Summary
Background
The conserved DOS motif proteins OSM-7 and OSM-11 function as co-ligands with canonical DSL ligands to activate C. elegans Notch receptors during development. We report herein that Notch ligands, co-ligands and the receptors LIN-12 and GLP-1 regulate two C. elegans behaviors: chemosensory avoidance of octanol and quiescence during molting lethargus.
Results
C. elegans lacking osm-7 or osm-11 are defective in their response to octanol. We find that OSM-11 is secreted from hypodermal seam cells into the pseudocoelomic body cavity and acts non-cell autonomously as a diffusible factor. OSM-11 acts with the DSL ligand LAG-2 to activate LIN-12 and GLP-1 Notch receptors in the neurons of adult animals,- thereby regulating octanol avoidance response. In adult animals, over-expression of osm-11 and consequent Notch receptor activation induces anachronistic sleep-like quiescence. Perturbation of Notch signaling altered basal activity in adults as well as arousal thresholds and quiescence during molting lethargus. Genetic epistasis studies revealed that Notch signaling regulates quiescence via previously identified circuits and genetic pathways including the egl-4 cGMP-dependent kinase.
Conclusions
Our findings indicate that the conserved Notch pathway modulates behavior in adult C. elegans in response to environmental stress. Additionally, Notch signaling regulates sleep-like quiescence in C. elegans suggesting Notch may regulate sleep in other species.
Introduction
Notch signaling is a highly conserved signaling pathway with well-characterized roles in development and cell fate specification. In the canonical Notch pathway, transmembrane Notch receptors are activated by binding to transmembrane ligands containing a highly conserved DSL (Delta, Serrate, and LAG-2) domain. Activated Notch receptors are cleaved by the gamma secretase/presenilin complex and the intracellular (IC) domain of the receptor translocates to the nucleus. Notch IC then acts in conjunction with a transcription factor called Suppressor of Hairless in Drosophila or LAG-1 in C. elegans, and transcription of target genes is activated [1]. The two C. elegans Notch receptors LIN-12 and GLP-1 play well-characterized roles during C. elegans development. For example, LIN-12 plays critical roles at multiple steps of vulval cell fate specification and differentiation [2], whereas GLP-1 signaling negatively regulates mitotic exit/meiotic entry in the germline [3].
The C. elegans proteins encoded by osm-7, osm-11, dos-1, dos-2 and dos-3 are predicted to encode secreted or transmembrane proteins containing an EGF repeat conforming to the DOS-motif consensus (Delta and OSM-11) [4]. DOS motifs are also found in canonical DSL ligands of other metazoans [4]. In C. elegans, OSM-11 acts with DSL ligands in vulval cell fate specification to help activate LIN-12 Notch [4]. As C. elegans DSL-domain ligands lack the DOS motif, they may function as bipartite ligands with DOS-motif proteins to activate C. elegans Notch receptors. DOS-motif proteins also play poorly understood non-developmental roles. Loss of OSM-11 or OSM-7 causes defects in osmotic avoidance behavior, decreases defecation rates, increases internal osmolyte levels, increases sensitivity to anoxia [5], and alters expression of specific innate immunity proteins [6–8].
Several lines of evidence suggest that Notch signaling plays a non-developmental role in adult nervous systems across species. Notch receptors are expressed in the adult nervous systems of mammals [9], Drosophila [10], and C. elegans [11, 12] (this study). Altering Notch signaling changes neuronal activity [13, 14] and this affects various processes including spatial learning, memory, long term potentiation and neuromuscular junction synaptic plasticity [10, 15–18]. In C. elegans, LIN-12 Notch receptor function is required in the RIG interneurons of adult C. elegans to regulate the rate at which animals spontaneously initiate backward locomotion (i.e., reversals) during forward locomotion [11].
Herein we describe roles for the Notch pathway in two C. elegans behaviors. First, Notch ligands function in adult animals to activate GLP-1 and LIN-12 Notch receptors expressed in neurons to modulate chemosensory avoidance of the odorant 1-octanol. Second, Notch ligands and receptors regulate quiescence during C. elegans molting lethargus, a sleep-like behavior. Decreased Notch signaling reduces arousal thresholds during the L4-to-adult molt and ectopic expression of OSM-11 in adult animals is sufficient to induce anachronistic quiescence, which is dependent on the cGMP-dependent kinase EGL-4 PKG and on the LIN-3/LET-23 EGF pathway. Combined, these and other results presented herein reveal hitherto unsuspected roles for Notch signaling regulating C. elegans behavior.
Results
The Notch co-ligand OSM-11 and related DOS-motif proteins are required for normal octanol response
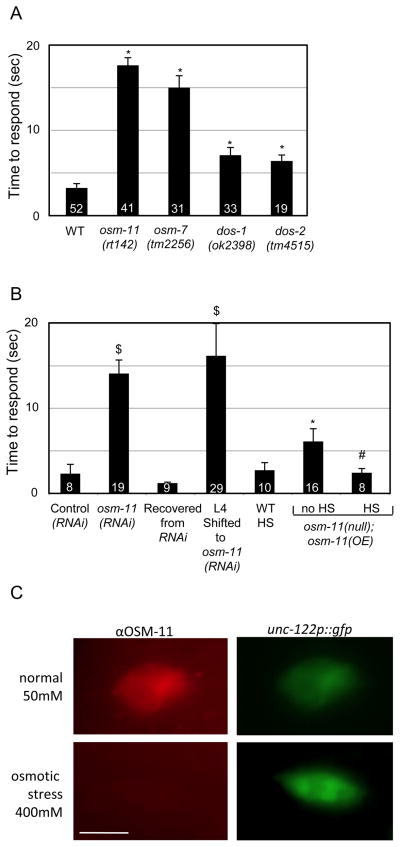
Wild type C. elegans avoid the noxious chemical odorant 1-octanol by rapidly initiating backward locomotion [19, 20]. We found that osm-11(rt142) complete loss of function (null) animals were defective in octanol response (Fig. 1A). Another DOS gene was required for octanol response; osm-7(tm2256) null animals were as defective in octanol response as animals lacking osm-11. By contrast, dos-1(ok2398) null and dos-2(tm4515) loss of function animals were only modestly defective (Fig. 1A). We confirmed these results by RNAi knockdown (see Supplemental data, Fig. S1A). Octanol response defects were not further exacerbated in osm-7;osm-11 double mutants (data not shown). Combined, these results suggest that osm-11 and osm-7 Notch co-ligand genes play essential and non-redundant roles in chemosensory avoidance of octanol; we focused here on the role of osm-11.
