Abstract
Induced pluripotent stem cells (iPSCs) can differentiate into mineralizing cells and thus have a great potential in application in engineered bone substitutes with bioactive scaffolds in regeneration medicine. In the current study we characterized and demonstrated the pluripotency and osteogenic differentiation of mouse iPSCs. To enhance the osteogenic differentiation of iPSCs, we then transduced the iPSCs with the potent transcription factor, nuclear matrix protein SATB2. We observed that in SATB2-overexpressing iPSCs there were increased mineral nodule formation and elevated mRNA levels of key osteogenic genes, osterix (OSX), Runx2, bone sialoprotein (BSP) and osteocalcin (OCN). Moreover, the mRNA levels of HoxA2 was reduced after SATB2 overexpression in iPSCs. The SATB2-overexpressing iPSCs were then combined with silk scaffolds and transplanted into critical-size calvarial bone defects created in nude mice. Five weeks post-surgery, radiological and micro-CT analysis revealed enhanced new bone formation in calvarial defects in SATB2 group. Histological analysis also showed increased new bone formation and mineralization in the SATB2 group. In conclusion, the results demonstrate that SATB2 facilitates the differentiation of iPSCs towards osteoblast-lineage cells by repressing HoxA2 and augmenting the functions of the osteoblast determinants Runx2, BSP and OCN.
1. Introduction
Successful repair of bone deficiencies continues to be a major concern and challenge to reconstructive surgeons. Autologous bone grafts provide the best clinical outcome to repair massive bone defects for their osteogenic and osteoinductive properties [1-3]. However, its collection is frequently associated with pain, risk of infection, hemorrhage, cosmetic disability, and nerve damage. Allogenic transplants are also used, but are often of poor quality, carrying the risk of infection and the transmission of disease from the donor to the recipient [4]. Due to these drawbacks, tissue engineering is a promising addition to establish an artificial biomaterial scaffold containing regenerating-competent cells. Bone marrow-derived mesenchymal stem cells are a potential cell source for such strategies, but having limited proliferative potential[5, 6]. Pluripotent stem cells are a possible alternative candidates because they are able to differentiate into all kinds of cells in the body and also self-renew indefinitely. Well-known pluripotent stem cells are embryonic stem (ES) cells, which are derived from the inner cell mass of the blastocyst and have the ability to grow indefinitely and differentiate into cells of all three germ layers [7, 8]. However, regardless of the different types of blastocysts used, isolation and establishment of ES cell lines involve embryo destruction and require oocytes. Moreover, ES cell-based therapy would be complicated by immune rejection due to the immunological incompatibility between patient and donor cells. These obstacles are circumvented by Takahashi and Yamanaka[9], who have reprogrammed fibroblasts to an ES cells-like state using four transcription factors: Oct3/4, Sox2, Klf4, and c-Myc. These so-called induced pluripotent stem cells (iPSCS) derived from mouse [10-12] or human[13-16] fibroblasts are indistinguishable from ES cells in morphology, proliferation abilities, gene expressions, epigenetic status of pluripotent cell-specific genes, and telomerase activity. The ultimate goal of somatic reprogramming is to generate functional cell types relevant for therapy. To date, iPSCs have been applied to study hematologic, cardiovascular, neurologic and metabolic diseases[17-23]. However, the study of iPSCS in bone disease is still in the initial stages.
Recent studies indicate that the osteogenic differentiation of iPSCs can be regulated by transforming growth factor beta1 (TGF-β1) and resveratrol [24, 25], suggesting their great potential for future application in musculoskeletal tissue repair and regeneration. However, before the cells can be clinically applied, investigations should be performed to identify bioactive factors that can enhance differentiation of these cells toward an osteogenic lineage. In our present study, we investigated the possibility of SATB2 as one such regulatory factor. The SATB2 gene is transcribed in a telomeric to centromeric direction and lies in a gene-poor region of 2q32–q33[26]; and the nuclear matrix protein SATB2 is a recently cloned member of the AT-rich binding protein family that binds to matrix attachment regions (MARs) and activates transcription in a MAR-dependant manner[27, 28]. Dobreva and colleagues report that SATB2 together with some regulatory proteins promotes osteoblast differentiation through repressing HoxA2 expression [29]. In the present study, we investigated the effect of SATB2 on the differentiation of iPSCs toward an osteogenic lineage both in vitro and in vivo. A series of real-time RT-PCR, radiographic, micro-computed tomographic, histological and immunohistochemical analyses were performed to determine if the SATB2 gene modified iPSCs enhanced repairs in murine calvarial critical-sized defects.
2. Materials and methods
2.1. Culture of mouse iPSCs
The mouse iPS cell clone ST-8 (a kind gift from Dr. G. Mostoslavsky, Boston University, Massachusetts, USA) was used in the present study. ST-8 iPS cells were generated by reprogramming tail-tip fibroblasts (TTFs) isolated from Sox2-green fluorescent protein (GFP)/R26-M2rtTA double knock-in mice through transduction with a single lentiviral Vector encoding 4 transcription factors including Oct4, Klf4, Sox2, and c-Myc[30] [31]. Freshly isolated murine embryonic fibroblast (MEF) were cultured to confluence and detached with trypsin/EDTA (0.25% w/v trypsin, 0.02% EDTA), and then subcultured at 1:5 dilutions. Cells at passage 2 were collected and resuspended in 3-5 milliliters MEF media for irradiation at 845 Rad/min for 6.4 min with a total exposure of about 5,500 Rad. Before inoculation, the inactivated cells were transferred into a plate that was pretreated with 0.1% gelatin for 5 min in the incubator, at a concentration of 8×104/ml. The mouse iPSCs were routinely cultured on the layer of inactivated MEF in leukemia inhibitory factor-containing mouse ES (embryonic stem) cell medium (http://www.bumc.bu.edu/stemcells/protocols/ ): 283 mL DMEM (Gibco11995, Invitrogen,CA, USA), 50 mL ESC qualified FBS (Hyclone SH30070.03E,Thermo Scientific,USA), 3.5 mL L-Glutamine (Gibco25030, Invitrogen,CA, USA), 3.5 mL Penicillin/ Streptomycin (Invitrogen,CA, USA), 380 μL 106 Units of LIF (ESG1106, Millipore,USA), and 2.381 μL of stock BME (0.1 mM final concentration) (Sigma Chemical CO., USA). All cell cultures were carried out within a humidified 5% CO2 incubator set at 37°C, and iPS cell lines were passaged every second day using 0.25% trypsin-EDTA (Invitrogen,CA, USA). Embryoid body (EB) formation from mouse iPSCs was induced using the hanging drop method.
2.2. Gene transduction of iPSCs
The mouse SATB2 cDNA was released from Vector pBs-SK-SATB2 (a kind gift from Dr. Rudolf Grosschedl, Max-Planck Institute of Immunobiology, Freiburg, Germany) [27] and ligated into the BamHI/EcoRI site of pBABE-hygro (Addgene), creating plasmid pBABE-hygro-SATB2. Briefly, retroviral Vector containing SATB2 cDNA and empty Vector (pBABE-hygro, Addgene) were transfected into HEK-293T cells using Lipofectamine transfection reagent (Invitrogen,CA,USA). Forty eight hours after transfection, the viral supernatant was collected and filtered through a 0.45 μm filter ((Millipore, MA,USA) and used to infect the iPs EB (embryoid body) with polybrene at a final concentration of 8 μg/ml for 6 hours at days 0, 2, and 5 after culture with a hanging drop.
