Summary
Cell motility driven by actin filament assembly demands the spatial and temporal coordination of numerous regulatory Actin Binding Proteins (ABPs) [1], many of which bind with affinities and kinetics that depend on the chemical state (ATP, ADP-Pi or ADP) of actin filament subunits. ADF/cofilin, one of three ABPs that precisely choreograph actin assembly and organization into “comet-tails” that drive motility in reconstituted in vitro systems [2], binds and stochastically severs “aged” ADP actin filament segments of de novo growing actin filaments [3]. Severing increases the density of filament ends from which subunits can add and dissociate, thereby increasing overall actin filament assembly dynamics. Deficiencies in methodologies to track in real time the nucleotide state of actin filaments as well as ADF/cofilin severing limits the molecular understanding of coupling between actin filament chemical and mechanical states and severing. We engineered a fluorescently labeled ADF/cofilin that retains actin filament binding and severing activities. Since ADF/cofilin binding depends strongly on the actin-bound nucleotide direct visualization of fluorescent ADF/cofilin binding serves as a marker of the actin filament nucleotide state and permits assessment of the “ATP/ADP-Pi cap” length of individual actin filaments during assembly and elongation. Bound ADF/cofilin allosterically accelerates Pi release from unoccupied filament subunits, which shortens the filament ATP/ADP-Pi cap length by nearly an order of magnitude. Rapid elongation far exceeds ADF/cofilin-acceleration of Pi release under in vivo conditions; thereby filament barbed end capping is required for efficient ADF/cofilin binding and severing. Real time visualization of filament severing indicates that fragmentation scales with and occurs preferentially at boundaries between bare and ADF/cofilin decorated filament segments, thereby controlling the overall filament length depending on the ADF/cofilin activity and filament binding density.
Results and Discussion
Direct visualization of ADF/cofilin binding to growing actin filaments
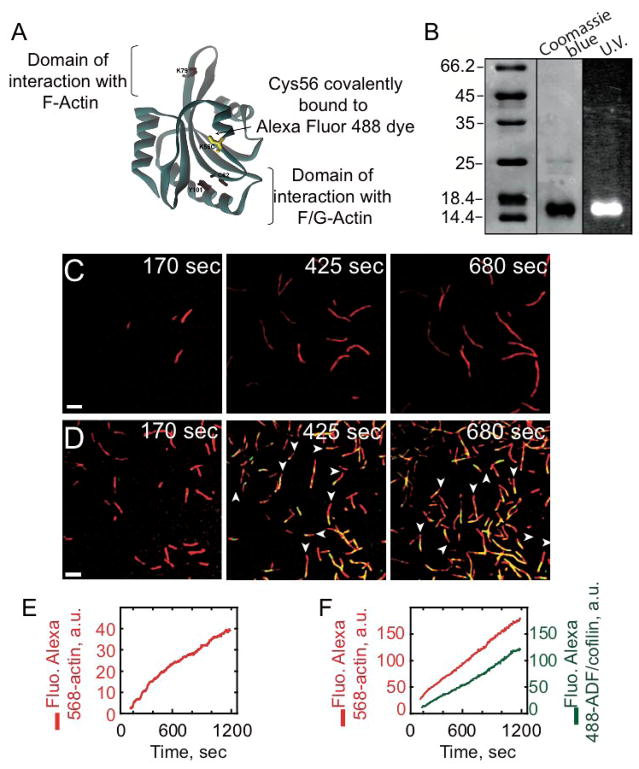
To follow in real time ADF/cofilin binding to actin filaments, we engineered a yeast ADF/cofilin mutant that could be specifically labeled with a fluorescent probe. Yeast ADF/cofilin contains a single cysteine residue that is buried in the protein structure, so we substituted K56, a solvent-exposed amino-acid residue positioned outside of the actin-binding site [4], to cysteine (Figure 1A) and labeled with Alexa-488 maleimide (Figure 1B). Labeled K56C ADF/cofilin retains strong actin filament binding (Figure S1A), severing (discussed forthcoming), and acceleration of spontaneous actin assembly activities (Figure S1B). Given the minimal perturbations of substitution and labeling, Alexa-488-labeled ADF/cofilin is a reliable tool to investigate the dynamic interaction with elongating actin filaments.