Figure 1.
C. elegans DOS-motif proteins are required for response to octanol. (A) Octanol responses of animals carrying the likely null alleles osm-11(rt142), osm-7(tm2256), dos-1(ok2398), and partial loss of function allele dos-2(tm4515). *p<0.0005 vs. wild type (WT). (B) osm-11 function is not required during development for octanol response in adults. $p<10−4 vs. control (RNAi). *p>0.05 vs. wild type, #p>0.05 vs. osm-11(null);osm-11(OE) no HS. (C) OSM-11 protein accumulates in coelomocytes. osm-11p::gfp reporter constructs do not drive GFP expression in coelomocytes [4]. Top left: Representative image of OSM-11 accumulation in a coelomocyte using αOSM-11 antisera [4]. Bottom left: Osmotic stress likely results in diminished OSM-11 secretion by seam cells into pseudocoelom. Top and bottom right: GFP expression (unc-122p:: gfp) in coelomocytes [53]. Scale bar=6 μm. See Supplemental Methods 1 for details.
We find that osm-11 acts in adult animals to modulate behavior. No morphological defects were observed in the octanol-sensing chemosensory amphid neurons in osm-11(null) animals (data not shown) consistent with a non-developmental role. To confirm that osm-11 function in adult animals was sufficient to regulate octanol response, osm-11 levels were manipulated in adult animals. First, wild type animals were reared on E. coli expressing double-stranded RNA corresponding to osm-11 (osm-11(RNAi)); these animals were severely defective in octanol response. However, when moved as adults to standard, non-RNAi E. coli, octanol response was fully restored (Recovered from RNAi, Fig. 1B). In a reciprocal experiment, wild type animals were transferred to plates containing E. coli expressing osm-11(RNAi) as L4 larvae; as adults, these animals were defective in octanol response (Fig. 1B). Finally, an inducible heat shock promoter was utilized to express osm-11 cDNA (hsp::osm-11) after cell fate specification was complete. Induction of osm-11 expression fully restored normal octanol response within hours (Fig. 1B). Combined, these results indicate that the neural circuitry required for response to octanol is intact in animals lacking osm-11 and that osm-11 function is required only in adult animals for response to octanol.
Previous work has shown that osm-11 animals have increased internal glycerol levels reminiscent of animals adapted to high external osmolarity [7]. To determine if osm-11 regulation of octanol response is dependent on glycerol accumulation, the octanol response in osm-11 animals lacking the glycerol biosynthesis genes gpdh-1 and gpdh-2 was examined. The loss of both gpdh-1 and gpdh-2 did not suppress the octanol response defect of osm-11 animals (Fig. S1B), suggesting that this behavioral defect may be independent of glycerol accumulation.
osm-11 acts non-cell autonomously to regulate octanol avoidance in adults
osm-11 is expressed in the hypodermal seam cells and spermathecae of adult animals, but is not expressed in neurons [4]. The seam cells lie along the lateral sides of the body contacting both the cuticle and the body cavity (pseudocoelom); they help secrete cuticle at each molt, but their roles in adult animals remain unexplored. Expression of osm-11 cDNA in seam cells and hypodermal tissues [21] restored octanol response in osm-11(null) animals (wrt-6p::osm-11, Fig. S1C). In tissue culture studies, OSM-11 is a secreted protein. Furthermore, in vivo OSM-11 can diffuse within the body cavity to activate Notch signaling at distant sites in developmental contexts [4]. We detected OSM-11 immunoreactivity in coelomocytes (Fig. 1C), which are endocytic cells that clear secreted proteins from the pseudocoelom. Coelomocyte clearance of secreted proteins requires the function of the cup-4 and cup-5 genes. Loss of cup-4 function dramatically decreases coelomocyte endocytosis while loss of cup-5 function results in aberrant accumulation of internalized proteins in enlarged endocytic vacuoles [22, 23]. OSM-11 immunoreactivity in coelomocytes was eliminated in cup-4 animals and accumulated aberrantly in cup-5 animals. However, OSM-11 immunoreactivity in seam cells was unchanged (data not shown). As the osm-11 promoter does not drive expression in coelomocytes [4], OSM-11 is likely secreted into the pseudocoelom, diffuses, and accumulates in coelomocytes.
Either prolonged exposure to high external osmolarity, loss of osm-11, or loss of osm-7 results in increased levels of the osmolyte glycerol and behavioral changes [5]. We found that when C. elegans are reared on standard plates containing 51 mM NaCl, all coelomocytes contained OSM-11 immunoreactivity (Fig. 1C), but rearing C. elegans on 400 mM NaCl plates decreased or eliminated OSM-11 immunoreactivity, whereas immunoreactivity in the seam cells was not altered (data not shown). This suggests that osmotic stress may diminish OSM-11 secretion by the seam cells into the body cavity. OSM-11 likely modulates octanol response, and perhaps physiological changes, in a non-cell autonomous manner. Additional rescue experiments support this conclusion (Fig. S1C). Taken together, these results suggest that OSM-11 is a secreted, diffusible protein released into the body cavity that modulates octanol response in a non-cell autonomous manner.
