB and PAUP* 4.0s (SinauerAssociates, Sunderland, Mass.) with only unambiguously aligned nucleotides.
For T-RFLP analysis, the bacterial 16S ribosomal RNA genes were amplified from community DNA as described above except that the 8F primer was labeled with the phosphoramidite dye 5-hexachlorofluoescein. Amplicons were purified with the Qiagen QIAquick PCR purification columns (Qiagen) and eluted in 50 μl of water. Purified PCR product (100 ng) was digested with 10 U of HaeIII and MspI (New England Biolabs) at 37°C for 4 h, followed by a deactivation step of 80°C for 15 min. Hi-Di formamide (10 μl) and Genescan-500 size standard ROX (1 μl) (Applied Biosystems, Warrington, United Kingdom) were added to each sample. The lengths of the terminal restriction fragments were determined with an Applied Biosystems 3100 genetic analyzer and Genotyper 3.6 software.
DNA extraction, primer design, and three-tube MPN-PCR in sediments.
Microbial community DNA for use in MPN-PCR was extracted directly from the sediment with the RNA-DNA simultaneous extraction protocol described by Hurt et al. (24). The DNA was purified twice with a Wizard column (Promega, Madison, Wis.) and resuspended in 50 μl of sterile water.
With the probe design function in the ARB software package (61), a primer set was designed to amplify Anaeromyxobacter-type sequences (60F, 5′-CGA GAA AGC CCG CAA GGG T-3′ , and 461R, 5′-ATT CGT CCC TCG CGA CAG T-3′ ). The primers were selected to optimize base pairing with target sequences. Primers used to amplify sequences within the Geobacteraceae family (494F and 825R) have already been developed (23). Optimum temperature and cycling parameters were determined to be an initial denaturation step of 94°C for 10 min, followed by 35 cycles of 95°C (30 s), 56.5°C (30 s), and 72°C (45 s), with a final extension step of 72°C for 10 min. To test primer specificity, each primer set was compared to the sequences available in the RDP (45) and GenBank databases with the Probe_Check (RDP) and Blast (2) algorithms.
The relative abundances of 16S rRNA gene sequences closely related to the Geobacter- and Anaeromyxobacter-type sequences were determined from DNA directly extracted from Field Research Center sediments with an MPN-PCR technique as previously described (64). Serial 10-fold dilutions of extracted DNA were made in sterile water, and 1-μl aliquots were used as the template in the PCR. The PCR conditions were as stated above. PCR products were analyzed by gel electrophoresis in 1% agarose gels, stained in an ethidium bromide bath, and visualized by UV transillumination. The highest dilution that yielded product was noted, and a standard three-tube MPN chart was consulted in order to determine the number of 16S rRNA gene copies per gram of sediment extracted.
RESULTS
Geochemical variables.
Groundwater chemistry showed substantial differences between the contaminated and background sites sampled, with the contaminated site showing lower pHs and higher nitrate and uranium concentrations (Table 1). The contrast between sites was more pronounced when comparing sediment parameters (pH and extractable nitrate concentrations). The pHs of background cores were on average 1.8 units higher than the pHs of the contaminated cores (Table 2). The extractable nitrate concentration was on average two to four orders of magnitude higher in the contaminated cores than in the background cores (Table 2). Therefore, the contaminated area of the Field Research Center is an “extreme” geochemical environment relative to the pristine background area.
TABLE 1.
Groundwater chemistry of contaminated and background subsurface sitesa
| Sample | Uranium(VI) (ug/liter) | Nitrate (mM) | pH |
|---|---|---|---|
| GW300 | — | 0.00255 | 7.6 |
| GW20 | 122 | NA | 5.2 |
| GW19 | 155 | 0.152 | 7.0 |
| GW30 | — | 99.7 | 3.8 |
| GW32 | 1.00 × 103 | 11.8 | 5.4 |
Background groundwater is sample number GW300, and contaminated groundwater samples are sample numbers GW20, GW19, GW30, and GW32. Data are averages of duplicate samples. —, concentration below the detection limit. NA, no data.
TABLE 2.
Sediment characteristics of contaminated and background subsurface sitesa
| Sample | Dry/wet ratio | Porosity | Density (g/cm3) | LOIb (%) | pH | Nitrate (μmol/cm3) | Fe-HCl extract (μmol/cm3) | Fe-dithionite extract (μmol/cm3) |
|---|---|---|---|---|---|---|---|---|
| 302-02 | 0.75 | 0.36 | 1.4 | 5.1 | 5.4 | 1.02-1.03 × 10−2 | 4.36 | 503.5 |
| 302-05 | 0.73 | 0.49 | 1.8 | 4.5 | 5.7 | 1.84 × 10−5-2.40 × 10−3 | 26.1 | 302.3 |
| 27 | 0.89 | 0.23 | 1.9 | 4.6 | 3.6 | 9.8-54.9 | 13.5 | 260.7 |
| 28 | 0.84 | 0.29 | 1.9 | 4.9 | 3.4 | 137-180 | 11.3 | 329.6 |
| 30 | 0.86 | 0.29 | 2 | 4.2 | 4.2 | 22.7-31.5 | 15.2 | 414.4 |
| 31 | 0.87 | 0.26 | 1.9 | 4.7 | 3.8 | 0.1-91.7 | 10.8 | 376.4 |
| 32 | 0.85 | 0.32 | 2.1 | 4.5 | 4.4 | 0.02-17.2 | 18.9 | 407.9 |
| 33 | 0.84 | 0.29 | 1.9 | 4.5 | 3.6 | 13.6-295 | 14.5 | 467.7 |
| 34 | 0.86 | 0.26 | 2 | 4.3 | 3.8 | 3.3-70.5 | 16.3 | 325.0 |
Background sediments are sample numbers 302-02 and 302-05 and were collected 61 to 344 cm below the surface, and contaminated sediments are sample numbers 27, 28, 30, 21, 32, 33, and 34 and were collected 308 to 576 cm below the surface. Values are averages of duplicate samples from two to four sediment cores or the range of values for duplicate samples from two to four sediment cores.
LOI, loss on ignition.