Figure 1. Direct visualization of ADF/coflin binding on elongating actin filaments by evanescent wave microscopy.
(A) Structure of S. cerevisiae cofilin (Protein Data Bank ID code COF1); its only ystein (C62) radical is buried in the wild type protein structure. We designed a mutant K56C-cofilin with a cystein therefore available for labeling by Alexa dyes. (B) 15% SDS-PAGE gel of purified Alexa-488 labeled K56C-cofilin revealed both by Coomassie blue staining and UV illumination. (C–F) Montage of timelapse TIRFM-images showing the polymerization of 0.8 μM Alexa-568-labeled actin with 2.4 μM profilin, (C, E) in absence or (D, F) in presence of 0.92 μM Alexa-488-cofilin. Alexa-568-actin filaments were colorized in red, Alexa-488-ADF/cofilin in green, and the decorated portions of filaments were yellow in the merged images (D). White arrowheads indicate the fast growing barbed ends of filaments. (E) shows the increase of the integrated intensity fluorescence over time along actin filaments (red curve) in (C), whereas (F) that of actin filaments (red curve) and bound Alexa-488-cofilin (green curve) in (D). Scale bars represent 5 μm.
We followed in real time using Total Internal Reflection Fluorescence microscopy (TIRFm) the interaction of ADF/cofilin with actin filaments as they spontaneously assembled form Alexa 568-labeled ATP-actin monomers (Figures 1C and 1E). Measurements were done in the presence of profilin to foster nucleotide exchange from actin monomers, thereby maintaining an ATP-actin monomer pool [5], and limiting ADF/cofilin binding to monomers in solution. The cumulative fluorescence of labeled actin in the evanescent field (proportional to polymer mass) increases linearly over time (Figure 1E), yielding a filament elongation rate of ≈ 5 subunits sec−1 (Figure 1E) in the absence of ADF/cofilin, consistent with previous determinations [6].
Using two-color TIRF microscopy, we simultaneously monitored in real time actin filament assembly and ADF/cofilin binding (Figures 1D and 1F and Movie S1). The density of bound ADF/cofilin scales with the increase in total polymer (Figure 1F). Remarkably, we detect only at the TIRFm resolution scale minor ADF/cofilin fluorescence before 170 seconds of actin assembly (Figure 1D), demonstrating that ADF/cofilin binding is delayed relative to actin polymerization, presumably due to the nucleotide state of filament subunits [5].
Bound ADF/cofilin dissociates slowly from actin filaments
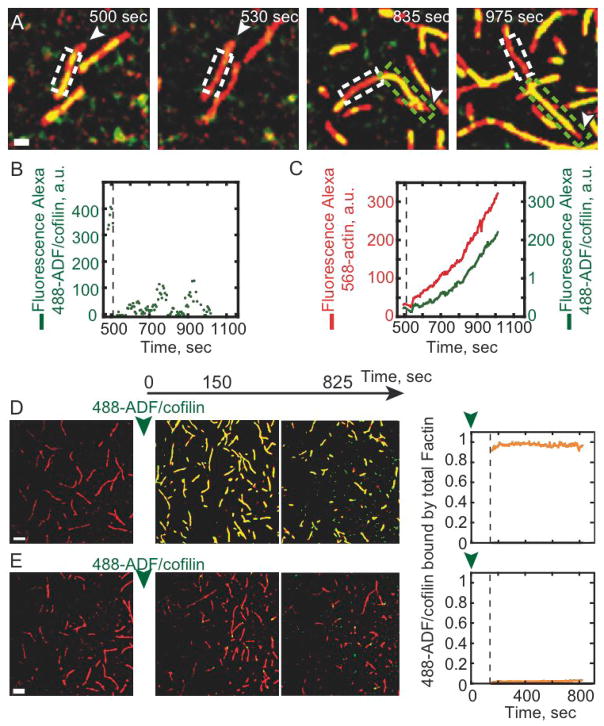
The lifetime and dissociation kinetics of bound ADF/cofilin was evaluated by Fluorescence Recovery After Photobleaching (FRAP). A defined segment of an Alexa-488-ADF/cofilin decorated filament was photobleached with an intense laser beam (Figure 2A, white box). Surprisingly, minimal fluorescence recovery associated with alexa-488-ADF/cofilin occurs within 500 seconds, indicating that the rate constant for yeast ADF/cofilin dissociation from filaments is very slow and negligible over the time courses of experimental visualization (Figure 2B and Movie S2). Locally bleached actin filaments elongate and bind Alexa-488-ADF/cofilin, thereby confirming that neither actin nor ADF/cofilin are limiting (Figure 2A, green box and 2C) and that the lack of ADF/cofilin recovery after photobleaching (Figures 2A and 2B) reflects slow ADF/cofilin dissociation.