Notch receptors are required for octanol avoidance
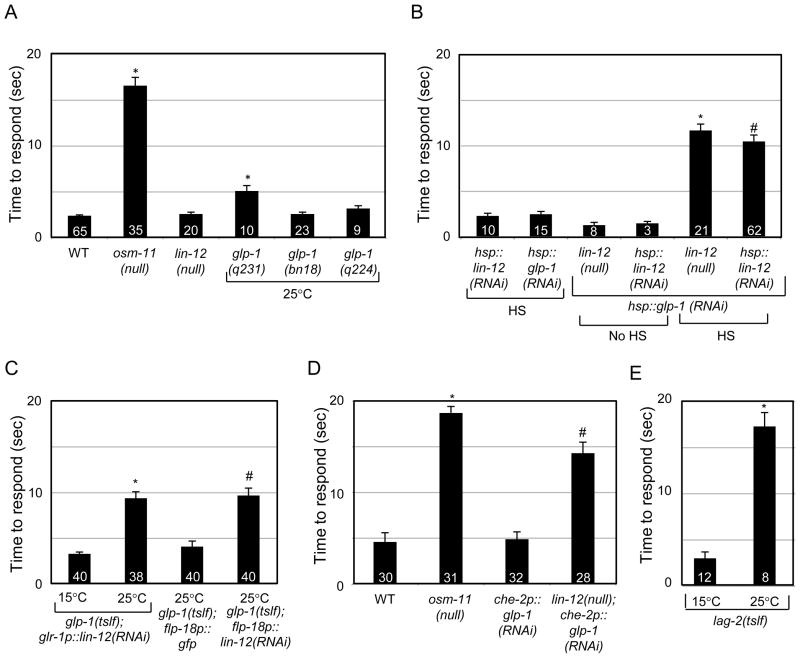
As OSM-11 is required for octanol response, the canonical Notch signaling pathway might play a hitherto unsuspected role in C. elegans chemosensory behavior. Therefore, the role of C. elegans Notch receptors LIN-12 and GLP-1 in octanol avoidance was examined using animals harboring the lin-12(n941) null allele or any of three glp-1 temperature sensitive, partial loss of function (tslf) alleles. lin-12(null) animals responded robustly to octanol. Two strains of animals carrying glp-1(tslf) alleles that were shifted to the restrictive temperature as adults had normal responses and animals carrying the third allele had only slightly impaired response (Fig. 2A). lin-12 and glp-1 are tightly linked (<22 kb apart) and loss of both Notch receptors results in embryonic lethality [24]. Therefore, a combination of transgene-based RNAi and/or mutant alleles was used to simultaneously decrease activity of both C. elegans Notch receptors.
Figure 2.
C. elegans Notch receptors and DSL ligand are required for normal octanol response. (A) Decreasing function of either Notch receptor gene alone does not dramatically impair octanol response. *p<10−11 vs. wild type (WT). (B) Knockdown of both Notch receptors impairs octanol response in adult animals #p<0.05 vs. hsp::lin-12(RNAi) HS, *p<0.05 vs. lin-12(null). (C) Notch receptors are required in neurons. lin-12 function is required in a subset of interneurons for normal octanol response. lin-12(RNAi) in glr-1 and flp-18-expressing neurons impairs octanol response. *p<10−11 vs. animals at 15°C. #p<10−4 vs. glp-1(tslf);flp-18p::gfp. (D) glp-1 functions in ciliated sensory neurons to regulate octanol response. glp-1(RNAi) in che-2-expressing ciliated sensory neurons impairs octanol response in lin-12(null) animals. *p<10−12 vs. WT; #p<10−8 vs. che-2p::glp-1(RNAi). (E) lag-2 function is required in adult animals for octanol response. *p<10−7. See Supplemental Methods 2 for details.
Induction of either glp-1 or lin-12 RNAi knockdown using a heat shock promoter in otherwise normal adult animals (hsp::glp-1(RNAi) and hsp::lin-12(RNAi), respectively) alone did not alter octanol response (Fig. 2B), consistent with the relatively normal response of lin-12 and glp-1 loss of function animals. However, RNAi knockdown of glp-1 in lin-12(null) animals dramatically impaired octanol response after heat shock induction. Similarly, RNAi knockdown of lin-12 in glp-1(tslf) animals impaired octanol response when animals were shifted to the restrictive temperature (Fig. 2C & data not shown). Additionally, transgenic animals in which lin-12 and glp-1 could be simultaneously knocked down in adults (hsp::glp-1(RNAi); hsp::lin-12(RNAi)) were defective in octanol response several hours after heat shock induction (Fig. 2B). These results suggest that both LIN-12 and GLP-1 Notch receptors play a role in adult animals modulating response to octanol, but the two receptors function redundantly.
Notch receptors act in non-overlapping subsets of neurons
Where might Notch receptors function to regulate octanol response? LIN-12 is required for proper development of the somatic gonad and GLP-1 is required for germ cell proliferation, but laser ablation of the gonad did not result in octanol response defects (Fig. S1D). We considered whether C. elegans Notch receptors might function in the nervous system to regulate behavior. lin-12p::gfp is expressed in RIG interneurons and glr-1p::lin-12(RNAi) knockdown of lin-12 in glr-1-expressing interneurons (which includes RIG) is sufficient to replicate lin-12 loss-of-function defects in reversal rates during locomotion [11]. glr-1p::lin-12(RNAi) in glp-1(tslf) animals at the permissive temperature (15°C) did not perturb octanol avoidance. However, glr-1p::lin-12(RNAi) significantly impaired octanol response in glp-1(tslf) animals raised at the restrictive temperature (25°C), consistent with lin-12 acting in glr-1-expressing neurons (Fig. 2C) [25, 26]. To further narrow down the site of lin-12 action, the flp-18 promoter was used to drive lin-12(RNAi) in AVA, RIM, AIY and RIG interneurons along with M2 and M3 pharyngeal neurons. While the control flp-18p::gfp transgene had no effect in glp-1(tslf) animals, the flp-18p::lin-12(RNAi) transgene perturbed octanol response in glp-1(tslf) animals raised at the restrictive temperature (25°C). Combined, these results indicate that lin-12 is likely required for octanol response in AVA, RIM, and/or RIG interneurons, which express both glr-1 and flp-18 (Fig. 2C).
Redundant function of the two Notch receptor genes could be explained by their expression in the same cells or by action in different parts of the response circuit. Our results suggest that the GLP-1 Notch receptor is required in a different set of neurons than LIN-12 for octanol response. We generated a glp-1p::gfp reporter construct that was expressed in many neurons, including the ciliated ASH, AWB and ADL sensory neurons that detect octanol (Fig. S2A). To determine the site of glp-1 function, glp-1 was knocked down by RNAi using the osm-10 promoter (for ASH, ASI, PHA and PHB), the gpa-11 promoter (for ASH and ADL [27]), and the che-2 promoter (for all ciliated sensory neurons including ASH, ADL, and AWB [28]). None of these promoters drive expression in AVA, RIM or RIG interneurons where lin-12 likely acts (see Fig. S6). RNAi knockdown of glp-1 in osm-10 and gpa-11 expressing neurons only modestly affected octanol response in lin-12(null) animals (Fig. S2B). However, RNAi knockdown in the larger set of che-2-expressing neurons dramatically impaired octanol response in lin-12(null) animals (Fig. 2D). These results suggest that glp-1 normally acts in che-2-expressing ciliated neurons to modulate octanol response and that lin-12 is primarily required in AVA, RIM, and/or RIG interneurons. The redundant function of Notch receptors is likely due to properties of the neural circuit and not due to co-expression of the two genes in the same neurons.