2.3. In vitro differentiation
Two days after culture with a hanging drop, the EBs were transferred into a Petri dish and maintained for 5 days in suspension culture in differentiation medium: DMEM (Gibco11995, Invitrogen,CA, USA), supplemented with 15% ESC qualified FBS (Hyclone SH30070.03E, Thermo Scientific,USA), 0.1 mM 2-mercaptoethanol (Sigma Chemical CO. USA), 1× non-essential amino acid (Gibco11140, Invitrogen,CA, USA), 100×nucleosides (Specialty Media , Millipore, USA), 2 mM L-glutamine (Gibco25030, Invitrogen,CA, USA) and Penicillin/Streptomycin (Invitrogen,CA, USA) [32-34]. The cells were transduced with pBABE- hygro-SATB2 or empty Vector at days 0, 2, and 5 after culture with a hanging drop as described above and plated on a gelatin-coated dish on day 7. For in vitro osteogenesis, the iPS EB cells were collected by brief centrifugation, gently dissociated by 0.25% trypsin-EDTA treatment, seeded as a single cell suspension onto a gelatin-coating plate, and cultured in osteoblastic differentiation mediums, containing osteogenic supplements (OS) as follows: 50μg/mL ascorbic acid (Sigma, USA), 10 nM dexamethasone (Sigma, USA), and 5 mM β-glycerophosphate (Sigma, USA), respectively. The osteogenic medium was changed every 3 days and the cells were used for study after 7, 14, 21 days.
2.4. Teratoma formation and histological analysis
Mouse iPSCs were transduced with pBABE-hygro-Vector for 6 hours. After the cells were cultured for 3 days, the mouse iPSCs were suspended at 2×107 cells/mL in PBS. Nude mice (6~8 weeks; Charles Revier Breeding Labs, Boston, USA) were anesthetized with Ketamine and Xylazine. 100 μl of the cell suspension (2×106 cells) were subcutaneously injected into the backs of nude mice. Five weeks later, the tumors were surgically dissected from the mice. Samples were washed and fixed in 10% formalin solution, then decalcified with 18% ethylenediaminetetraacetic acid (pH 7.0-7.4) for one week, and embedded in paraffin wax. After sectioning 5 μm thick, the tissue was dewaxed in ethanol, rehydrated and stained with hematoxylin and eosin.
2.5. Biochemical assays
Alkaline phosphatase staining
Mouse iPSCs (1 × 106 cells) were plated on 6-well plates. On the following day, cells were transduced with pBABE-hygro-SATB2 or empty Vector for 6 hours. After the cells were cultured for 3 days, alkaline phosphatase staining was performed using an Alkaline Phosphatase Detection Kit (Chemicon, Temecula, CA) according to the manufacturer’s instructions[34].
Alizarin Red S staining and von Kossa assay
iPSCs plated in triplicate for different groups in 6-well plates were fixed in 70% ethanol for 14 days in differentiation medium after gene transduction. Alizarin Red S Staining was performed to analyze the newly formed nodules. Briefly, cells were washed with PBS twice and fixed with 10% formalin for 10 min at room temperature followed by several washes with distilled H2O. Cultures were then stained with 2% (w/v) Alizarin Red (Sigma-Aldrich, USA) at room temperature for 5 min followed by several washes with distilled H2O and viewed under a light microscope. Cells were stained with Von Kossa silver and placed under ultraviolet light for 10 min. Cells were then treated with 5% NaS2O3 for 2 min, and washed with distilled water. Calcium nodules with a diameter greater than 1 mm were counted and analyzed.
ALP activity assay
For the measurement of alkaline phosphatase (ALP) activity, cells were lysed in 10 mM Tris-HCl (pH 7.5) containing 1 mM MgCl2 and 0.1% Triton X-100, and the lysates were then used for assay. ALP activity was measured using the LabAssay ALP kit (Bioassay Systems, CA, USA) according to the manufacturer’s instructions. The protein concentration of the lysates was determined using a Bio-Rad assay kit (Bio-Rad laboratories, Hercules, CA), and ALP activity was then normalized by protein concentration.
2.6. Total RNA isolation, Reverse transcription-mediated polymerase chain reaction (RT-PCR) and real-time quantitative PCR (qRT-PCR) analysis
For both RT-PCR and qRT-PCR, total RNA of different cell groups were extracted using a RNeasy Kit (Qiagen, CA, USA) following the manufacturer’s instructions. RT-PCR was performed to determine the expression of genes regulating pluripotency at 0, 3, 5, and 10 days after differentiation: Nanog, Oct 3/4, and GFP. cDNA was synthesized using the SuperScript II reverse transcriptase (Invitrogen, Carlsbad, USA) and the oligo(dT) primer according to the manufacturer’s instructions. Primer sequences were shown in Table 1. The product was assessed by 2% agarose gel electrophoresis followed by ethidium bromide staining.
TABLE 1.
The sequences of the primers for RT-(q)PCR in the experiment.
| Primer | Sequence |
|---|---|
| Oct-3/4 | forward: 5′-GTTTGCCAAGCTGCTGAAGC-3′ |
| reverse: 5′-TCTAGCCCAAGCTGATTGGC-3′ | |
| Nanog | forward : 5′-ATGGTCTGATTCAGAAGGGC-3′ |
| reverse : 5′-TTCACCTCCAAATCACTGGC-3′ | |
| GFP | forward: 5′-CACATGAAGCAGCACGACTT-3′ |
| reverse: 5′-TGCTCAGGTAGTGGTTGTCG-3′ | |
| SATB2 | forward : 5′-GAGATGAGTTGAAGAGGGCTAGTG--3′ |
| reverse : 5′-CCCTGTGTGCGGTTGAAT -3 | |
| OSX | forward: 5′-ATGGCGTCCTCTCTGCTTG-3′ |
| reverse: 5′-TGAAAGGTCAGCGTATGGCTT-3 | |
| Runx2 | forward: 5′-CAGTCACCTCAGGCATGTCC-3′ |
| reverse: 5′-GTGCTGCTGGTCTGGAAGG-3′ | |
| BSP | forward: 5′-CAGGGAGGCAGTGACTCTTC-3′ |
| reverse: 5′-AGTGTGGAAAGTGTGGCGTT-3′ | |
| OCN | forward : 5′-GCGCTCTGTCTCTCTGACCT-3′ |
| reverse : 5′-GCCGGAGTCTGTTCACTACC-3′ | |
| HoxA2 | forward: 5′-CGGAGAATGAAGCATAAGAG-3′ |
| reverse : 5′-GCAGTTAGGAACAGTGGGT-3′ | |
| GAPDH | forward: 5′-AGGTCGGTGTGAACGGATTTG-3′ |
| reverse: 5′-TGTAGACCATGTAGTTGAGGTCA--3′ |
qRT-PCR was performed at 7, 14, and 21 days after differentiation, to compare the expression levels of genes associated with osteogenic differentiation, such as Osterix, Runx2, bone sialoprotein (BSP), Osteocalcin (OCN), and HoxA2. 1μg of RNA was reverse transcribed with a SuperScript first-strand synthesis system (Invitrogen, CA, USA) following the manufacturer’s recommendations, then set up to RT-PCR reactions utilizing iQ SYBR Green Supermix (Bio-Rad Laboratories, USA). Primer sequences were shown (Table 1), and GAPDH gene expression was detected for normalization purposes.