Figure 2. The ADF/cofilin turnover on actin filaments is limited by its low off-rate constant.
The polymerization of 0.8 μM Alexa-568-actin monomers in presence of 2.4 μM profilin and 1.8 μM Alexa-488-ADF/cofilin (A) or 2 μM Alexa-488-ADF/cofilin (D) was followed by TIRF microscopy. Fluorescence signals were colorized as in Figure 1. (A–C) Fluorescence Recovery After Photobleaching assays were performed on Alexa-488-ADF/cofilin in interaction with growing actin filaments. (A) After 500 seconds of actin polymerization, Alexa-488-ADF/cofilin fluorescence was bleached (dashed white box) and the fluorescence recovery was followed over a period of an additional 500 seconds of actin assembly. The actin filament was still elongating outward the bleached box by its fast growing barbed end (white arrowhead) and was decorated by Alexa-488-ADF/cofilin (green box). After photobleaching, (B) the integrated Alexa-488-ADF/cofilin fluorescence over time in the bleached area (dashed white box) remained negligible compared to its initial value; however, in (C) the integrated fluorescence intensity of both Alexa-568-actin filaments and Alexa-488-ADF/cofilin outside the bleached area still increase over time (green box). (D) Pulse-chase experiments. 2 μM Alexa-488-ADF/cofilin was added to 0.8 μM Alexa-568-actin saturated with 2.4 μM profilin after 3 mn of polymerization. (E) same that D but actin polymerization occurred in presence 2 μM unlabeled ADF/cofilin before addition of Alexa-488-ADF/cofilin. The time zero corresponds to the addition of 2 μM Alexa-488-ADF/cofilin. The rightmost graphs show that the integrated fluorescence intensity of Alexa-488-ADF/cofilin bound along the bare actin filament (D) and along the actin filament pre-incubated with unlabeled-ADF/cofilin (E). Scale bars represent 2 μm in (A) and 5 μm in (D).
To ensure that slow yeast ADF/cofilin dissociation is not a consequence of labeling or photobleaching procedures, we competed bound unlabeled ADF/cofilin with Alexa-labeled ADF/cofilin (Figure 2E and Movie S2). Undetectable levels of Alexa-ADF/cofilin incorporate into actin filaments decorated with unlabeled ADF/cofilin filaments within 800 seconds (Figure 2E), thereby confirming that slow ADF/cofilin dissociation is an intrinsic biochemical property of yeast ADF/cofilin that contributes to a high overall binding affinity [7]. Note that Alexa-488-ADF/cofilin binds rapidly to bare actin filaments (Figure 2D).