Notch ligands act via Notch receptors to regulate octanol avoidance
If LIN-12 and GLP-1 Notch receptors are activated by OSM-11 to regulate octanol response, then increasing Notch signaling should ameliorate osm-11 loss of function defects. We utilized two Notch gain of function alleles: lin-12(n137n460), a cold-sensitive gain of function (csgf) allele, and glp-1(ar202), a temperature-sensitive gain of function (tsgf) allele to test this hypothesis. The gain of function alleles had little or no effect on octanol response in otherwise normal animals regardless of temperature (Fig. S2C). Both the receptor gain of function alleles partially suppressed the octanol response defect of osm-11(null) animals at the restrictive temperature (Fig. S2C), suggesting OSM-11 acts via the Notch receptors.
Canonical DSL domain Notch ligands, such as LAG-2, activate Notch receptors in concert with DOS-motif proteins in developmental contexts [4]. The cellular expression pattern of a lag-2p::gfp reporter [29] suggested that lag-2 is expressed in many neurons of adult C. elegans including the AVA command interneurons that regulate locomotion and help initiate reversals in response to aversive stimuli (Fig. S2D). We found that octanol response requires lag-2 function. lag-2(q420tslf) animals were defective in their response to octanol at the restrictive temperature, but responded normally when reared at the permissive temperature (Fig. 2E). Taken together, these results suggest that the DSL domain ligand LAG-2 and the DOS-motif co-ligand OSM-11 both contribute to activation of neuronal Notch receptors in adult animals to modulate octanol response.
Identification of neuronal Notch targets
Direct transcriptional targets of the Notch pathway that are expressed in neurons would be excellent candidates for downstream targets of Notch signaling in octanol avoidance pathways, but few functional targets of Notch receptors beyond HLH transcription factors have been identified. A previous study identified a list of 163 likely Notch target genes with 4 or more LAG-1 consensus binding sites in upstream or intronic regulatory sequences [30]. Of these, only 22 were likely or definitely expressed in neurons based on known expression patterns or predicted protein function. Two of these genes egl-4 and lst-1, acted downstream of Notch receptors based on genetic criteria.
egl-4 encodes a cGMP-dependent protein kinase (PKG) that is expressed in many head neurons and has been previously implicated in several C. elegans behaviors [31–34]. egl-4(n479) loss of function animals respond normally to octanol (data not shown). However, the gain-of-function egl-4(ad450) allele partially suppressed the octanol response defect of osm-11(null) animals in double mutant animals (Fig. S2E). This result is consistent with egl-4 acting, directly or indirectly, as one of several downstream targets of Notch receptor activation.
lst-1 is a direct transcriptional target of lin-12 Notch signaling in vulval cell fate specification and encodes a protein lacking vertebrate orthologs [30]. lst-1p::gfp is expressed in many C. elegans head neurons, including the octanol-sensing AWB neurons [35]. Animals lacking lst-1 were as defective in their octanol response as osm-11(null) animals (Fig. S2E). As the lin-12(csgf) allele suppressed the octanol response defect of lst-1(null) animals, we conclude that lin-12 likely acts via additional target genes to regulate octanol response.
OSM-11 over-expression induces quiescence in adults via Notch receptors
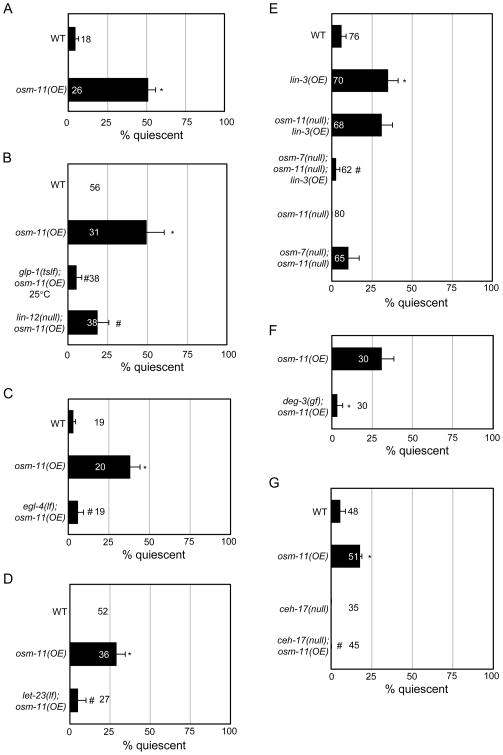
Adult C. elegans over-expressing osm-11 (osm-11(OE)) via the hsp::osm-11 transgene exhibited uncharacteristic spontaneous and transient bouts of quiescence after heat shock induction, during which locomotion and pharyngeal pumping ceased for several minutes regardless of the presence of food (Fig. 3A). Control animals did not display noticeable changes in behavior after a one hour recovery from heat shock. Between quiescent bouts, osm-11(OE) animals moved with coordinated sinusoidal locomotion and had normal pharyngeal pumping rates.
Figure 3.
OSM-11 over-expression induces quiescence in adults. In all panels, osm-11 was expressed under the heat shock promoter (hsp::osm-11). (A) OSM-11 over-expression (OE) induces spontaneous, transient and anachronistic quiescence in adult animals. *p<10−5 vs. WT. (B) osm-11(OE)-induced adult quiescence requires Notch receptor function. glp-1(tslf) or lin-12(null) suppress osm-11(OE) induced quiescence. *p<0.05 vs. WT; #p<0.05 vs. osm-11(OE). (C) OSM-11 over-expression induced quiescence is suppressed by egl-4 (lf). *p<0.0001 vs. wild type (WT); #p<0.05 vs. osm-11(OE). (D) Loss of LET-23 EGF receptor function (let-23(lf)) suppresses osm-11(OE)-induced quiescence. *p<0.001 vs. WT; #p<0.05 vs. osm-11(OE). (E) Simultaneous loss of osm-7 and osm-11 suppresses LIN-3 EGF over-expression (lin-3(OE))-induced adult quiescence. *p<0.01 vs. WT; #p<0.05 vs. lin-3(OE). (F) deg-3-induced neurodegeneration suppresses osm-11(OE)-induced quiescence. *p<0.05. As the locomotion of deg-3(u662) animals is severely uncoordinated, pharyngeal pumping was used to assess quiescence. (G) Defective axon outgrowth in ceh-17-expressing neurons suppresses osm-11(OE)-induced quiescence. ceh-17(null) affects processes of ALA and a subset of other neurons. *p<0.05 vs. WT; #p<0.001 vs. osm-11(OE). See Supplemental Methods 3 for details.