2.7. Silk Scaffold preparation and cell seeding
The water based silk fibroin scaffolds (pore size 500-600 microns, disk-shaped, 4 mm diameter and 2 mm thick) were prepared by our previously published procedures[35, 36]. For cell seeding, 14 days cultured in differentiation medium after gene transduction, iPSCs were released from the culture substratum using trypsin/EDTA (0.25% w/v trypsin, 0.02% EDTA) and concentrated to 2×107 cells/ml in serum-free medium. Then iPSCs were seeded onto the silk scaffold by pippetting the iPSCs suspension onto the materials. The iPSCs/SS (Silk Scaffold) construct was incubated for an additional 4 hours to allow cell attachment in vitro before implantation. In a parallel experiment, 3 mm×3 mm×3 mm cuboids were prepared and seeded with iPSCs at the same cell density. The extent of cell attachment and growth was assessed 4 hours and 5 days after cell seeding. The constructs were fixed in 2.5% glutaraldehyde (Sigma, USA) for 24 h at room temperature. After thoroughly washing with PBS, the cells adhered to the scaffold section, then were dehydrated in an ethanol-graded series (50-100%) for 10 min each and allowed to dry on a clean bench at room temperature[37] . The samples were cut into two halves, and then characterized by scanning electron microscopy (Philips SEM XL-30, Amsterdam, Netherlands) after platinum coating.
2.8. Surgical procedure
Six- to 8-week-old nu/nu mice (Charles Revier Breeding Labs, Boston, USA) were maintained and used in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals, prepared by the Institute on Laboratory Animal Resources, National Research Council (DHHS Publ. NIH 86-23, 1985). The Institutional Animal Use and Care Committee at the Tufts–New England Medical Center, in Boston, MA, approved the animal protocol[38].
The nu/nu mice were anesthetized, and a 4-mm-diameter calvarial critical-sized defect was created on each side of the calvarial bone using a dental bur attached to a slow-speed hand piece with minimal invasion of the Dura mater (Figure 4A). The critical-sized defects in mice were randomly divided into 5 groups to receive the following implants: (1) SS (Silk Scaffold) alone (n=7); (2) SS with iPSCs w/o (without) OS (n=7); (3) SS with iPSCs with OS (n=7); (4) SS with iPSCs transduced with empty Vector (n=7); (5) SS with iPSCs transduced with SATB2 (n=7).
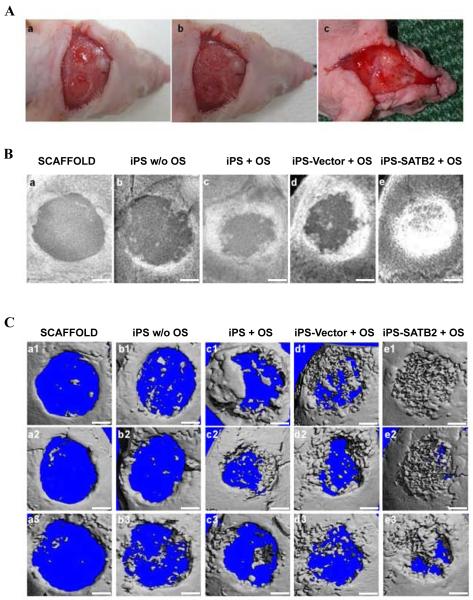
Figure 4. SATB2 promotes bone healing in the skull assay.
(A) Surgical procedure. (a) A non-healing full thickness defect of 4 mm diameter in the either side of the cranial bone was made; (b) the cranial defect was filled with a silk scaffold seeded with gene-modified iPSCs; (c) Representative picture of the healing wound 5 weeks after implantation. (B) Representative pictures showed large defined radiopacities at the defect sites in scaffold alone group and little calluses observed restrict to the defect margin in untransduced iPSCs derived from EB without OS, while an obviously superior extent in untransduced iPSCs derived from EB with OS group, and Vector-transduced iPSCs with OS group. Moreover, SATB2 transduction into iPSCs further closed the defects. (C) Microcomputer tomography (microCT) images of cranial bony defects taken 5 weeks after implantation. Scale bar =1 mm.
2.9. qRT-PCR of newly formed tissue
The mice were sacrificed 5 weeks post-surgery. Ten cranial samples were frozen immediately in liquid nitrogen and kept at −80°C for qRT-PCR. Tissues were homogenized in TRIzol solution (Invitrogen,CA, USA), followed by total RNA isolation procedure according to the manufacturer’s instructions. Freshly isolated RNAs were reverse transcribed into cDNAs then real-time quantitative RT-PCR (qRT-PCR) was performed, as we described previously [39]. The sequences of the primers were shown in Table 1.
2.10. Radiographic analysis and micro-CT measurement
Healing of calvarial defects was examined by a radiological exam 5 weeks after iPSCs implantation. Radiographs were taken with a Faxitron radiographic inspection unit (Faxitron X-ray Corp., Buffalo, IL). The kVP (230 V, 8 mA) and exposure time (1.8s) were kept consistent for all samples. The morphology of the reconstructed cranium was assessed using a micro-CT system (mCT-80, Scanco Medical, Bassersdorf, Switzerland). The CT settings were used as follows: pixel matrix, 1024×1024; slice thickness, 20μm; slice thickness, 20μm. After scanning, the micro-CT images were segmented using a nominal threshold value of 225 as reported previously[40], and a three dimensional (3D) histomorphometric analysis was performed automatically. The parameters of bone volume fraction (Bone volume/total volume, BV/TV), volume of newly regenerated bone, and bone mineral densities (BMD) were used for comparison in this study.
2.11. Histology and immunohistochemistry
For histological examination, cranial specimens were fixed in 10% formalin solution and then decalcified with 18% ethylenediaminetetraacetic acid (pH 7.0-7.4) for one week. Samples were then dehydrated in an ascending series of ethanol and embedded in paraffin wax. Five-μm sections were cut using a microtome and mounted on glass slides. Three randomly selected cross sections from each implant were stained with hematoxylin and eosin. H&E staining was performed using standard methods as previously described[38] .For immunohistochemical studies, expressions of BSP, SATB2, and HoxA2 were detected using immunohistochemical staining., Followed by counter-staining in hematoxylin with a Histostain SP kit (Invitrogen, CA, USA) for BSP, or with 3,3′-diaminobenzidine for SATB2 and HoxA2 and a BSP monoclonal antibody (1:25, Chemicon International, Temecula, CA), SATB2 polyclonal antibody (1:200, Santa Cruz Biotechnology, Inc., CA), and HoxA2 polyclonal antibody (1:100, Santa Cruz Biotechnology, Inc., CA) as previously described [38, 41]. H&E staining and immunostaining sections were examined and photographed with a Nikon Eclipse E600 microscope and Spot Advanced software (Diagnostic Instruments, Sterling Heights, MI). Newly formed bone in H&E-stained sections was quantified in 4 sections of at least 4 different defects for each treatment at 40× magnification using Spot Advanced software (Diagnostic Instruments, Sterling Heights, MI). New bone formation on each section was expressed as a percentage of the total area of the defect. The localization of BSP, SATB2 and HoxA2 staining was studied on transverse sections of the cranium as previously described[38]. All slides were coded to prevent the introduction of examiner bias.