ADF/cofilin shortens the ATP/ADP-Pi cap length of actin filaments by allosterically accelerating Pi release
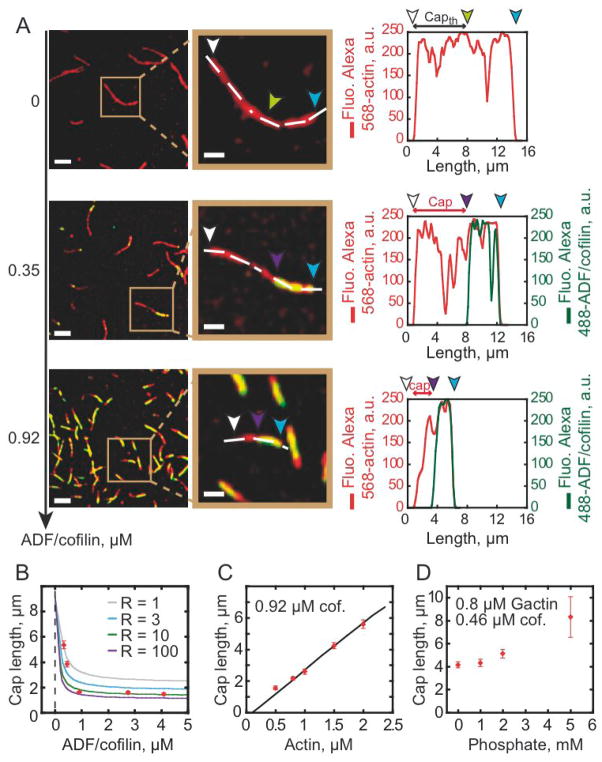
ADF/cofilin binds 40-fold more strongly to ADP-actin filament subunits than to ATP-or ADP-Pi subunits, and weakens Pi binding by accelerating release from ADP-Pi subunits through thermodynamic and kinetic linkage [5]. Labeled ADF/cofilin therefore serves as an effective marker to directly probe the nucleotide composition of individual actin filaments. TIRF microscopy reveals that ADF/cofilin does not decorate filament barbed end segments, even at high [ADF/cofilin] (Figure 3A, middle and bottom panels and Movie S3), which we interpret as weak binding to “ATP/ADP-Pi cap” at filament barbed ends (Figures 3A). We note that the filament is comprised predominantly of ADP-Pi subunits at these [actin] and in the absence of ADF/cofilin (Figure 3A, top panel; [8, 9]) to which ADF/cofilin binds very weakly [5, 10]. ADF/cofilin must therefore accelerate Pi release from filaments, as reported for assays done with bulk filament populations [5], to decorate with such high efficiency (Figure 3A, middle and bottom panels). In addition, observation of multiple ADF/cofilin clusters along individual actin filaments favors a random ATP hydrolysis mechanism for filament subunits over a vectorial mechanism. (Movies S1 and S3).
Figure 3. Tight coupling between binding and effect on nucleotide state of actin filaments modulates ADF/cofilin-actin interaction.
(A) 0.8 μM Alexa-568-actin monomers were polymerized in presence of 2.4 μM profilin and ADF/cofilin as indicated. TIRFM images were taken at 800 seconds. Fluorescence signals were colorized as in Figure 1. The images in middle column are zooms of the boxed areas in the first column. Arrowheads indicate pointed ends (blue), barbed ends (white), and the ATP/ADP-Pi “cap” length (white to purple). In the absence of ADF/cofilin, the theoretical position of the interface between ADP-Pi and ADP zones (green) was determined according to the slow phosphate release, whose half life time is ~6 minutes [5]. In presence of ADF/cofilin, the cap length is determined by the absence of fluorescence in the green channel (Figure A middle and bottom panels). Graphs in the rightmost column quantified the fluorescence intensity of Alexa-568-actin and Alexa-488-ADF/cofilin along actin filament length marked by a dashed line. Scale bars represent 5 μm and 2 μm respectively for the left and middle column. (B) Allosteric effect of ADF/cofilin on ATP/ADP-Pi cap length. Experimental data (dots) were fitted by a kinetic model (lines, see supplemental information) as a function of Alexa-488-ADF/cofilin concentrations. We varied in the model the “R” value, which represents how Pi release is propagated allosterically from ADF/cofilin-occupied sites to 1 (grey), 3 (blue), 10 (green), or 100 (purple) vacant subunits along the filament. (C) The experimental ATP/ADP-Pi cap (dots) increases linearly with the concentration of actin monomers in solution as predicted by the model (line). (D) Variation of the ATP/ADP-Pi cap length in presence of an increasing concentration of inorganic phosphate in the medium.
Since the ATP/ADP-Pi cap size can be limited both by slow ADF/cofilin binding [11, 12]; and/or Pi release, we investigated the variation in cap length as a function of ADF/cofilin and actin monomer concentrations. Statistical analysis reveals that the mean cap length depends on the ADF/cofilin concentration, reaching a minimum of 1.6 μm at saturating ADF/cofilin and 0.8 μM actin (Figures 3B). Higher ADF/cofilin concentrations do not shorten the cap length (Figure 3B), which remains stationary over time while the aged zone of the filament is decorated with ADF/cofilin (Figure S1C).