The quiescence of osm-11(OE) animals was similar to the sleep-like quiescence observed in C. elegans during larval molting lethargus [36, 37] in four ways. First, the quiescent state was transient. Second, animals entered and exited the quiescent state spontaneously. Third, quiescent osm-11(OE) animals could be “woken” by prodding with a metal wire. This perturbation elicited short bursts of coordinated sinusoidal locomotion. Fourth, osm-11(OE) animals exhibited increased arousal thresholds to mechanosensory stimulation during quiescent bouts, but in between quiescent bouts they responded normally. During ectopic quiescence, osm-11(OE) animals failed in 47±9% of trials to respond to body touch. By contrast, only 10±0% of heat-shocked wild type or 8±4% of non-heat-shocked osm-11(OE) animals failed to respond (n=30, p<0.01 for osm-11(OE) HS vs. WT or no HS). Similar results were observed using dilute octanol as a stimulus (data not shown). The diminished sensory responses during quiescence suggested that over-expression of OSM-11 might anachronistically activate the quiescence observed during C. elegans molting lethargus.
We did not observe inappropriate quiescence in transgenic adult animals overexpressing Notch receptors. To confirm that osm-11 over-expression induced inappropriate quiescence by activating Notch receptors, osm-11(OE) quiescence was examined in animals lacking Notch receptor function. We found that either loss of the lin-12 Notch receptor or diminished function of the glp-1 Notch receptor suppressed osm-11(OE) induced locomotion and pumping quiescence (Fig. 3B). These results suggested that increasing osm-11 levels inappropriately activates Notch signaling resulting in behavioral quiescence.
OSM-11 induced quiescence requires previously defined quiescence pathways
Does osm-11 over-expression induce quiescence through signaling pathways previously implicated in C. elegans molting lethargus quiescence? C. elegans quiescence requires the function of the EGL-4 protein kinase G/cGMP dependent kinase (PKG) [36, 37]. We found that loss of egl-4 suppressed osm-11(OE) induced quiescence (Fig. 3C); this is consistent with OSM-11 over-expression leading to inappropriately increased egl-4 activity with consequent quiescence in adult animals.
Endogenous C. elegans molting quiescence also requires LIN-3 EGF and LET-23 EGF receptor function [37]. LET-23 receptor function is required in the ALA neuron. Loss of let-23, the ALA neuron, or ALA processes is sufficient to decrease quiescence induced by LIN-3 EGF ligand over-expression [37]. Loss of the LET-23 EGF receptor suppressed osm-11(OE)-induced locomotion and pumping quiescence in adult animals (Fig. 3D), suggesting that EGF receptor signaling is required for Notch-induced quiescence. We also examined the relationship between osm-11 and lin-3. Over-expression of lin-3 (lin-3(OE)) in transgenic adult animals using the heat shock promoter (hsp::lin-3) causes anachronistic quiescence [37] reminiscent of quiescence induced in osm-11(OE) animals. We found that loss of osm-11 was not sufficient to suppress lin-3OE-induced quiescence, but simultaneous loss of both osm-11 and osm-7 dramatically suppressed lin-3OE-induced quiescence (Fig. 3E). Thus, osm-7 and osm-11 may play redundant roles in lin-3(OE)-induced anachronistic quiescence.
The role of the ALA neuron in osm-11 over-expression quiescence was examined using two strategies. First, the ALA neuron (among others) was genetically ablated in deg-3(u662) animals [38]. Inappropriate osm-11-induced quiescence was suppressed in deg-3(u662);osm-11(OE) animals (Fig. 3F). Second, ALA (and SIA) processes were perturbed in the ceh-17(np1) background [39], which also suppressed osm-11(OE)-induced quiescence (Fig. 3G). Combined, these results suggest that Notch-induced quiescence in adult C. elegans requires the ALA neuron and EGF signaling that normally regulate quiescence during molting lethargus.
Notch signaling alters quiescence during molting lethargus
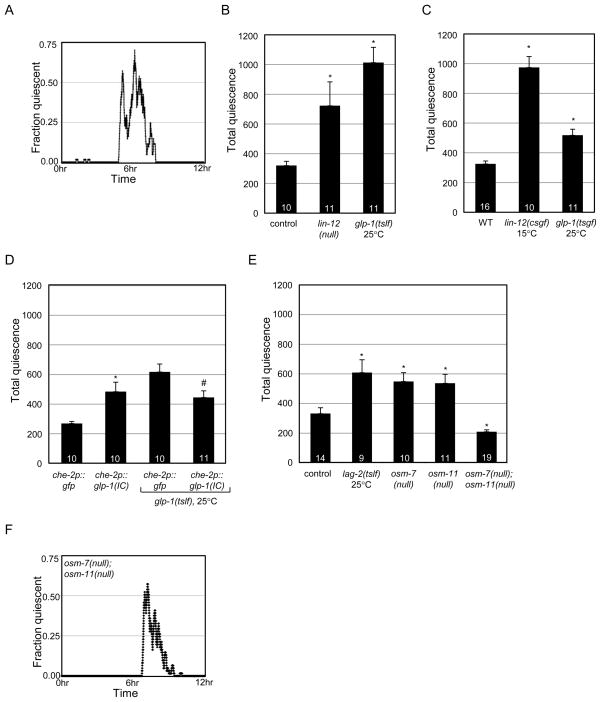
To determine if altering Notch signaling affects quiescence during L4-to-adult (L4/A) molting lethargus, we developed a simple microfluidics-based approach to assess quiescence in multiple animals simultaneously during L4/A lethargus (Fig. S3). Total quiescence was determined for L4/A lethargus by loading mid-L4 stage larvae into microfluidic chambers and recording activity into adulthood (Fig. 4A). Both lin-12(null) and glp-1(tslf) animals had increased total quiescence versus control animals (Fig. 4B). Also, Notch receptor gain of function alleles resulted in increased total quiescence (glp-1(ar202tsgf) and lin-12(n137n460csgf) animals, Fig. 4C). Similar results were observed in animals over-expressing LIN-12 (lin-12p::lin-12, Fig. S4A). Increasing GLP-1 receptor activity in ciliated sensory neurons was sufficient to increase total quiescence in wild type animals (che-2p::glp-1IC, Fig. 4D) and expression of GLP-1IC in ciliated neurons partially restored quiescence in glp-1(tslf) animals (Fig. 4D).
Figure 4.