Between the two sample sites, similar ranges in porosity (0.23 to 0.49), wet density (1.4 to 2.1 g cm−3), and organic carbon content (4.2 to 5.1%) were observed (Table 2). Solid-phase Fe concentrations extractable in HCl ranged from 4 to 26 μmol cm−3, with the majority of HCl-Fe being oxidized (Table 2). The reduced iron within these HCl extractions ranged from negligible to 5.7 μmol cm−3 (data not shown). Dithionite-extractable Fe concentrations were an order of magnitude higher than HCl-Fe, ranging from 260.7 to 503.5 μmol cm−3 (Table 2). At similar depths below the surface, extractable Fe concentrations showed very similar ranges between the contaminated and background sites.
Iron(III)-reducing bacteria enumeration experiments.
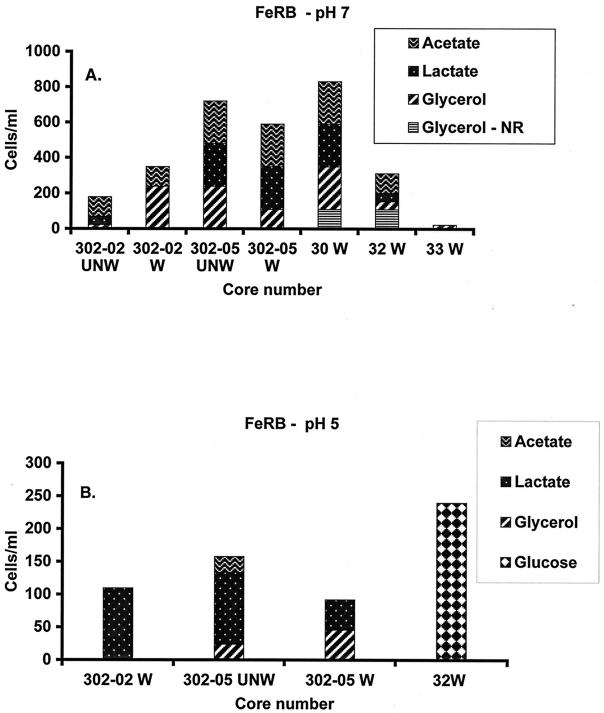
The MPN data are summarized in Fig. 2 for all cores and carbon sources tested where cell counts greater than or equal to 20 cells ml−1 were observed. Background MPN counts were of the same magnitude as contaminated MPN counts. Iron(III) was completely reduced, as indicated by quantification of the accumulation of 25 to 35 mM Fe(II) in all of the positive Fe(III)-reducing enrichment cultures, including primary enrichments and successive transfers. In neutrophilic enrichments, the utilization of organic acids (acetate or lactate) coupled to Fe(III) reduction was also confirmed. By comparison, little or no Fe(III) reduction occurred in killed controls or control cultures to which no carbon substrate was added.
FIG. 2.
MPN counts of Fe(III)-reducing bacteria (FeRB) at pH 7 (A) and pH 4 to 5 (B) from the contaminated and background sites. W, washed sediment; UNW, unwashed sediment. Background cores are labeled 302-02 and 302-05, and contaminated cores are labeled 30, 32, and 33. Only counts greater than or equal to 20 cells/ml are included in this graph.
At the background site, growth was detected in 87% of Fe(III)-reducing enrichments at pH 7 and 53% of Fe(III)-reducing enrichments at pH 4 to 5. In background enrichments at pH 7, all cores tested showed some growth, and counts were typically higher than in the pH 5 enrichments (Fig. 2). In pH 4 to 5 enrichments, the highest counts were observed from enrichments utilizing lactate as the carbon source (Fig. 2B). In these Fe(III)-reducing enrichments with lactate as the sole carbon source, acetate was produced from lactate at close to a 1:1 ratio. No other organic acids were produced during lactate utilization. No substantial difference was observed for the growth or activity of background enrichments between washed and unwashed sediment treatments.
In contrast, a general decrease in viability was detected as a lack of activity in enrichments from acidic, contaminated sediments, and nitrate removal had a large influence on the abundance of iron(III)-reducing bacteria cultivated from contaminated sediments. Growth was detected in only 37% of all pH 7 enrichments from acidic, contaminated sediment. No growth was detected at pH 7 from cores 28 and 31 with glycerol, lactate, or acetate as a carbon substrate. The highest counts from neutrophilic enrichments were obtained from contaminated sediment cores 30 and 32. When nitrate was removed from contaminated sediments by washing, substantially higher counts were observed in comparison to those from unwashed sediment (only enrichments from washed sediments are depicted in Fig. 2A because all unwashed sediment enrichments yielded counts of <20 cells ml−1).
As determined for cultures from background sediments, the utilization of organic acids in neutrophilic Fe(III)-reducing enrichments from contaminated sediments paralleled the accumulation of Fe(II). Also, when lactate was the carbon source, acetate was produced in a 1:1 ratio with lactate utilization, and no other organic acids were detected. All pH 4 to 5 enrichments from contaminated cores with acetate as a buffer and glycerol, lactate, or acetate as a carbon source showed negligible growth. Without the acetate buffer, pH 4 to 5 contaminated enrichments amended with glucose (core 32) yielded growth in 44% of the unwashed enrichments and 100% of the washed enrichments.
Direct counts.
Direct counting of both the background and the contaminated sediments yielded similar numbers of bacteria from both sites, ranging from 1.58 × 107 to 8.33 × 107 cells/ml.
Screening for U(VI) reduction.
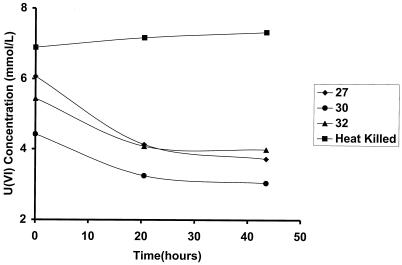
Three neutral Fe(III)-reducing enrichment cultures from the contaminated Field Research Center subsurface were tested for uranium(VI) reduction. Enrichments from contaminated sites 27, 30, and 32 showed substantial reduction of U(VI) in comparison to heat-killed controls (Fig. 3). Up to 50% of 4 to 6 mM uranium(VI) was reduced in less than 48 h. Addition of approximately 6 mM uranyl acetate changed the growth medium in the test tubes to a yellowish color. In cultures containing live cells, this yellow color turned clear over the course of the experiment, and the color change was accompanied by the formation of a brown-black precipitate suggestive of the removal of soluble U(VI) from solution and transformation into insoluble U(IV). The cultures containing the heat-killed cells did not change color or form additional precipitate, suggesting no reduction of U(VI) to U(IV).