A kinetic model in which the nucleotide-linked equations of actin filament nucleation, elongation, random ATP hydrolysis, Pi release, and ADF/cofilin binding are explicitly accounted was used to fit the experimental cap length data (Figures 3B, 3C and supplementary information). The [ADF/cofilin]-dependence of the cap length is well described by a model in which bound ADF/cofilin increases Pi release from ADP-Pi subunits by an order of magnitude from 0.0019 s−1 to 0.013 s−1 (Figure 3B; [5]). The fit to the data, however, is significantly improved if acceleration of Pi release is propagated allosterically from ADF/cofilin-occupied sites to ≥10 vacant subunits along the filament (i.e. non-nearest neighbor effects), as predicted from long-range effects on filament subunit torsional dynamics [13].
The actin filament ATP/ADP-Pi cap size (at a given [actin]) is determined by the maximum Pi release rate constant, even though it is accelerated allosterically by ADF/cofilin binding. This behavior predicts that the cap length increases linearly with [actin] and also with inclusion of Pi in the medium, as is observed (Figures 3C and 3D). Similarly, if filament barbed end elongation is stopped with capping protein, the ATP/ADP-Pi cap disappears and ADF/cofilin decorates the entire filament (Figure S1D). Taken together, these results demonstrate that the ATP/ADP-Pi cap length reflects a tight balance between filament elongation, random ATP hydrolysis, ADF/cofilin binding, and allosteric acceleration of Pi release from vacant filament subunits.
Actin filament severing occurs at low ADF/cofilin binding densities and preferentially at boundaries of bare and ADF/cofilin-decorated segments
Direct, real time visualization of ADF/cofilin binding to actin filaments also permits evaluation of the sites of severing and identifying how they correlate with filament occupancy. Of particular importance is identifying the site(s) of preferential filament fragmentation. That is, whether it occurs preferentially at junctions of bare and decorated regions [14], or internally within homogenous (bare or ADF/cofilin-decorated) segments.
ADF/cofilin binding alters the average structure [15, 16] and dynamics [7, 13, 17, 18] of actin filaments such that they are more flexible than native filaments (Figure S2A and [18, 19]). It is hypothesized that shear stress associated with thermal induced fluctuations accumulates locally at boundaries of mechanical asymmetry, thereby leading to preferential severing at junctions of bare and decorated filament segments [3, 12, 14, 19, 20].
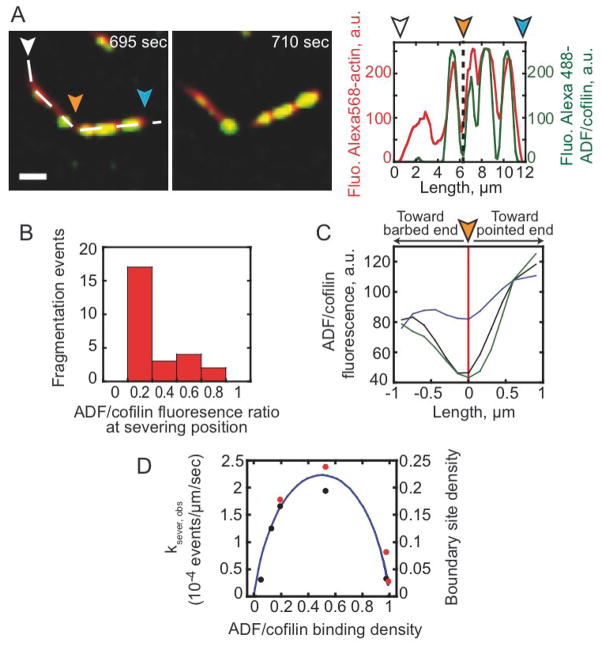
To test the prediction of preferential severing at boundaries of bare and decorated segments, we quantified the severing events occurring during spontaneous assembly of ATP-actin filaments. Linescans of fluorescence intensity along actin filaments reveal that fragmentation is statistically favored at sites of low ADF/cofilin binding density and occurs exclusively outside the ATP/ADP-Pi cap (Figures 4A and 4C). Note that severing is not obligatory with ADF/cofilin binding, but the frequency of severing events correlates with the position (Figure 4A) and density (Figure 4D) of bare and ADF/cofilin-decorated boundaries, consistent with preferential severing at or near these boundaries on filaments (Movie S4).
Figure 4. ADF/cofilin severing occurs between regions of bare and ADF/cofilin-decorated actin filaments.