Notch signaling regulates L4-to Adult molting quiescence. (A) Representative fractional quiescence graph. Mid-L4 animals exhibit L4/A molting lethargus quiescence for a few hours before adulthood. (B) Loss of LIN-12 receptor or decreased GLP-1 receptor function increased total L4/A quiescence. *p<0.02 vs. control (mgIs49; see Experimental Procedures for details). (C) Increased LIN-12 or GLP-1 function increased total L4/A molting quiescence. *p<10−4 vs. wild type (WT). (D) Increasing GLP-1 receptor activity in ciliated sensory neurons increased total L4/A quiescence in wild type animals (che-2p::glp-1(IC)). Expression of che-2p::glp-1(IC) transgene restored total quiescence in glp-1(tslf) animals to normal levels. *p<0.003 vs. che-2p::gfp. #p<0.02 vs. glp-1(tslf);che-2p::gfp. (E) Notch ligands/co-ligands regulate total L4/A quiescence. *p<0.002 vs. control (mgIs49; see Experimental Procedures for details). (F) Representative fractional quiescence graph of osm-7(null);osm-11(null) for comparison with (A) is presented. See Supplemental Methods 4 for details.
Next, we examined the impact of Notch ligands. Decreasing lag-2 function increased total quiescence (Fig. 4E, lag-2(q420tslf)). Similarly, loss of either osm-7 or osm-11 increased total quiescence (Fig. 4E). However, simultaneous loss of function of both osm-7 and osm-11 genes resulted in decreased total quiescence (Fig. 4E), which was unexpected given the increased quiescence of single mutant animals. Combined, these results suggest that Notch receptors and ligands regulate L4/A quiescence in a complex fashion.
Notch signaling impacts the duration of L4/A lethargus
The decreased total quiescence of osm-7(null);osm-11(null) animals could be caused by a shorter L4/A lethargus. Therefore, the impact of Notch signaling on the duration of lethargus was determined. The L4/A lethargus was significantly shorter for osm-7(null);osm-11(null) animals (Table S1A). The decreased lethargus period of osm-7(null);osm-11(null) suggested that these animals were defective in lethargus entry or maintenance. There was no difference in the time at which osm-7(null);osm-11(null) animal entered lethargus (Fig. S4D). However, osm-7(null);osm-11(null) animals exited lethargus early (Fig. S4E & Fig. 4F) suggesting that premature exit from L4/A lethargus caused decreased total quiescence.
To reconcile the complex effects of Notch pathway manipulation on L4/A quiescence, we delineated the impact of Notch signaling on two other components of quiescence: basal locomotion activity (hereafter referred as basal activity) and arousal threshold.
Notch signaling regulates basal activity and arousal thresholds
One factor that could influence quiescence maintenance is basal activity, which was assessed as body bends per minute [40]. Animals lacking both osm-7 and osm-11 had twice as many body bends per minute compared to control animals at either the adult stage or in between L4/A quiescent bouts (Table S1B). The dramatically increased basal activity likely contributes to the decreased total quiescence of osm-7;osm-11 animals.
During quiescence, animals have high arousal thresholds as they respond slowly or less robustly to sensory stimulation [36]. To assess arousal threshold, we used the mechanosensory body touch (Mec) assay [41]. Wild type adult animals respond to light touch 100% of trials, but quiescent L4/A animals respond in roughly 50% of trials (Table 1) reflecting an increased arousal threshold. Decreasing Notch signaling lowered arousal thresholds during L4/A quiescence; quiescent lin-12(null) or glp-1(tslf) animals responded more frequently than control quiescent animals (Table 1). Conversely, increasing Notch receptor function increased arousal thresholds; glp-1(tsgf), lin-12(csgf), che-2p::glp-1IC, and lin-12(OE) animals had increased arousal thresholds compared to their respective controls (Table 1). Also, decreasing lag-2 function or loss of osm-7 and/or osm-11 decreased arousal thresholds. Overall, Notch signaling correlated with arousal thresholds during L4/A quiescence (Table 1 & Table. S1C). Low arousal thresholds may reflect poor quality quiescence. We speculate that poor quality quiescence leads to compensatory increases in quiescence, as observed in both total L4/A quiescence and the duration of lethargus (Fig. 4, Table S1A). Overall, our results indicate that the Notch signaling pathway regulates basal activity, duration of lethargus, arousal thresholds, and quiescence in C. elegans.
Table 1. Notch activity regulates arousal during L4/A molting quiescence.
(A) Notch activity regulates arousal thresholds during L4/A molting quiescence. % of animals failing to respond to body touch with a hair during quiescent bouts is reported. Notch activity correlates with arousal thresholds. High Notch activity results in animals with high arousal thresholds, and vice versa.
| % failed to respond | n | |
|---|---|---|
| Wild type | 50±2 | 221 |
| osm-11(null) | 29±5* | 71 |
| osm-7(null) | 25±5* | 71 |
| osm-7(null);osm-11(null) | 44±4* | 76 |
| lag-2(tslf), 25°C | 31±1^ | 35 |
| glp-1(tslf), 25°C | 27±3# | 49 |
| glp-1(tsgf), 25°C | 70±5@ | 31 |
| lin-12(null) | 40±1# | 51 |
| lin-12(csgf), 15°C | 65±1@ | 37 |
| lin-12p::gfp | 45±2 | 40 |
| lin-12p::lin-12 | 62±3$ | 43 |
| che-2p::gfp | 46±2 | 31 |
| che-2p::glp-1(IC) | 61±3& | 35 |
ANOVA analysis between the genotypes: p<10−3.
p<0.002 vs. wild type.
p<0.003 for lin-12p::lin-12 compared to transgenic control animals expressing lin-12p::gfp in pha-1 rescue.
p<0.008 for che-2p::glp-1(IC) compared to transgenic control animals expressing che-2p::gfp in wild type background. See supplemental methods for Table 1 for details. Similar results were obtained in osm-11(OE) -induced quiescent adult animals (see text).
Discussion
Like sleep, quiescence is a complex behavior dependent on multiple processes including arousal threshold and cessation of activity. We propose a model (Fig. 5) wherein Notch signaling levels directly correlate with arousal levels during the L4/A molt. Increased arousal thresholds resulted in either inappropriate or increased quiescence. The consequences of decreased Notch activity are more complicated. Increased basal activity and/or decreased arousal thresholds resulted in homeostatic compensation in which animals increased quiescence to compensate for poor quiescence quality. Further decreases in Notch signaling left osm-7(null);osm-11(null) animals unable to maintain quiescence due to dramatically increased basal activity and decreased arousal thresholds. The results presented here are consistent with Notch signaling playing a major, albeit complex, role in the regulation of behavioral quiescence. Notch pathway regulation of L4/A quiescence is specific, as the Notch signaling pathway genes had no impact on satiety quiescence (You and Avery, unpublished results).