2.12. Statistical analysis
All results are expressed as mean ± S.D. of the mean of 3 or more independent experiments. Statistically significant differences (p<0.05) between the various groups were measured using ANOVA. All statistical analysis was carried out using an SAS 6.12 statistical software package (SAS, Cary, NC, USA).
3. Results
3.1. No effect on the undifferentiated state and pluripotency of iPSCs by gene transduction
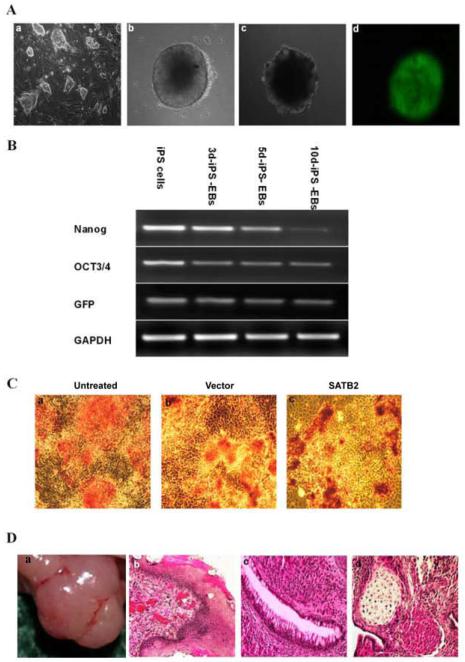
Mouse iPS cell clone ST-8 was cultured in the conventional conditions of ES medium, on the top of an MEF feeder cell layer. Embryoid body (EB) formation occurred from mouse iPSCs, after 48-h of drop-suspension and 7-d of suspension culture, which exhibited a regular round shape (Figure 1A). To investigate the properties of iPSCs under the present culture conditions, we first observed the level of cellular marker genes of iPSCs (Figure 1B). Semi-quantitative RT-PCR analysis showed strong expressions of Nanog and Oct-3/4 in iPSCs. Upon EB formation, the expression levels of Oct-3/4, Nanog, and GFP in iPSCs were decreased. Given that the Nanog promoter drove the green fluorescent protein GFP expression, the high level of GFP in the undifferentiated iPSCs was also reduced after EB formation. These results showed that the gene expression patterns of iPSCs were indistinguishable from those of ES cells.
Figure 1. Gene expression patterns of mouse iPS cells.
(A) Representative pictures of (a) mouse induced pluripotent (iPS) cells (ST-8) with the layer of inactivated MEF cells (200×); (b) Two days after culture with a hanging drop, representative pictures of mouse iPS embryoid body formation (EBs) (200×); (c) the EBs were transferred into a Petri dish and maintained for 5 days in suspension culture, and plated on gelatin-coated dish in differentiation medium on day 7(200×); (d) iPS cell-derived embryoid bodies expressed green fluorescent protein by fluorescence view(200×). The EBs were transduced with pBABE-hygro-SATB2 at days 0, 2, and 5 during suspension culture in the Petri dish. (B) Total RNA was isolated from mouse iPSCS (lane 1), 3d-iPS-EBs (lane 2), 5d-iPS-EBs (lane 3), or 10d-iPS-EBs (lane 4), and semi-quantitative RT-PCR was carried out using primers for Nanog, Oct-3/4, GFP and GAPDH as described in the Materials and Methods. (C) Mouse iPSCs were transduced with pBABE-hygro-Vector or pBABE-hygro-SATB2. Three days later, alkaline phosphatase (ALP) staining was observed by light microscopy(100×). (a) untreated; (b) Vector; (c) SATB2. (D) Teratomas formation derived from Vector-transduced iPSCs were prepared, the tumor was resected (a), and sections were stained with hematoxylin and eosin: (b), ectoderm (epidermis); (c), endoderm (gut epithelium); and (d), mesoderm (cartilage and muscle) (200×).
To confirm that the undifferentiated state and pluripotency in iPSCs were still maintained after gene transduction, we observed the alkaline phosphatase (ALP) expression in iPSCs on day 3 after transduction with pBABE-Hygro. As shown in Figure 1C, the staining of ALP in transduced cells (Figure1C-b and c) was similar as that in non-transduced cells (Figure1C-a). We also examined the pluripotency of pBABE-Hygro-transduced iPSCs by teratoma formation. Mouse iPSCs were transduced with pBABE-Hygro-Vector, and were then injected subcutaneously into the backs of nude mice. After subcutaneous transplantation, we obtained teratomas containing epidermis, cartilage, muscle and gut epithelial tissues (Figure 1D). These observations demonstrated that properties of iPSCs were not changed after transduction.
3.2. Osteogenic differentiation of iPSCs
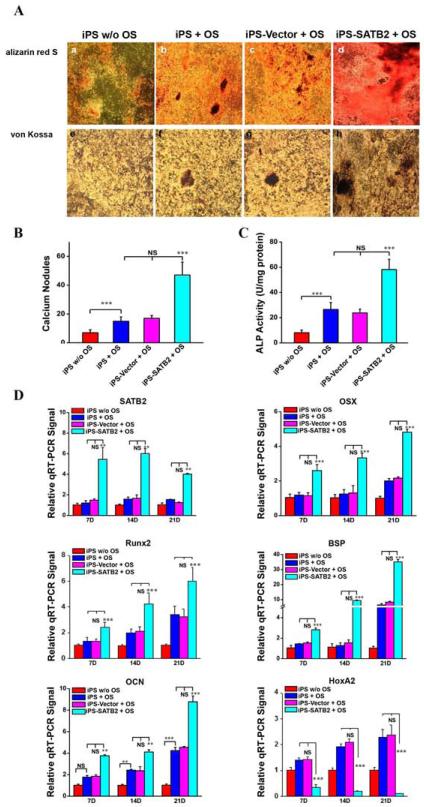
It has been known that iPSCs have the potential to differentiate toward mineralizing cells in vitro and SATB2, a nuclear matrix protein, acts as a molecular node in a transcriptional network regulating skeletal development and osteoblast differentiation. To investigate the effect of SATB2 on the iPSCs differentiation, iPSCs were transduced in triplicate with pBABE--Hygro-Vector or pBABE--Hygro-SATB2, and were cultured with or without osteogenic supplements (OS). We initially examined the mature osteoblast differentiation. Matrix mineralization in the cells was detected by alizarin red S staining (Figure 2A a-d) and von Kossa staining (Figure 2A e-h). The increase of alizarin red S staining and von Kossa staining in iPSCs by OS treatment was further enhanced by SATB2 transfection, suggesting that SATB2 transduction with OS treatment enhanced the matrix mineralization. Moreover, after SATB2 transduction, both of the calcium nodules and the ALP activity were enhanced in iPSCs (Figure 2B and 2C). The amount of calcium nodules in pBABE--Hygro-SATB2-transduced iPSCs was almost 3-fold more than that of non-transduced or pBABE--Hygro-Vector-transduced cells (Figure 2B). These results indicated that early osteoblast differentiation of iPSCs was promoted by SATB2. Real-time quantitative PCR (qRT-PCR) analysis also showed that the mRNA levels of SATB2, Osterix, Runx2, BSP, and OCN were up-regulated in the iPSCs transduced with SATB2, while the mRNA levels of HoxA2 decreased in vitro (Figure 2D). The decreased mRNA levels indicated that the osteogenic potential in iPSCs, and that efficient osteoblast differentiation from iPSCs could be achieved by exogenous SATB2 expression using optimized pBABE-Hygro-Vectors.