FIG. 3.
Uranium(VI) reduction by Fe(III)-reducing enrichment cultures from contaminated Field Research Center subsurface sediments.
Microbial community composition of Fe(III)-reducing enrichments.
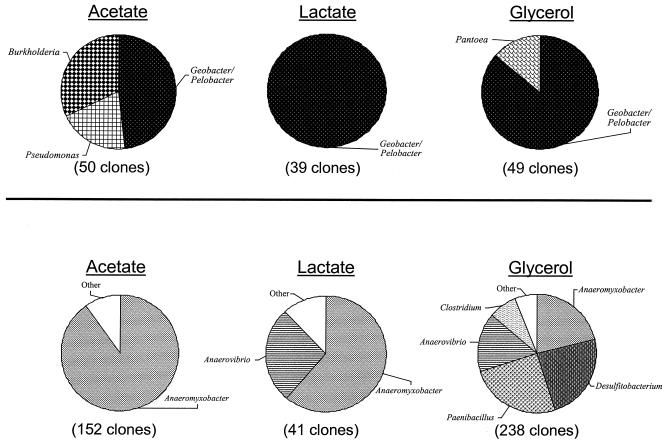
There were marked differences in the phylotypes observed in Fe(III)-reducing bacterial enrichments from background compared to contaminated sediments (Fig. 4). From the background sediment enrichments cultivated at neutral pH, 76% of all the 16S rRNA gene sequences obtained from three enrichments were closely related to the family Geobacteraceae within the delta proteobacteria. Three groups of clone sequences were found in the Geobacteraceae family, with 15% sequence variability between them (Fig. 5). All of the 16S rRNA gene sequences obtained from lactate-amended enrichments and 48% of the 16S rRNA gene sequences from acetate-amended enrichments fell within the Geobacteraceae family. The remaining sequences from acetate-amended enrichments were related to the common soil proteobacteria Pseudomonas rhodesiae and Burkholderia graminis. In the enrichments with glycerol as a carbon source, 86% of the cloned sequences fell within the Geobacteraceae family, while the remaining sequences were closely related to Pantoea agglomerans, a facultative anaerobe capable of dissimilatory metal reduction (19).
FIG. 4.
Phylogenetic affiliations of 16S rRNA cloned genes obtained from pH 7 enrichment cultures of background (top) and contaminated (bottom) sediments.
FIG. 5.
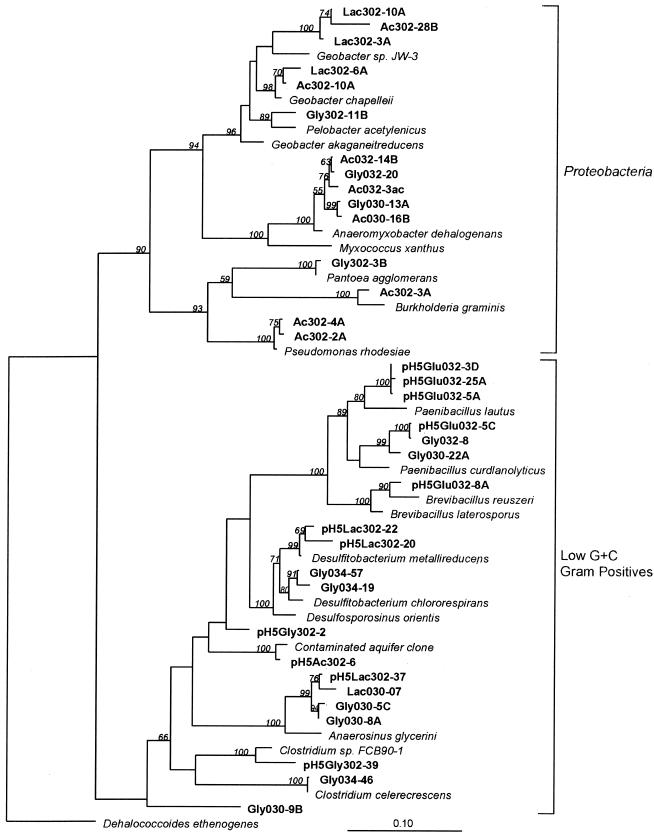
Phylogenetic tree of 16S rRNA genes cloned from Fe(III)-reducing enrichments from background and contaminated subsurface sediments cultivated at pH 7 and pH 4 to 5. The scale bar equals a 10% difference in nucleotide sequence. The cloned genes are named according to the pH, carbon source, and sediment source. Names containing the numbers 302 are from background sediment, while those with numbers 030, 032, and 034 are from contaminated sediment.
In contrast to the neutral-pH enrichments from background sediments, no Geobacteraceae-related sequences were detected in contaminated neutral pH sediment enrichments. Forty-four percent of all 16S rRNA gene sequences obtained from seven enrichments were greater than 96% similar to Anaeromyxobacter dehalogenans (Fig. 4 and 5), a dissimilatory Fe(III)-reducing bacterium recently isolated from surficial aquatic sediments (21, 55) and rice paddy soils (66). Ninety percent of the sequences obtained from neutral acetate-amended enrichments were related to A. dehalogenans, while only 61% were related to A. dehalogenans in lactate enrichments. The remaining 16S rRNA gene sequences obtained from the pH 7 lactate enrichments were related to the gram-positive genus Anaerosinus, an obligate anaerobe previously isolated from freshwater sediments (56, 60). Enrichments at pH 7 with glycerol as a carbon source were the most diverse (Fig. 4). In addition to 16S rRNA sequences closely related to A. dehalogenans, there were also sequences related to several gram-positive organisms such as Desulfitobacterium frappieri and Desulfitobacterium chlororespirans, Paenibacillus curdlanolyticus, and Clostridium celerecrescens (Fig. 4, 5).