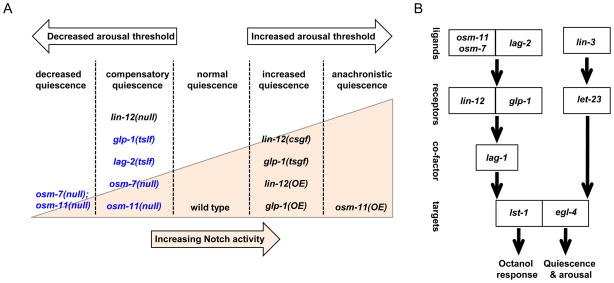
Figure 5.
Model for Notch signaling regulating multiple aspects of L4-to-adult molting quiescence. (A) Schematic model summarizing the impact of various Notch pathway genes. Gene name and type of allele is listed; Notch activity increases left to right (illustrated by peach wedge). For each genotype, the impact of altered Notch signaling on total L4-to-adult quiescence is listed above the wedge; OSM-11 co-ligand over-expression also induced anachronistic quiescence in adult animals. Decreased Notch signaling decreased arousal threshold during L4-to-adult molting lethargus while increased Notch signaling increased arousal thresholds (white arrows). Decreased glp-1 Notch receptor function, decreased DSL ligand function or loss of DOS co-ligands increased C. elegans basal locomotion activity (blue text) while loss of lin-12 Notch receptor decreased basal activity. (B) Genetic pathway illustrating relationships between C. elegans genes regulating octanol response and quiescence. egl-4 PKG also contributes, but is not required for octanol response; the role of the EGF pathway in octanol response has not been addressed. Neuronal Notch receptors act redundantly and in different populations of neurons in this behavior: lin-12 in a subset of interneurons and glp-1 in ciliated sensory neurons. Notch receptors may also act elsewhere to regulate quiescence. Activation of either EGF or Notch pathways by ligand over-expression induces anachronistic quiescence and these pathways are both required for normal L4-to-adult molting quiescence. EGF and Notch pathways may act in parallel converging on egl-4 to regulate molting quiescence (illustrated) and/or there may be functional interrelationships between these signaling pathways.
The presence of sleep-like states in Drosophila and C. elegans permits the dissection of conserved pertinent pathways [42]. In Drosophila, CREB has been implicated in sleep homeostasis [43] and a novel GPI-anchored protein (with a possible homolog in C. elegans) encoded by sleepless regulates sleep via an interaction with the Shaker K+ channel [44]. cAMP and EGFR signaling have been implicated in sleep regulation in both Drosophila and C. elegans [36, 37, 43, 45]. Interestingly, the mammalian OSM-11 ortholog DLK1 is expressed in the ventral tegmental area, substantia nigra pars compacta, and Raphe nuclei of adult rat and human brains [46], regions of the brain that have been implicated in sleep-wake cycle regulation [47].
Previous studies identified two signaling pathways that regulate quiescence in C. elegans: EGF and PKG [36, 37]. egl-4 PKG functions downstream of EGF signaling, as loss of egl-4 prevents inappropriate quiescence caused by lin-3 EGF over-expression [37]. We found that loss of egl-4 PKG function also prevents OSM-11-induced anachronistic quiescence, suggesting that egl-4 also acts downstream of Notch signaling. However, loss of osm-7 and osm-11 suppressed LIN-3 EGF-induced quiescence suggesting that there may be cross talk/feedback regulation between the Notch and EGF signaling pathways.
Multiple lines of evidence suggest that OSM-11 is expressed in hypodermal seam cells [4], but is secreted and acts non-cell autonomously. OSM-11 likely acts on sensory neurons in the head, including those that co-express GLP-1 and EGL-4 PKG and on the RIG interneurons that express LIN-12. However, other neurons are likely also involved. The ALA neuron is required for quiescence [37]; interestingly, ALA is one of three C. elegans neurons with processes directly underlying seam cells. OSM-11 secreted from the seam cells may act directly on the ALA neuron. Or, as Notch receptor expression has not been observed in ALA, the ALA neuron may regulate OSM-11 secretion from seam cells. There are two other classes of neurons with processes underneath the seam cells: CAN and PVD. The function of the CAN neurons remains unclear, although they associate with the excretory canal cell that regulates C. elegans osmotic balance [48]. The PVD neurons are involved in mechanical touch response [49], which might occur during osmotic swelling and/or shrinking. A functional interaction of PVD and/or CAN with the seam cells would be consistent with a role for OSM-11 in osmotic stress [7].
Our results support an ethological model in which OSM-11 and Notch signaling modulate neuronal function, behavior and adaptation to environmental stress. Adaptation to high external osmolarity or loss of DOS co-ligands osm-7 or osm-11 cause both physiological and behavioral changes in C. elegans. Increased external osmolarity decreases OSM-11 levels in the pseudocoelom, consistent with osmotic regulation of OSM-11 secretion from the seam cells. Either osmotic adaptation or loss of DOS co-ligands causes physiological adaptation to environmental high osmolarity via an unknown pathway that induces gpdh-1 expression and consequent glycerol osmolyte synthesis [50]. Additionally, either osmotic adaptation or DOS gene loss causes behavioral changes including decreased response to high osmolarity and octanol. These behavioral changes are dependent on Notch receptor signaling in the neurons of adult animals and are not dependent on gpdh-1 up-regulation. It is tempting to speculate that these physiological and behavioral changes are adaptive and are coordinated by global regulation of Notch receptor signaling. Humoral OSM-11 may play a key role in regulating Notch signaling in various tissues to coordinate physiological and behavioral adaptation to osmotic stress. As Notch receptor signaling has been recently been implicated in regulation of C. elegans heterochronic genes [51, 52], humoral OSM-11 acting on Notch receptors may also play a pivotal role in the temporal regulation of physiological and behavioral events at the larval molt, including quiescence.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health NIH R01 GM078171 and NS055813 (A.C.H.), NIH MH051383 and RC1AI087059 (S.L.), as well as the M.G.H. Medical Discovery Fund and NIH R15 DA021667-01 (M.Y.C.). We are grateful for advice and/or reagents from the C. elegans knockout consortia and numerous members of the C. elegans community. We thank Leon Avery and Young-jai You for unpublished results. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Komudi Singh, Email: komudi_singh@brown.edu.