Figure 2. Enhanced osteoblast differentiation from mouse iPS cells by the transfection of SATB2 gene.
(A) Matrix mineralization in the cells was detected by Alizarin Red S and von Kossa assay in Vector-, SATB2- transduced cells and untransduced control cells after 14 days culture (a-h, 100×) with or without osteogenic supplements (OS). Quantitative analysis of calcium nodules (B) present and alkaline phosphatase (ALP) activity (C) in the four groups. A significant increase is seen in the SATB2 group when compared with the Vector and untransduced control groups. (D) Total RNA was isolated, and real-time quantitative PCR (RT-qPCR) was carried out using primers for SATB2, OSX, Runx2, BSP, OCN, HoxA2 and GAPDH. iPS w/o OS: non-treated iPS-EBs; iPS + OS: iPS-EBs with OS; iPS-Vector + OS: iPS-EBs with OS plus Vector; iPS-SATB2 + OS: iPS-EBs with OS plus SATB2; w/o: without. These data are expressed as the mean ± S.D. (n=3). ** p < 0.01, *** p < 0.001 compared with non-transduced or Vector -transduced iPS-EB cells. NS: no significant.
3.3. Growth of iPSCs on Silk Scaffold
Next, we investigated the iPS cellular attachment and interaction within 3D silk scaffolds through electron microscope. As shown in Figure 3A, cells were attached to the inner surface of the scaffold in vitro after the iPSCs were seeded onto the scaffold for 4-hours. Moreover, after 5 days of being cultured in media, the iPSCs were suspended among the backbones of scaffolds and grew along the pores of the scaffolds. The cells grew tightly to each other and reached confluence with abundant fibril networks of extracellular matrix deposited on the scaffolds (Figure. 3B). These results indicated that the biomaterial was suitable for the following in vivo studies given that it facilitated iPSCs initial attachment onto the surface, spreading and subsequent growth.
Figure 3. Scanning electron microscopic evaluation of the silk scaffold microstructure and cells state.
(A) 4 h after the iPS cells were seeded onto the scaffold; cells were seen attached to the inner surface of the scaffold. (B) 5 days after seeding, the iPSCs grew tight to each other and reached confluence with abundant fibril networks of extracellular matrix deposited on the scaffolds. Scale bar =30 μm.
3.4. Enhancement of bone healing by transduction of SATB2 in mice
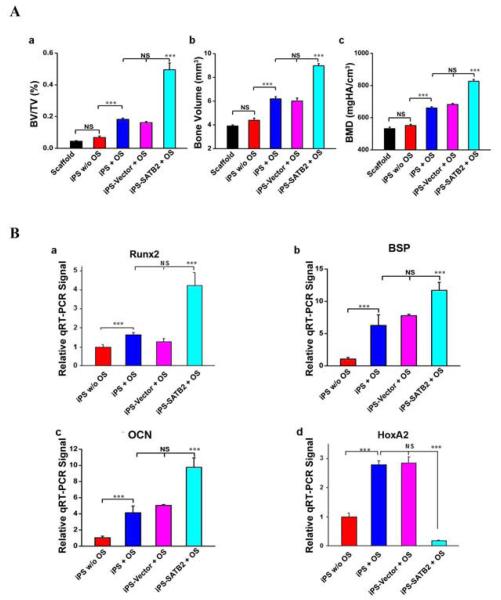
A non-healing full thickness defect of 4 mm in diameter was made in both sides of the cranial bone and filled with a silk scaffold seeded with gene-modified iPSCs (Figure 4A). To evaluate new bone formation and the development of bone unions within the defects, X-ray images were taken at 5 weeks after explantation of the crania. As shown in Figure 4B, a larger defined radio-opaque mass representing newly formed mineralized tissue was observed to fill in the defects of SATB2-transduced-iPSCs group when compared with the empty Vector-transduced or untransduced groups. Defects filled with scaffolds alone or iPSCs culturing without OS had little osseous healing and failed to show any appreciable new bone formation (Figure. 4B-a and b). A healing response and some mineralization within the defect were observed in the group transplanted with untransduced iPSCs culturing with OS (Figure. 4B-c) and empty Vector-transduced group (Figure. 4B-d), but to a lower extent when compared with that in the SATB2-transduced iPSCs group (Figure. 4B-e). The morphology of the newly formed bone was reconstructed using microCT imaging, and the representative images from each group were shown in Figure 4C. The majority of the cranial defects were filled with substantial newly formed bone tissue in the defect site treated with the SATB2-transduced iPSCs implants 5 weeks post-operation (Figure 4C-e). The implantation of untransduced iPSCs and the empty Vector-transduced iPSCs culturing with OS-seeded scaffolds also showed the formation of scattered new bone in the defect sites, while less than that in the SATB2-transduced iPSCs group (Figure 4C-c and d). No obvious evidence of new bone growth was observed in the defects treated with iPSCs culturing without OS or scaffold alone groups, and minor evidence of new bone was visible in the periphery of the defect edges 5 weeks post-operation (Figure 4C-a and b). To quantify the new bone regeneration within the calvarial defects, the ratio of bone volume to total volume (BV/TV), volume of regenerated bone and local bone mineral density (BMD) were measured and compared with normal calvarial bone (removed from the same parts of three 12-week-old nude mice without surgery). As shown in Figure 5A-a, BV/TV, as an indicator of the relative amount of newly formed bone, was significantly higher for the SATB2-transduced iPSCs group when compared to all other groups. Significantly more bone volume was observed in the SATB2-transduced iPSCs group, and BMD for SATB2- transduced iPSCs group were also higher than the other four groups (Figure 5A-b and c). Moreover, to further determine the effect of transduced iPSCs in transplantation during new bone formation, total RNA was isolated from five experimental groups, and qRT-PCR was performed to measure mRNA expression levels of Runx2, BSP, OCN, and HoxA2. As shown in Figure 5B, the Runx2, BSP or OCN mRNA expression showed a higher level, but HoxA2 mRNA expression showed a lower level in the defects with SATB2- transduced iPSCs group than the other three groups 5 weeks post-operation. These results demonstrated that iPSCs transduced with SATB2 and seeded on silk scaffold could efficiently enhance cranial critical-size bone healing in nude mice.
Figure 5. Analysis of new bone formation in mice at 5 weeks post-operation.
(A) (a) Morphometric analysis (BV/TV) of new bone formation, (b) volume of newly regenerated bone; (c) local bone mineral density (BMD) analysis by microCT of five groups at 5 weeks post-operation. Quantitative micro-CT analysis revealed that implantation of SATB2 transduced iPSCs/SS constructs achieved the most amount of new bone formation, volume of newly regenerated bone and the highest local BMD than other four groups, ***p<0.001, NS: no significant. (B) Total RNA was isolated from newly formed tissue, and RT-qPCR was carried out using primers for Runx2, BSP, OCN, HoxA2 and GAPDH.