All 16S rRNA sequences retrieved from background and contaminated low-pH enrichments were most closely related to members of the gram-positive bacteria. In the contaminated sediment enrichments amended with glucose, only two types of organisms were detected. The cloned 16S rRNA gene sequences detected were closely related to the gram-positive genera Paenibacillus and Brevibacillus (Fig. 5). The sequences showed less than 13% difference in sequence from P. lautus and P. curdlanolyticus and less than 6% difference in sequence from Brevibacillus reuszeri.
In the pH 4 to 5 enrichments from background sediments, the gram-positive organisms that were detected differed depending on the carbon substrate added to the enrichments. In the acetate-amended enrichments, 76% of the 16S rRNA gene sequences obtained were very closely related (1.4% different) to a previously isolated sequence obtained from hydrocarbon- and chlorinated solvent-contaminated aquifers (12). In the lactate-amended enrichments, 78% of the sequences exhibited a 97% similarity to Desulfitobacterium metallireducens and 72% of the glycerol-amended background enrichments showed 93% sequence similarity to Desulfosporosinus orientis (Fig. 4 and 5).
Although sulfate concentrations from groundwater surrounding the tested sediment were in the range of 0.6 to 71.8 mg liter−1 (background = 6.21 mg liter−1), it is unlikely that adequate sulfate for electron accepting capability was transferred through the dilutions and transfers tested (NABIR website, January 1999, http://www.lbl.gov/NABIR/). Furthermore, no sulfide was detected in any of the enrichment cultures.
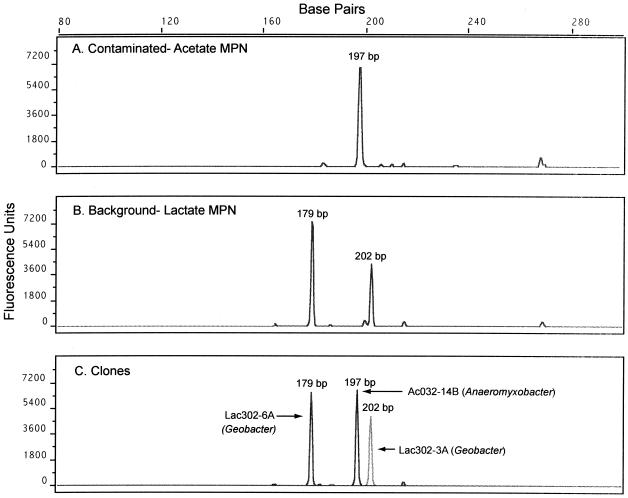
T-RFLP patterns based on amplified 16S rRNA genes were used to estimate diversity and to compare culturable iron(III)-reducing bacteria within the Fe(III)-reducing enrichment cultures, including successive transfers from background and contaminated sediment. The T-RFLP results agreed well with the cloning results described above and suggest very little diversity in the contaminated enrichments with lactate or acetate as a carbon source (Fig. 6). All identifiable terminal restriction fragments from contaminated enrichments amended with lactate or acetate matched the terminal restriction fragments of sequences closely related to the genus Anaeromyxobacter. In contrast, only Geobacter-type 16S rRNA gene sequences were detected in background enrichments with lactate as the carbon source (Fig. 6).
FIG. 6.
Electropherograms of the 5′-terminal restriction fragments of HaeIII-digested 16S rRNA genes amplified from contaminated and background Fe(III)-reducing MPN tubes (A and B) and cloned 16S rRNA genes (C).
More diversity was detected in glycerol-amended contaminated enrichments, including terminal restriction fragments matching Anaeromyxobacter-, Paenibacillus-, and Anaerovibrio-type sequences. Glycerol-amended enrichments from the background sediment were also more diverse, with terminal restriction fragments matching Pantoea- and Pelobacter-type sequences. Glucose-amended enrichments cultured at low pH revealed terminal restriction fragments matching Paenibacillus- and Brevibacillus-type 16S rRNA sequences, corresponding exactly to the previously described cloning and sequencing results. Screening of successive transfer cultures from original Fe(III)-reducing enrichment cultures revealed nearly identical T-RFLP patterns.
Quantification of Fe(III) reducers with a cultivation-independent approach.
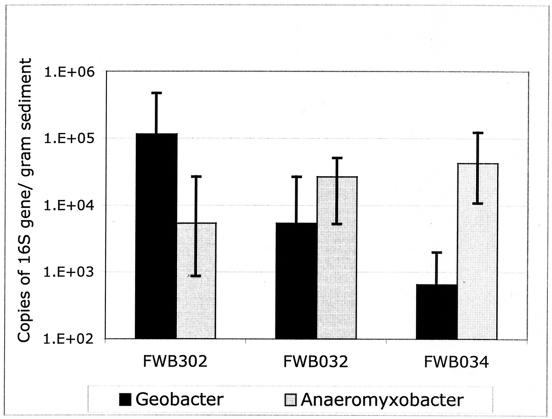
To support the cultivation-dependent results, a cultivation-independent quantitative MPN-PCR technique was employed to amplify the gene sequences of known Fe(III)-reducing bacteria from contaminated and background Field Research Center sediments. DNA was extracted directly from the Field Research Center sediments, and primer sets were developed to detect organisms that predominated in Fe(III)-reducing enrichments. Results from the quantitative MPN-PCR method agreed well with cloning and sequencing results from Fe(III)-reducing enrichment cultures. Geobacteraceae 16S rRNA gene sequences were one to two orders of magnitude more abundant in pristine background sediment (FWB302) compared to contaminated sediments (FWB032 and FWB034; Fig. 7). In contrast, Anaeromyxobacter 16S rRNA gene sequences were more abundant in contaminated sediments (Fig. 7).
FIG. 7.
Quantification of Anaeromyxobacter and Geobacter 16S gene sequences extracted from sediments at contaminated (FWB032, FWB034) and background (FWB302) sites by MPN-PCR.
DISCUSSION
Dissimilatory Fe(III)-reducing bacteria are capable of rapid reduction and immobilization of U(VI) and therefore are considered target organisms for bioremediation strategies directed toward U(VI) contamination in the subsurface. In this study, we observed that sediment geochemical parameters impact the growth and community composition of Fe(III)-reducing consortia from contaminated subsurface sediments. In acidic subsurface sediments of the U.S. Department of Energy's Field Research Center, acid tolerance and competition with nitrate as an electron acceptor are suggested to be important variables controlling the metabolism of Fe(III)-reducing bacteria. The majority of organisms cultivated from contaminated Field Research Center sediments were closely affiliated with previously cultured isolates that produce spores or sporelike bodies, suggesting an adaptation to extreme environmental conditions.