(A) The polymerization of 0.5 μM Alexa-568-actin monomers in presence of 1.5 μM profilin and 0.3 μM Alexa-488-cofilin was followed by TIRF microscopy. The distribution of Alexa-568-actin and Alexa-488-cofilin along the filament was quantified using linescans of their respective fluorescence (dashed line). Fluorescence signals were colorized as in Figure 1. The arrowheads (orange) indicate the position of actin filament’s severing site. Barbed end (white arrowhead) and pointed end (blue arrowhead). (B) The histogram quantified the frequency of fragmentation events as a function of the ratio of Alexa-488-ADF/cofilin over actin filament. (C) Statistics of the Alexa-488-ADF/cofilin fluorescence ratio, calculated as in (B), along fragmented filaments, which were centered on their fragmentation site (red line). The curves give the average of the fluorescence ratio (n = 28) for 0.5 μM (black), 0.9 μM (blue), and 2.8 μM Alexa-488-ADF/cofilin (green curve). (D) ADF/cofilin severing activity (red dots for labeled ADF/cofilin and black dots for unlabeled ADF/cofilin) scales with the density of boundaries between bare and ADF/cofilin-decorated filament segments (solid line). The ADF/cofilin binding density (cofilins bound per actin subunit) and the fractional site density of boundaries between bare and ADF/cofilin-decorated segments (solid line) was calculated from the Alexa-488 labeled ADF/cofilin or unlabeled ADF/cofilin binding parameters determined in equilibrium binding measurements (Figure S1A; [20]). The boundary density reaches a maximum of ~22% total sites at ~50% filament occupancy. (A) Scale bars represent 2 μm.
Concluding Remarks
A. ADF/cofilin modulates the nucleotide composition of growing actin filaments
The age and stability of actin filaments is linked to the chemical state of the bound adenine nucleotide. ATP bound to monomers is rapidly hydrolyzed after incorporation into filaments such that freshly polymerized filaments are comprised of subunits with bound ATP or ADP-Pi, whereas older filament subunits release Pi slowly and have bound ADP. The actin binding activities of many ABPs including ADF/cofilin are sensitive to the chemical state of the actin-bound nucleotide, so the filament nucleotide composition dramatically influences the organization, stability and dynamics of cellular actin-based structures.
ADF/cofilin “ages” filaments by accelerating Pi release over an order of magnitude. This effect is allosteric, and propagates to distal sites unoccupied by ADF/cofilin, presumably through allosteric modulation of filament twist and dynamics [13, 21, 22]. Therefore, a kinetic competition between monomer addition, intrinsic random ATP hydrolysis [23] and Pi release [5], ADF/cofilin binding [5, 11, 12] and allosteric ADF/cofilin-mediated acceleration of Pi release (Figure 3B) exists during assembly and network growth.
Pi release, though accelerated allosterically by ADF/cofilin, remains considerably slower than filament elongation (up to 500 subunits sec−1) at high in vivo actin concentrations, which yields a large filament ATP/ADP-Pi cap (ca. 100 μm in length) that precludes ADF/cofilin binding and severing. Even if the Pi release is faster for yeast actin [24], this behavior is difficult to reconcile with the observation that ADF/cofilin binds growing cellular filaments only 0.2 – 1 μm away from their nucleation sites [25–27]. We favor a mechanism in which filament barbed ends must be rapidly capped - to stop rapid elongation - for significant ADF/cofilin binding to occur. Such a mechanism would account for co-localization of ADF/cofilin and capping protein in actin networks [26] and modulation of ADF/cofilin severing efficiency by capping protein. Subsequent ADF/cofilin binding to stochastically emerging ADP subunits of capped filaments allosterically accelerates Pi release, thereby promoting Arp2/3 complex dissociation [10] and network remodeling. Therefore, although ADF/cofilin-mediated acceleration of Pi release minimally affects the ATP/ADP-Pi cap length of rapidly elongating filaments in vivo, it rapidly ages filaments and networks by allosterically accelerating Pi release once they are capped and stop elongating.