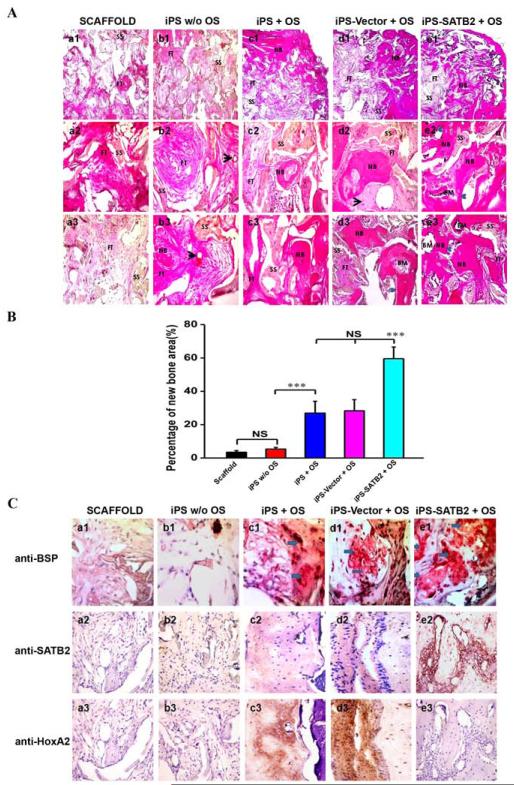
3.5. Histological analysis of bone regeneration
Histological evidence further supported the radiographic findings, indicating that the specimens from the SATB2 transduced group showed nearly complete osseous closure of the defect, where the newly formed bone had a typical organized and mature bone morphology with noticeable marrow spaces and similar to native bone (Figure 6A-e). In contrast, histological analysis indicated a small amount of irregularly arranged bone tissue and less bone formation in empty Vector-transduced iPSCs (Figure 6A-d) or untransduced iPSCs culturing with OS group (Figure 6A-c). Defects were filled with fibrous connective tissue and no obvious bone formation was found in the iPSCs culturing without OS group (Figure 6A-b) or scaffold alone group (Figure 6A-a). The percentage of new bone area after 5 weeks was 59.58±7.00% in the SATB2-transduced iPSCs implants group, 28.34±6.66% in the empty Vector-transduced iPSCs group, 26.94±7.10% in the untransduced iPSCs culturing with OS-seeded scaffolds group, 5.37±1.01% in the iPSCs culturing without OS group, and 3.43±1.12% in the scaffold alone group, respectively. The percentage of new bone area in the SATB2 group was significantly higher than the other four groups 5 weeks post-operation (Figure 6B). To further analyze the extent of de novo bone mineralization, immunohistochemistry was performed to determine BSP, SATB2, and HoxA2 expression levels. As shown in Figure 6C, immunohistochemistry exhibited strong expression for BSP or SATB2 in areas of new bone formation within the defect region from samples treated with the SATB2-transduced iPSCs, whereas BSP or SATB2 stainings were visible in empty Vector-transduced iPSCs and untransduced iPSCs culturing with OS groups, but much weaker. Moreover, the reduced HoxA2 staining was observed in SATB2-transduced iPSCs group compared with untransduced or Vector- transduced iPSCs group. No obvious positive staining was observed in the iPSCs culturing without OS group or scaffold alone group (Figure 6C).
Figure 6. Histological and Immunohistochemical analysis of new bone formation.
(A) The photomicrograph of the histological images of the implants represented the differences among five groups. The cross-section images of representative slices in (a1–a3) silk scaffold alone group, (b1–b3) untransduced iPSCs without OS, (c1–c3) untransduced iPSCs with OS, (d1–d3) Vector-transduced iPSCs with OS, and (e1–e3) SATB2 transduced iPSCs with OS. The whole images of representative slices in each group (a1~e1, 40×); The center photomicrograph of the defect sites in each group (a2~e2, 200×); The edge photomicrograph of the defect sites in each group (a3~e3, 200×). BM: bone marrow; FT: fibrous tissue; NB: new bone; SS: silk scaffold. Black arrow represents blood vessels. Blue arrows show the ingrowth of osteoblasts. (B) Histomorphometrical analysis of the bone formation for five groups at 5 weeks post-operation. ***p < 0.05. (C) Immunohistochemical analysis of new bone formation in each group at 5 weeks post-operation (a-e, 400×). BSP, SATB2 and HoxA2 staining demonstrates no obvious positive staining in scaffold alone (a1~3), untransduced iPSCs without OS group (b1~3), while a weak BSP and SATB2 staining in untransduced iPSCs +OS group (c1~2) and Vector- transduced iPSCs group (d1~2), and a stronger BSP and SATB2 expression was observed in both of the bone and surrounding fibroblastic-like tissue (arrows) from SATB2-transduced iPSCs group (e1~2). However, the HoxA2 positive staining was reduced in SATB2-transduced iPSCs group (e3) compared with untransduced iPSCs +OS group (c3) or Vector- transduced iPSCs group (d3).
4. Discussion
In the present study, we showed that SATB2 transduction increased the matrix mineralization, the calcium nodule formation, and the ALP activity of iPSCs in vitro. Moreover, five weeks after the implantion of SATB2-transduced iPSCs within 3D silk scaffolds, the newly formed bone showed 2 times as great as that in the empty Vector-transduced group, and 20 times as great as that in the scaffold group, suggesting that the overexpression of SATB2 facilitates the osteogenic differentiation of iPSCs in vitro and in vivo.
In the bone research field, tissue-engineering approaches are attempting to heal bone lesions above a critical size by using resorbable scaffolds supplemented with regeneration-competent cells [42]. The biodegradability, distinguishing mechanical properties, and low inflammatory response of silk fibroin make it one of the promising scaffolds for osteogenic applications [43, 44]. Except for osteoconductive scaffolds, both of osteoprogenitor cells and osteoinductive factors are the key elements for forming tissue-engineered bone. The aim of this research is to find the appropriate osteogenic cell type and osteoinductive factors.
The ground-breaking study of Takahashi and Yamanaka demonstrated that the reprogrammed somatic cells to a pluripotent state can be achieved by a simple retroviral over expression of specific transcription factors [9]. The generation of iPSCs and the latest development showing the production of iPSCs without intergrating factors [15, 45, 46], open up new opportunities for the establishment of clinically autologous stem cell lines. Recently, iPSCs have been shown to have the potential to differentiate into osteogenic lineage [24, 25, 34]. In the present study, the iPSCs treated with OS media also showed osteoblast differentiation in vitro (Figure 2). Compared with the group without OS treatment, OS-treated iPSCs showed more matrix mineralization and a higher expression level of the osteogenic genes, such as osterix, Runx2, BSP and OCN. Moreover, the newly formed bone was observed after implanting the OS-treated iPSCs into bone defects (Figure 4). This data suggests that iPSCs are valuable for bone cell replacement therapy. Firstly, the production of iPSCs is more convenient and flexible. Secondly, patient-specific iPSCs prevent the immunological rejection after cell engraftment. Thirdly, the generation of iPSCs no longer requires early embryos or germ cells, solving the ethical issues associated with ES cells. Recent reports have shown that iPSCs can be generated without using viral Vectors, which further bring iPSCs one step closer to clinical application [47, 48]. Although there are still other obstacles before iPSCs can be used for clinical application, studies directed to understanding factors that control differentiation of these cells toward osteogenic lineage are required for their future application.