Geochemical analysis showed that the Field Research Center subsurface was an extreme environment which differed from most previously studied subsurface systems due to low pH in conjunction with high levels of nitrate and U(VI). The correlation between high nitrate and low pH was most likely due to nitric acid leaching from the S-3 ponds into the surrounding areas, as nitric acid was disposed into ponds after the cleaning of U-contaminated machinery at Y-12. The heterogeneity of the sediment within the contaminated site was reflected in the wide range of uranium, nitrate, and iron concentrations between and within coring locations. Concentrations of solid-phase Fe from the dithionite extractions were two orders of magnitude higher than that of Fe from HCl extractions (Table 2). This indicates that while poorly crystalline Fe(III) minerals were present, crystalline Fe(III) minerals constituted the dominant electron acceptor available to iron(III)-reducing bacteria. Crystalline Fe(III) minerals, such as goethite and Fe(III)-containing aluminosilicates, have been shown to be effective electron acceptors for the growth of iron(III)-reducing bacteria in laboratory studies (27, 29, 53), although their importance in situ has not been quantified. The Fe(III) mineral content also did not vary substantially across the large geochemical gradients observed between background and contaminated subsurface sediments.
Impacts of geochemical parameters on the growth and activity of iron(III)-reducing bacteria.
Nitrate concentrations in the contaminated sediments (Table 2) likely favored a preferential utilization of nitrate as an electron acceptor by some metal-reducing bacteria (10, 15). Thus, it was not surprising that nitrate appeared to inhibit growth, as determined by potential Fe(III) reduction activity in MPN serial dilutions. Nitrate removal by sediment washing alleviated this inhibition in contaminated enrichments yet had little effect in background enrichments where nitrate concentrations were very low initially (Fig. 2). In addition to nitrate, the washing step likely removed other toxic metals in solution, thus preventing effects such as pathway blockage, substitution of metals for other functional units, and disturbance of membrane integrity or enzyme function (50, 63). Therefore, the numbers of iron(III)-reducing bacteria detected in washed sediment enrichments from the contaminated site could be overestimations of actual in situ potential. Bacterial counts from over 2,000 iron(III)-reducing bacteria enrichment cultures were comparable to the low-end range of iron(III)-reducing bacteria counts obtained from other subsurface Department of Energy sites in Gunnison, Colo. (2,300 cells per ml of sediment) and at Shiprock, N.Mex. (430 cells per ml of sediment) (J. E. Kostka et al., unpublished results).
Low pH inhibited the growth of iron(III)-reducing bacteria, and a synergy was observed between culture pH and carbon source. Iron(III) reducers were most abundant in organic acid-amended enrichments when cultured at neutral pH (Fig. 2). In contrast, when cultured at in situ pH, the number of positive enrichments was largely diminished, and growth was only observed with glucose as the carbon substrate in cultures from acidic sediments (Fig. 2). A likely explanation for this is that at low pH, most short-chain organic acids such as acetate are present as undissociated acids which can pass through the cell membrane, dissociate, and uncouple the proton motive force (4, 25, 30, 32, 44, 54). Thus, iron(III)-reducing bacteria might not be able to utilize electron donors such as acetate at low pH in the Field Research Center subsurface. Recent investigations conducted in parallel at the Field Research Center site corroborate our observations. For example, low counts of culturable, aerobic heterotrophs have been observed in acidic compared to background Field Research Center sediments (D. L. Balkwill and T. Marsh, unpublished results).
Iron(III)-reducing bacteria that are capable of coupling the complete oxidation of glucose to the reduction of Fe(III) have been isolated from sediments exposed to acid mine drainage (30). Fe(III) reduction was also determined to be the predominant terminal-electron-accepting process in lake sediments exposed to acid mine drainage, and organic acids (lactate, acetate) were not used as electron donors under acidic (pH 3 to 4) conditions (51). Therefore, our results suggest that the contaminated subsurface sediments that we studied may be analogous to those exposed to acid mine drainage. In such acidic environments, organic acids such as acetate and lactate may be poor carbon substrates, whereas sugars (such as glucose) or alcohols may be preferentially used by anaerobic bacteria.
Impact of geochemical parameters on the community composition of Fe(III) reducers.
Few past studies have enumerated or characterized Fe(III)-reducing microbial populations from subsurface sediments (15, 23, 37, 38), and none of these has included acidic environments, which are commonly associated with uranium contamination. In this study, geochemical parameters appeared to control the community composition as well as the growth of Fe(III)-reducing microorganisms. Vast differences were observed in the culturable Fe(III)-reducing communities enriched from the contaminated site compared to background subsurface sediments, and the pH of the culture medium was a dominant factor in these differences. Given the fact that only a small fraction of microorganisms can be routinely cultivated from natural environments, we used cultivation-independent, quantitative PCR to corroborate our observations regarding known Fe(III) reducers.
The majority of culturable Fe(III) reducers detected in neutrophilic enrichments from background sediments were closely related to the members of the Geobacteraceae family (Fig. 4 to 6). In agreement with the cultivation results, quantitative analysis of 16S rRNA gene sequences with MPN-PCR indicated that Geobacteraceae sequences were one to two orders of magnitude more abundant in pristine background compared to contaminated Field Research Center sediments (Fig. 7). Members of the Geobacteraceae family are well known Fe(III)-reducing organisms and have been isolated from many neutrophilic sedimentary environments, including the subsurface (37). However, to our knowledge, no cultured member of the Geobacteraceae family has been shown to reduce Fe(III) under acidic conditions. Other sequences detected in background enrichments were related to the genera Burkholderia, Pseudomonas, and Pantoea, which are all considered ubiquitous soil bacteria (19, 62).