B. ADF/cofilin preferentially severs ADP-actin filaments at boundaries of bare and cofilin-decorated segments
Quantitative analysis of filament binding [5, 9, 11, 12, 20, 28] and severing [3, 29, 30] indicates that ADF/cofilin severing activity scales with the density of boundaries between bare and ADF/cofilin-decorated filament segments [14]. It has been hypothesized that asymmetry originating from discontinuities in filament topology and mechanics (i.e. bending and twisting elasticity) generates a local accumulation of shear stress [19], thereby leading to preferential fragmentation at or near these boundaries [14]. This hypothesis relies on three important observations: (a) severing occurs at low ADF/cofilin binding densities and small cluster sizes [20, 29, 30]; (b) cofilin-decorated filaments display significantly different mechanical properties than bare filaments [13, 18, 19]; (c) partially ADF/cofilin-decorated filaments are considerably less stable than bare or ADF/cofilin-saturated filaments [21, 22].
The prediction that ADF/cofilin-mediated severing occurs at bare and decorated boundaries lacks direct proof, and is best evaluated by direct, real time visualization of ADF/cofilin binding and filament severing, as performed in this study. Severing is not obligatory with ADF/cofilin binding, but the frequency of severing events scales with the boundary density and also occurs at or near these boundaries. These observations lend credence to the hypothesis that shear stress accumulates at a mechanical asymmetry presented at boundaries of bare and ADF/cofilin-decorated filament segments, thereby promoting severing. A challenge for future investigations will be to determine how others actin binding proteins including coronin and AiP1 [31] modulate this mechanism to promote actin disassembly.
Supplementary Material
Acknowledgments
This work was supported by grants from Agence Nationale de la Recherche to L.B. and J.L.M (ANR-08-BLANC-0022 and ANR-08-SYSC-013), the American Heart Association (0940075N awarded to E.M.D.L.C.; U.S.A.), the National Institutes of Health (GM071688 and GM071688-03S1 awarded to E.M.D.L.C, U.S.A.), and the Institute of Complex Systems IXXI, Rhône-Alpes (awarded to J.L.M.; France). E.M.D.L.C. is an American Heart Association Established Investigator, an NSF-CAREER Award recipient (MCB-0546353) and Hellman Family Fellow. We thank Pekka Lappalainen and Bruce Goode for the ADF/cofilin K56 construct.
Footnotes
Supplemental Data include supporting text, two figures, four movies and supplemental references can be found with this article on line at http://www.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollard TD, Blanchoin L, Mullins RD. Molecular Mechanisms Controlling Actin Filament Dynamics in Nonmuscle Cells. Annu Rev Biophys. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 3.Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 6.Achard V, Martiel JL, Michelot A, Guerin C, Reymann AC, Blanchoin L, Boujemaa-Paterski R. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 20:423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 7.Bobkov AA, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. J Mol Biol. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 8.Vavylonis D, Yang Q, O’Shaughnessy B. Actin polymerization kinetics, cap structure, and fluctuations. Proc Natl Acad Sci USA. 2005;102:8543–8548. doi: 10.1073/pnas.0501435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roland J, Berro J, Michelot A, Blanchoin L, Martiel JL. Stochastic severing of actin filaments by ADF/cofilin controls the emergence of a steady dynamical regime. Biophys J. 2007;94:2082–2094. doi: 10.1529/biophysj.107.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 12.De La Cruz EM, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys J. 2010;98:1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prochniewicz E, Janson N, Thomas DD, De la Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. J Mol Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 14.De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhlrad A, Kudryashov D, Michael Peyser Y, Bobkov AA, Almo SC, Reisler E. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J Mol Biol. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Pfaendtner J, De La Cruz EM, Voth GA. Actin filament remodeling by actin depolymerization factor/cofilin. Proc Natl Acad Sci USA. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough BR, Blanchoin L, Martiel JL, De la Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 21.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 22.Dedova IV, Nikolaeva OP, Safer D, De La Cruz EM, dos Remedios CG. Thymosin beta4 induces a conformational change in actin monomers. Biophys J. 2006;90:985–992. doi: 10.1529/biophysj.105.063081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchoin L, Pollard TD. Hydrolysis of bound ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- 24.Ti SC, Pollard TD. Purification of actin from fission yeast Schizosaccharomyces pombe and characterization of functional differences from muscle actin. J Biol Chem. 2011;286:5784–5792. doi: 10.1074/jbc.M110.199794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasa JH, Mullins RD. Spatial and Temporal Relationships between Actin-Filament Nucleation, Capping, and Disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 29.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.