Enhanced osteogenic differentiation of iPSCs induced by genetic modification could potentially generate a number of cell. Such genetic modifications have frequently been achieved by over-expression of individual regulatory proteins. SATB2, one of the nuclear matrix proteins and contains two central nuclear matrix attachment region (MAR) domains, may be such one. During skeletogenesis, SATB2 regulates other genes’ activities in osteoblast differentiation by binding to the specific MAR element[27-29]. Null mouse model and human mutations of SATB2 established that the protein is involved in craniofacial development and osteoblast differentiation. More recent data indicated that SATB2 was involved with the regulatory network for the miR cluster 23a~ 27a~24-2 in both progression and maintenance of the osteocyte phenotype [49]. To investigate the role of SATB2 in osteogenic differentiation of iPSCs, we over expressed SATB2 in iPSCs. After SATB2 transduction, the alizarin red S staining and von Kossa staining in iPSCs were increased (Figure 2A and B). At the same time, the ALP activity, the marker of early osteogenic differentiation, as well as mineralized nodules in iPSCs was also enhanced by SATB2 (Figure 2C and D). These data suggest SATB2 facilitates the differentiation of iPSCs to an osteogenic lineage. The previous experiments of chromatin immunoprecipitation and transactivation [29] showed that SATB2 could bind to and regulate Runx2, BSP and OCN genes, the critical components in osteoblast formation. Knockdown of SATB2 significantly reduced the expression of Runx2, BSP, and OCN [29]. Consistent with it, the levels of Runx2, BSP, and OCN genes were increased in SATB2-transfected iPSCs (Figure 2E). In another study about iPSCs, it was also reported that Runx2 expression was constantly expressed at higher levels for up to 3 weeks after iPSCs were drive to osteogenic lineage [25, 50]. Recent data from Koromi showed that Runx2 induces the differentiation of multipotent mesenchymal cells into immature osteoblasts, directing the formation of immature bone, while also inhibiting osteoblast maturation and mature bone formation [51]. In our previous study, we also observed that Runx2 enhances osteoblast differentiation at an early stage and inhibits the late stage of osteoblast maturation in bone marrow–derived mesenchymal stem cells (BMSCs) [38, 39]. The elevated level of Runx2 in the present study and other iPSC studies, may be due to the delayed maturation of osteoblasts derived from iPSCs. Moreover, we observed reduced HoxA2 mRNA in SATB2 group (Figure 2E, Figure 5B). Several studies have verified that SATB2 and HoxA2 are expressed in craniofacial tissues at the key stage of mouse lip and palate embryogenesis [26, 52]. Furthermore, SATB2 was reported to promote osteoblast differentiation through repressing HoxA2 expression [29] and HoxA2 has been shown to antagonize bone formation, including in the craniofacial region [53]. Therefore, we favor the view that SATB2 is a molecular node of a transcriptional network underlying skeletal development by regulating the expression or activity of multiple key determinants of this process, such as HoxA2, Runx2, BSP and OCN.
Importantly, we demonstrated that SATB2 transduction significantly enhanced osteogenic differentiation of transplanted iPSCs (Figure 4). The implantation of SATB2-transduced iPSCs-seeded scaffolds into the defects resulted in newly formed bone that was well integrated with the host bone. In addition, the bone volumes per trabecular volume (BV/TV) were higher, volume of regenerated bone and the local bone mineral density was increased compared with other four groups. Moreover, mature lamellar bone with Haversian systems and bone marrow was observed in the SATB2-transduced iPSCs group. In contrast, in the Vector-transduced group and the untransduced group, the newly formed bone was composed of irregular trabecular with large bone lacuna. These data suggest that SATB2-modified iPSCs promote new bone formation and maturation in calvarial defects created in the mice. The implantation of SATB2-transduced iPSCs did not form tumors in the syngenic mice, which indicates a stable phenotype and no reversion to an embryonic stage of differentiation. However, we observed the tumor formation in 2 mice among 7 mice implanted with iPSCs w/o OS. These data also suggest that the obstacle should be overcome before clinical application of iPSCs.
To date, three strategies are typically used to augment massive bone defect repair [54]. The first is to engraft mesenchymal stem cells (MSCs) onto a graft or a biosynthetic matrix to provide a viable osteoinductive scaffold material for segmental defect repair. The second strategy is to introduce critical factor(s), for example, BMPs, in the form of bone-derived or recombinant proteins onto the graft or matrix directly. The third strategy uses the targeted delivery of therapeutic genes that either transduce host cells in vivo or stably transduce cells in vitro for subsequent implantation in vivo. Our study indicated that MSCs-like cells can be generated from iPSCs transduced with SATB2 in the present of OS, which solving the immunological and ethical problem faced by ES cells researcher. These may contribute to the future application of iPSCs for musculoskeletal tissue repair and regeneration. The differences between MSC-like cells derived from iPSCs and bone-marrow derived MSCs (BM-MSCs) provide an argument for the use of iPSCs engineered cells. Lian et al. demonstrated that iPSCs derived MSC could proliferate to 120 population doublings while maintaining a normal karyotype, had a 10-fold greater level of telomerase activity and had more of a protective effect in a rodent model of hindlimb ischemia than BM-MSCs [55]. The difference between cell types was attributed to the increased ability of iPSCs derived MSCs to survive, engraft and promote de novo vasculogenesis and muscle differentiation [34, 56, 57].
5. Conclusion
In summary, our data demonstrate that ex vivo gene therapy of SATB2-modified iPSCs with silk scaffold can effectively promote bone regeneration in critical-sized calvarial defects. Although further studies are needed to elucidate the cell and molecular mechanisms by which SATB2 promotes bone formation. Our study provides a strong rationale for the development of ex vivo therapies using SATB2 to repair bone defects. In future, iPSCs may be applied to the treatment of osteoporosis and other orthopedic diseases by SATB2-induced osteogenic differentiation.