In the low-pH enrichments from the background sediment, no cloned 16S rRNA gene sequences related to Geobacteraceae were detected. A different consortium of organisms was detected, all of which were gram-positive. Lactate-amended enrichments yielded 16S rRNA gene sequences closely related to Desulfitobacterium metallireducens, a known Fe(III)-respiring microorganism isolated from neutrophilic uranium-contaminated aquifer sediments. D. metallireducens is capable of the incomplete oxidation of lactate to acetate and reduction of U(VI), Mn(IV), Co(III), and chlorinated compounds (15). Similar to the metabolism of D. metallireducens, the incomplete oxidation of lactate to acetate was demonstrated in our lactate-amended, Fe(III)-reducing enrichments. However, D. metallireducens has thus far been shown to reduce only soluble Fe(III) forms (15), whereas our enrichment cultures reduced solid-phase Fe(III). Metal reducing isolates of Desulfitobacterium species have also been shown to be capable of spore formation (49).
A combination of cultivation-based and cultivation-independent results also pointed to very different Fe(III)-reducing microbial communities in the contaminated Field Research Center subsurface. None of the culturable organisms detected in contaminated sediment enrichments were closely related to the most commonly cultured Fe(III)-reducing bacteria, such as Shewanella or Geobacter, suggesting that low pH and high nitrate concentrations may prevent their survival. All cloned 16S rRNA gene sequences retrieved from contaminated sediment enrichments showed high sequence similarity to gram-positive genera or to a single cultivated isolate from the gram-negative Proteobacteria, Anaeromyxobacter dehalogenans (Fig. 4 and 5). Sequences similar to A. dehalogenans were only detected under neutrophilic culture conditions. However in community DNA extracted directly from sediments, quantitative analysis of 16S rRNA gene sequences with MPN-PCR showed that Anaeromyxobacter-type sequences were more abundant in acidic, contaminated sites compared to pristine sites of the Field Research Center subsurface (Fig. 7).
Anaeromyxobacter has recently been characterized as a facultative anaerobe capable of using Fe(III), nitrate, fumarate, and chlorophenolic compounds as terminal electron acceptors for growth on acetate (55, 66). Though A. dehalogenans is a member of the delta proteobacteria, it has a very different phylogeny from the Geobacteraceae and has been grouped in a new order of the Myxococcales. In pure-culture studies of A. dehalogenans, optimal growth was shown to occur at a neutral pH with low electron donor concentrations (<1 mM), and the organism was observed to form refractile sporelike bodies (55). To our knowledge, this the first time that Anaeromyxobacter has been detected in subsurface sediments, and no previous studies have quantified Anaeromyxobacter sequences with cultivation-independent methods. Since the recent isolation and description of this new Fe(III) reducer, all studies have been conducted with pure cultures isolated from surficial soils and sediments (21, 55, 66).
All other organisms detected in Fe(III)-reducing enrichments from the contaminated sediments were closely related to gram-positive genera. The 16S rRNA gene sequences obtained from the contaminated sediment enrichments cultured at low pH were all closely related to Paenibacillus and Brevibacillus. The genus Paenibacillus, containing nitrate-reducing organisms previously isolated from sediment, can utilize acetate as a carbon source (48). Brevibacillus has been found to utilize glucose as a carbon source, degrade poly(β-hydroxyalkanoate), and be incapable of nitrate reduction (58, 62). Both organisms also form spores (8, 57, 58). To our knowledge, no published studies on dissimilatory Fe(III) reduction by Paenibacillus or Brevibacillus which ferment glucose are available. These organisms could be fermenting rather than respiring glucose, thereby shunting electrons to Fe(III). Although results from our enrichment culture and quantitative PCR approaches provide strong evidence for identifying microbial community members catalyzing metal reduction in the Field Research Center subsurface, confirmation of our conclusions will require isolation of such organisms and further characterization of their capacity for dissimilatory Fe(III) reduction.
The growth and phylogenetic affiliations of enriched organisms, along with quantitative PCR results, suggest that Fe(III) reducers are adapted to the low pH as well as to available growth substrates (electron donors and acceptors) in the Field Research Center subsurface. Only low G+C gram-positive sequences were obtained from low-pH enrichments, suggesting that gram-positive organisms are better adapted to moderately acid conditions and thus are more important organisms at low pH. The predominant sequences detected from contaminated acidic sediments (gram-positives, Anaeromyxobacter) were most closely related to organisms that are known to produce spores or spore-like structures, further suggestive of adaptation to extreme conditions in the acidic subsurface.
The majority of Fe(III) reducers detected in contaminated Field Research Center sediments were closely affiliated with cultured organisms capable of coupling the reduction of nitrate or halogenated compounds to growth (Geobacter, Anaeromyxobacter, Desulfitobacterium) (15, 21, 37, 48, 49, 55). This provides further evidence in support of the adaptation of metal-reducers to the ecological niche of contaminated Field Research Center sediments, which contain high concentrations of nitrate (Table 2) and halogenated organic compounds (see Field Research Center website http://public.ornl.gov/nabirfrc/dataindex.cfm).
To date, members of the Shewanellae and Geobacteraceae families within the proteobacteria have been used as model metal-reducing organisms for the development of bioremediation strategies for radionuclide and metal contaminants. However, radionuclide and heavy metal contamination often occurs in acidic sedimentary environments where these organisms do not appear to thrive. Thus, we suggest that other model metal-reducing organisms, such as those adapted to environmental extremes, would be more appropriate for the development of these strategies in acidic sediments. Furthermore, because recent field experiments have shown that U(VI) reduction is stimulated upon pH neutralization in the acidic subsurface (J. Istok, 2002, Field Research Center workshop proceedings, http://public.ornl.gov/nabirfrc/workshop2002.cfm), the ability of the neutrophilically cultured organisms from acidic sediment to reduce large amounts of U(VI) to U(IV) (Fig. 3) could be of great importance to the development of future U(VI) bioremediation strategies.
Acknowledgments
This research was funded by the Natural and Accelerated Bioremediation Research (NABIR) program, Biological and Environmental Research (BER), U.S. Department of Energy (grant DE-FG02-00ER62986).
We thank Harold J. Adams, Jr., Dava Dalton, Hayley Skelton, and Afonso Souza for technical assistance. We also thank David Watson, Jack Istok, Lee Krumholz, Susan Pfiffner, and Barry Kinsall for sediment and groundwater sampling and shipment.
REFERENCES
- 1.Achtnich, C., A. Schuhmann, T. Wind, and R. Conrad. 1995. Role of interspecies H2 transfer to sulfate and ferric iron-reducing bacteria in acetate consumption in anoxic paddy soil. FEMS Microbiol. Ecol. 16:61-70. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1969. Standard methods for the examination of water and wastewater, including bottom sediments and sludge. American Public Health Association, Washington, D.C.
- 4.Baronofsky, J. J., W. J. A. Schreurs, and E. R. Kashket. 1984. Uncoupling by acetic acid limits growth of and acetogenesis by Clostridium thermoaceticum. Appl. Environ. Microbiol. 48:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braman, R. S., and S. A. Hendrix. 1989. Nanogram nitrate and nitrite determination in environmental and biological materials by vanadium(III) reduction with chemiluminescence detection. Anal. Chem. 61:2715-2718. [DOI] [PubMed] [Google Scholar]
- 6.Brina, R., and A. G. Miller. 1992. Direct detection of trace levels of uranium by laser-induced kinetic phosphorimetry Anal. Chem. 64:1413-1418. [Google Scholar]
- 7.Chang, Y., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. A. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claus, D., and R. C. W. Berkeley. 1986. Genus Bacillus Cohn 1872, p. 1105-1140. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams and Wilkins Co., Baltimore, Md.
- 9.Department of Energy. 1997. Linking legacies report Department of Energy/EM-319. U.S. Department of Energy, Washington, D.C.
- 10.DiChristina, T. J. 1992. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J. Bacteriol. 174:1891-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dollhopf, S. L., S. A. Hashsham, F. B. Dazzo, R. F. Hickey, C. S. Criddle, and J. M. Tiedje. 2001. The impact of fermentative organisms on carbon flow in methanogenic systems under constant low substrate conditions. Appl. Microbiol. Biotechnol. 56:531-538. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, A., S. Huang, S. Seston, J. Xing, R. F. Hickey, C. Criddle, C., and J. M. Tiedje. 1999. How stable is stable? Function versus community stability. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finneran, K. T., R. T. Anderson, K. P. Nevin, and D. R. Lovley. 2002. Potential for bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sed. Contam. 11:339-357. [Google Scholar]
- 15.Finneran, K. T., H. M Forbush, C. Gaw VanPraagh, and D. R. Lovley. 2002. Desulfitobacterium metallireducins sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids, as well as chlorinated compounds. Int. J. Syst. Evol. Microbiol., http://dx.doi.org/10.1099/ijs.0.02121-0. [DOI] [PubMed]
- 16.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 17.Fortin, D., and T. J. Beveridge. 1997. Microbial sulfate reduction within sulfidic mine tailings: formation of diagenetic Fe sulfides. Geomicrobiol. J. 14:1-21. [Google Scholar]
- 18.Fortin, D., M. Roy, J. Rioux, and P. Thibault. 2000. Occurrence of sulfate-reducing bacteria under a wide range of physico-chemical conditions in Au and Cu-Zn mine tailings. FEMS Microbiol. Ecol. 33:197-208. [DOI] [PubMed] [Google Scholar]
- 19.Francis, C. A., A. Y. Obraztsova, and B. M. Tebo. 2000. Dissimilatory metal reduction by the facultative anaerobe Pantoea agglomerans SP1. Appl. Environ. Microbiol. 66:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorby, Y. A., and D. R. Lovley. 1992. Enzymatic uranium precipitation. Environ. Sci. Technol. 26:205-207. [Google Scholar]
- 21.He, Q., and R. Sanford. 2003. Characterization of Fe(III) reduction by chlororespiring Anaeromxyobacter dehalogens. Appl. Environ. Microbiol. 69:2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbie, J. E. R. J. Daley, and S. Jasper. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteriaceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, D. B. 1998. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol. Ecol. 27:307-317. [Google Scholar]
- 26.Johnson, D. B., and S. McGinness. 1991. Ferric iron reduction by acidophilic heterotrophic bacteria. Appl. Environ. Microbiol. 57:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostka, J. E., D. D. Dalton, H. Skelton, S. Dollhopf, and J. W. Stucki. 2002. Growth of iron(III)-reducing bacteria on clay minerals as the sole electron acceptor and comparison of growth yields on a variety of oxidized iron forms. Appl. Environ. Microbiol. 68:6256-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostka, J. E., and G. W. Luther III. 1994. Partitioning and speciation of solid-phase iron in saltmarsh sediments. Geochem. Cosmochem. Acta 58:1701-1710. [Google Scholar]
- 29.Kostka, J. E., and K. H. Nealson. 1995. Dissolution and reduction of magnetite by bacteria. Environ. Sci. Technol. 29:2535-2540. [DOI] [PubMed] [Google Scholar]
- 30.Küsel, K., T. Dorsch, G. Acker, and E. Stackebrandt. 1999. Microbial reduction of Fe(III) in acidic sediments: isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. App. Environ. Microbiol. 65:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Küsel, K. U. Roth, and H. Drake. 2002. Microbial reduction of Fe(III) in the presence of oxygen under low pH conditions. Environ. Microbiol. 4:314.1-314.8. [DOI] [PubMed] [Google Scholar]
- 32.Küsel, K. U. Roth, T. Trinkwalter, and S. Peiffer. 2001. Effect of pH on the anaerobic microbial cycling of sulfur in mining-impacted freshwater lake sediments. Environ. Exp. Bot. 46:213-223. [Google Scholar]
- 33.Langworthy, T. A. 1978. Microbial life in extreme pHs, p. 279-315. In D. J. Kushner (ed.), Microbial life in extreme environments. Academic Press Inc., New York, N.Y.
- 34.Liger, E., L. Charlet, and P. Van Chappellen. 1999. Surface catalysis of uranium(VI) reduction by iron(II). Geochim. Cosmochim. Acta 63:2939-2955. [Google Scholar]
- 35.Liu, Y., D. L. Balkwill, H. C. Aldrich, G. R. Drake, and D. R. Boone. 1999. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov., and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 49:545-556. [DOI] [PubMed] [Google Scholar]
- 36.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimalatory metal reduction. metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 37.Lovley, D. R. 2000. Fe(III) and Mn(IV) reduction, p. 3-30. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washinton, D.C.