Acknowledgment
We appreciate the technical assistance of Dr. G.P Liu for micro-CT scanning, C. Preda for silk scaffold, and Dana Murray for her assistance in article preparation. The study was supported by NIH grant DE14537 and DE16710 to J.Chen, National Natural Science Foundation of China (30801301) and Jiangsu Government Scholarship for Overseas Studies to J.H-Ye, National Natural Science Foundation of China (30772430) to C.P-Zhang. National Key Basic Research Program of China (2011CBA01104) and National Natural Science Foundation of China (30871264) to J. Gao.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Damien CJ, Parsons JR. Bone graft and bone graft substitutes: a review of current technology and applications. J Appl Biomater. 1991;2(3):187–208. doi: 10.1002/jab.770020307. [DOI] [PubMed] [Google Scholar]
- 2.Bauer TW, Muschler GF. Bone graft materials. An overview of the basic science. Clin Orthop Relat Res. 2000;(371):10–27. [PubMed] [Google Scholar]
- 3.Khan SN, Cammisa FP, Jr., Sandhu HS, Diwan AD, Girardi FP, Lane JM. The biology of bone grafting. J Am Acad Orthop Surg. 2005;13(1):77–86. [PubMed] [Google Scholar]
- 4.Kainer MA, Linden JV, Whaley DN, Holmes HT, Jarvis WR, Jernigan DB, et al. Clostridium infections associated with musculoskeletal-tissue allografts. N Engl J Med. 2004;350(25):2564–2571. doi: 10.1056/NEJMoa023222. [DOI] [PubMed] [Google Scholar]
- 5.Stenderup K, Justesen J, Clausen C, Kassem M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33(6):919–926. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Quarto R, Thomas D, Liang CT. Bone progenitor cell deficits and the age-associated decline in bone repair capacity. Calcif Tissue Int. 1995;56(2):123–129. doi: 10.1007/BF00296343. [DOI] [PubMed] [Google Scholar]
- 7.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 8.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 11.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 13.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105(8):2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 16.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 17.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 18.Xu D, Alipio Z, Fink LM, Adcock DM, Yang J, Ward DC, et al. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci U S A. 2009;106(3):808–813. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460(7251):53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A. 2008;105(15):5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104(4):e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, Matsuoka S, et al. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118(5):498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- 23.Ebert AD, Yu J, Rose FF, Jr., Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao CL, Tai LK, Chiou SH, Chen YJ, Lee KH, Chou SJ, et al. Resveratrol promotes osteogenic differentiation and protects against dexamethasone damage in murine induced pluripotent stem cells. Stem Cells Dev. 2010;19(2):247–258. doi: 10.1089/scd.2009.0186. [DOI] [PubMed] [Google Scholar]
- 25.Fei G, Raehal K, Liu S, Qu MH, Sun X, Wang GD, et al. Lubiprostone reverses the inhibitory action of morphine on intestinal secretion in guinea pig and mouse. J Pharmacol Exp Ther. 2010;334(1):333–340. doi: 10.1124/jpet.110.166116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12(19):2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- 27.Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17(24):3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21(3):658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- 29.Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125(5):971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. stem cells. 2009;27(3):543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2(3):230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dani C, Smith AG, Dessolin S, Leroy P, Staccini L, Villageois P, et al. Differentiation of embryonic stem cells into adipocytes in vitro. J Cell Sci. 1997;110(Pt 11):1279–1285. doi: 10.1242/jcs.110.11.1279. [DOI] [PubMed] [Google Scholar]
- 33.Kawaguchi J, Mee PJ, Smith AG. Osteogenic and chondrogenic differentiation of embryonic stem cells in response to specific growth factors. Bone. 2005;36(5):758–769. doi: 10.1016/j.bone.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Tashiro K, Inamura M, Kawabata K, Sakurai F, Yamanishi K, Hayakawa T, et al. Efficient adipocyte and osteoblast differentiation from mouse induced pluripotent stem cells by adenoviral transduction. stem cells. 2009;27(8):1802–1811. doi: 10.1002/stem.108. [DOI] [PubMed] [Google Scholar]
- 35.Meinel L, Fajardo R, Hofmann S, Langer R, Chen J, Snyder B, et al. Silk implants for the healing of critical size bone defects. Bone. 2005;37(5):688–698. doi: 10.1016/j.bone.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Kim HJ, Kim UJ, Kim HS, Li C, Wada M, Leisk GG, et al. Bone tissue engineering with premineralized silk scaffolds. Bone. 2008;42(6):1226–1234. doi: 10.1016/j.bone.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh SH, Park IK, Kim JM, Lee JH. In vitro and in vivo characteristics of PCL scaffolds with pore size gradient fabricated by a centrifugation method. Biomaterials. 2007;28(9):1664–1671. doi: 10.1016/j.biomaterials.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Tu Q, Valverde P, Li S, Zhang J, Yang P, Chen J. Osterix overexpression in mesenchymal stem cells stimulates healing of critical-sized defects in murine calvarial bone. Tissue Eng. 2007;13(10):2431–2440. doi: 10.1089/ten.2006.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu B, Zhang J, Brewer E, Tu Q, Yu L, Tang J, et al. Osterix enhances BMSC-associated osseointegration of implants. J Dent Res. 2009;88(11):1003–1007. doi: 10.1177/0022034509346928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cacciafesta V, Dalstra M, Bosch C, Melsen B, Andreassen TT. Growth hormone treatment promotes guided bone regeneration in rat calvarial defects. Eur J Orthod. 2001;23(6):733–740. doi: 10.1093/ejo/23.6.733. [DOI] [PubMed] [Google Scholar]
- 41.Zhang JH, Wang J, Tang J, Barnett B, Dickson J, Hahsimoto N, et al. Bone sialoprotein promotes bone metastasis of a non-bone-seeking clone of human breast cancer cells. Anticancer Res. 2004;24(3a):1361–1368. [PubMed] [Google Scholar]
- 42.Petite H, Viateau V, Bensaid W, Meunier A, de Pollak C, Bourguignon M, et al. Tissue-engineered bone regeneration. Nat Biotechnol. 2000;18(9):959–963. doi: 10.1038/79449. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Kim HJ, Vunjak-Novakovic G, Kaplan DL. Stem cell-based tissue engineering with silk biomaterials. Biomaterials. 2006;27(36):6064–6082. doi: 10.1016/j.biomaterials.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Jiang X, Zhao J, Wang S, Sun X, Zhang X, Chen J, et al. Mandibular repair in rats with premineralized silk scaffolds and BMP-2-modified bMSCs. Biomaterials. 2009;30(27):4522–4532. doi: 10.1016/j.biomaterials.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 46.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25(10):1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 47.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107(46):19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bilousova G, Jun D Hyun, King KB, Delanghe S, Chick WS, Torchia EC, et al. Osteoblasts Derived from Induced Pluripotent Stem Cells form Calcified Structures in Scaffolds both in vitro and in vitro. stem cells. 2010 doi: 10.1002/stem.566. doi: 10.1002/stem.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komori T. Regulation of osteoblast differentiation by runx2. Adv Exp Med Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 52.Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, et al. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet. 2006;79(4):668–678. doi: 10.1086/508214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanzler B, Kuschert SJ, Liu YH, Mallo M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development. 1998;125(14):2587–2597. doi: 10.1242/dev.125.14.2587. [DOI] [PubMed] [Google Scholar]
- 54.Awad HA, Zhang X, Reynolds DG, Guldberg RE, O′Keefe RJ, Schwarz EM. Recent advances in gene delivery for structural bone allografts. Tissue Eng. 2007;13(8):1973–1985. doi: 10.1089/ten.2006.0107. [DOI] [PubMed] [Google Scholar]
- 55.Lian Q, Zhang Y, Zhang J, Zhang HK, Wu X, Lam FF, et al. Functional mesenchymal stem cells derived from human induced pluripotent stem cells attenuate limb ischemia in mice. Circulation. 2010;121(9):1113–1123. doi: 10.1161/CIRCULATIONAHA.109.898312. [DOI] [PubMed] [Google Scholar]
- 56.Mizuno Y, Chang H, Umeda K, Niwa A, Iwasa T, Awaya T, et al. Generation of skeletal muscle stem/progenitor cells from murine induced pluripotent stem cells. FASEB J. 2010;24(7):2245–2253. doi: 10.1096/fj.09-137174. [DOI] [PubMed] [Google Scholar]
- 57.Taura D, Noguchi M, Sone M, Hosoda K, Mori E, Okada Y, et al. Adipogenic differentiation of human induced pluripotent stem cells: comparison with that of human embryonic stem cells. FEBS Lett. 2009;583(6):1029–1033. doi: 10.1016/j.febslet.2009.02.031. [DOI] [PubMed] [Google Scholar]