- 38.Lovley, D. R., and R. T. Anderson. 2000. The influence of dissimilatory metal reduction on the fate of organic and metal contaminants in the subsurface. Hydrogeol J. 8:77-88. [Google Scholar]
- 39.Lovley, D. R., and E. J. P. Phillips. 1992. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 26:2228-2234. [Google Scholar]
- 40.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 42.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 43.Lovley, D. R., P. K. Widman, J. C. Woodward, and E. J. P. Phillips. 1993b. Reduction of uranium by cytochrome c3 of Desulfovibrio vulagris. App. Environ. Microbiol. 59:3572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luli, G. W., and W. R. Strohl. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1996. The ribosomal database project (RDP). Nucleic Acids Res. 24:82-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLean, E. O. 1982. Soil pH and lime requirement, p. 199-209. In Methods of soil analysis, part 2, chemical and microbiological properties. Agronomy Monograph 9, 2nd ed. ASA-SSSA, Madison, Wis.
- 47.Mohagheghi, A., D. M. Updegraff, and M. B. Goldhaber. 1985. The role of sulfate reducing bacteria in the deposition of sedimentary uranium ores.“ Geomicrobiol. J. 4:153-173. [Google Scholar]
- 48.Nakamura, L. K. 1984. Bacillus amylolyticus sp. nov., nom. rev., Bacillus lautus sp. nov., nom. rev., Bacillus pabuli sp. nov., nom. rev., and Bacillus validus sp. nov., nom. Rev. Int. J. Syst. Bacteriol. 34:224-226. [Google Scholar]
- 49.Niggemyer, A., S. Spring, E. Stackebrandt, and R. F. Rosenzweig. 2001. Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl. Environ. Microbiol. 67:5568-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ochiai, E. I. 1987. General principles of biochemistry of the elements. Plenum Press, New York, N.Y.
- 51.Peine, A., A. Tritschler, K. Küsel, and S. Peiffer. 2000. Electron flow in an iron-rich acidic sediment — evidence for an acidity driven iron cycle. Limnol. Oceanogr. 45:1077-1087. [Google Scholar]
- 52.Proctor, L. M., and A. C. Souza. 2001. Method for enumeration of 5-cyano-2, 3-ditoyl tetrazolium chloride (CTC)-active cells and cell-specific CTC activity of benthic bacteria in riverine, estuarine and coastal sediments. J. Microbiol. Methods 43:213-222. [DOI] [PubMed] [Google Scholar]
- 53.Roden, E. E., and J. M. Zachara. 1996. Microbial reduction of crystalline Fe(III) oxides: influence of oxide surface area and potential for cell growth. Environ. Sci. Technol. 30:1618-1628. [Google Scholar]
- 54.Russell, J. B. 1991. Intratcellular pH of acid-tolerant ruminal bacteria. Appl. Environ. Microbiol. 57:3383-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanford, R. A., J. R. Cole, and J. M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schauder, R., and B. Schink. 1989. Anaerovobrio glycerini sp. nov., an anaerobic bacterium fermenting glycerol to propionate, cell matter, and hydrogen. Arch. Microbiol. 152:473-478. [Google Scholar]
- 57.Shida, O., H. Takagi, K. Kadowaki, and K. Komagata. 1996. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int. J. Syst. Bacteriol. 46:939-946. [DOI] [PubMed] [Google Scholar]
- 58.Shida, O., H. Takagi, K. Kadowaki, L. K. Nakamura, and K. Komagata. 1995. Proposal of Bacillus reuszeri sp. nov., Bacillus formosus sp. nov., nom. rev., and Bacillus borsstelensis sp. nov., nom. rev. Int. J. Syst. Bacteriol. 45:93-100. [Google Scholar]
- 59.Stackebrandt, E., C. Sproer, F. A. Rainey, J. Burghardt, O. Pauker, and H. Hippe. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int. J. Syst. Bacteriol. 47:1134-1139. [DOI] [PubMed] [Google Scholar]
- 60.Strompl, C., B. J. Tindall, G. N. Jarvis, H. Lunsdorf, E. R. B. Moore, and H. Hippe. 1999. A re-evaluation of the taxonomy of the genus Anaerovobrio, with the reclassification of Anaerovobrio glycerini as Anaerosinus glycerini gen. nov., comb. nov., and Anaerovobrio burkinensis as Anaeroarcus burkinensis [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:1861-1872. [DOI] [PubMed] [Google Scholar]
- 61.Strunk, O., and W. Ludwig. 1997. ARB: software for phylogenetic analysis. Technical University of Munich, Munich, Germany.
- 62.Suyama, T., Y. Tokiwa, P. Ouichanpagdee, T. Kanagawa, and Y. Kamagata. 1998. Phylogenetic affiliation of soil bacteria that degrade aliphatic polyesters available commercially as biodegradable plastics. Appl. Environ. Microbiol. 64:5008-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki, Y., and J. F. Banfield. 1999. Geomicrobiology of uranium, p. 393-432. In P. C. Burns and R. Finch (ed.), Uranium: mineralogy, geochemistry and the environment. Reviews in Mineralogy vol. 38. Mineralogical Society of America, Washington, D.C.
- 64.Sykes, P. J., S. H. Neoh, M. J. Brisco, E. Hughes, J. Condon, and A. A. Morley. 1992. Quantitation of targets for PCR by use of limiting dilution. BioTechniques 13:444-449. [PubMed] [Google Scholar]
- 65.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol Lett. 162:193-198. [Google Scholar]
- 66.Truede, N., D. Rosencrantz, W. Liesack, and S. Schnell. 2003. Strain Fac12, a dissimilatory iron-reducing member of the Anaeromyxobacter subgroup of Myxococcales. FEMS Microbiol. Lett 44:261-269. [DOI] [PubMed]
- 67.Truex, M. J., B. M. Peyton, N. B. Valentine, and Y. A. Gorby. 1996. Kinetics of U(VI) reduction by a dissimilatory Fe(III)-reducing bacterium under nongrowth conditions. Biotechnol. Bioeng. 55:490-496. [DOI] [PubMed] [Google Scholar]
- 68.Widdel, F., and F. Bak. 1992. gram negative mesophilic sulfate reducing bacteria, p. 3352-3378 In A. Balows et al. (ed.), The prokaryotes. a handbook on the biology of bacteria: ecophysiology, isolation, identification, and applications, 2nd edition, vol. 2. Springer-Verlag, New York, N.Y.