Michael Y. Chao, Email: mchao@csusb.edu.
Gerard A. Somers, Email: gerard.somers@yale.edu.
Hidetoshi Komatsu, Email: Komatsu_Hidetoshi@takeda.co.jp.
Mark E. Corkins, Email: markcork@yahoo.com.
Jonah Larkins-Ford, Email: j_lford@yahoo.com.
Tim Tucey, Email: ttucey@ucsd.edu.
Heather M. Dionne, Email: dionneh@janelia.hhmi.org.
Melissa B. Walsh, Email: melissa_hoh@brown.edu.
Emma K. Beaumont, Email: eb273@bath.ac.uk.
Douglas P. Hart, Email: dphart@mit.edu.
Shawn Lockery, Email: shawn@chinook.uoregon.edu.
Anne C. Hart, Email: anne_hart@brown.edu.
References
- 1.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Developmental Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg PW. Vulval development. WormBook: the online review of C elegans biology. 2005:1–28. doi: 10.1895/wormbook.1.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimble J, Crittenden SL. Germline proliferation and its control. WormBook: the online review of C elegans biology. 2005:1–14. doi: 10.1895/wormbook.1.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komatsu H, Chao M, Larkins-Ford J, Corkins M, Somers G, Tucey T, Dionne H, White J, Wani K, Boxem M, et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. Plos Biol. 2008;6:e196. doi: 10.1371/journal.pbio.0060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frazier HN, Roth MB. Adaptive sugar provisioning controls survival of C. elegans embryos in adverse environments. Current biology: CB. 2009;19:859–863. doi: 10.1016/j.cub.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci USA. 2006;103:12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler J, Thomas J. Identification of a novel gene family involved in osmotic stress response in Caenorhabditis elegans. Genetics. 2006;174:1327–1336. doi: 10.1534/genetics.106.059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, Ewbank JJ. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008;4:e1000105. doi: 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berezovska O, McLean P, Knowles R, Frosh M, Lu FM, Lux SE, Hyman BT. Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience. 1999;93:433–439. doi: 10.1016/s0306-4522(99)00157-8. [DOI] [PubMed] [Google Scholar]
- 10.Ge X, Hannan F, Xie Z, Feng C, Tully T, Zhou H, Xie Z, Zhong Y. Notch signaling in Drosophila long-term memory formation. Proc Natl Acad Sci USA. 2004;101:10172–10176. doi: 10.1073/pnas.0403497101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao M, Larkins-Ford J, Tucey T, Hart A. lin-12 Notch functions in the adult nervous system of C. elegans. BMC Neurosci. 2005;6:45. doi: 10.1186/1471-2202-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouellet J, Li S, Roy R. Notch signalling is required for both dauer maintenance and recovery in C. elegans. Development. 2008;135:2583–2592. doi: 10.1242/dev.012435. [DOI] [PubMed] [Google Scholar]
- 13.Costa RM, Drew C, Silva AJ. Notch to remember. Trends in Neurosciences. 2005;28:429–435. doi: 10.1016/j.tins.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 15.Costa RM, Honjo T, Silva AJ. Learning and memory deficits in Notch mutant mice. Curr Biol. 2003;13:1348–1354. doi: 10.1016/s0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Chan SL, Miele L, Yao PJ, Mackes J, Ingram DK, Mattson MP, Furukawa K. Involvement of Notch signaling in hippocampal synaptic plasticity. Proc Natl Acad Sci USA. 2004;101:9458–9462. doi: 10.1073/pnas.0308126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Q, Sun K, Shuai Y, Lin R, You W, Wang L, Zhong Y. Suppressor of Hairless is required for long-term memory formation in Drosophila. J Neurogenet. 2009;23:405–411. doi: 10.3109/01677060903096133. [DOI] [PubMed] [Google Scholar]
- 18.de Bivort BL, Guo HF, Zhong Y. Notch signaling is required for activity-dependent synaptic plasticity at the Drosophila neuromuscular junction. J Neurogenet. 2009;23:395–404. doi: 10.3109/01677060902878481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao M, Komatsu H, Fukuto HS, Dionne H, Hart A. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci USA. 2004;101:15512–15517. doi: 10.1073/pnas.0403369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 21.Hao L, Aspöck G, Bürglin TR. The hedgehog-related gene wrt-5 is essential for hypodermal development in Caenorhabditis elegans. Developmental Biology. 2006;290:323–336. doi: 10.1016/j.ydbio.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 23.Patton A, Knuth S, Schaheen B, Dang H, Greenwald I, Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 24.Lambie EJ, Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. 1991;112:231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- 25.Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- 26.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 27.Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara M, Ishihara T, Katsura I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development. 1999;126:4839–4848. doi: 10.1242/dev.126.21.4839. [DOI] [PubMed] [Google Scholar]
- 29.Blelloch R, Anna-Arriola SS, Gao D, Li Y, Hodgkin J, Kimble J. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev Biol. 1999;216:382–393. doi: 10.1006/dbio.1999.9491. [DOI] [PubMed] [Google Scholar]
- 30.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- 32.L’Etoile ND, Coburn CM, Eastham J, Kistler A, Gallegos G, Bargmann C. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- 33.Raizen DM, Cullison KM, Pack AI, Sundaram MV. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics. 2006;173:177–187. doi: 10.1534/genetics.106.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hukema R, Rademakers S, Dekkers M, Burghoorn J, Jansen G. Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. EMBO J. 2006;25:312–322. doi: 10.1038/sj.emboj.7600940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuppen E, Van Der Linden A, Jansen G, Plasterk RH. Proteins Interacting With Caenorhabditis elegans Galpha Subunits. Comp Funct Genom. 2003;4:479–491. doi: 10.1002/cfg.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raizen D, Zimmerman J, Maycock M, Ta U, You Y, Sundaram M, Pack A. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 37.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 38.Treinin M, Chalfie M. A mutated acetylcholine receptor subunit causes neuronal degeneration in C. elegans. Neuron. 1995;14:871–877. doi: 10.1016/0896-6273(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 39.Pujol N, Torregrossa P, Ewbank JJ, Brunet JF. The homeodomain protein CePHOX2/CEH-17 controls antero-posterior axonal growth in C. elegans. Development. 2000;127:3361–3371. doi: 10.1242/dev.127.15.3361. [DOI] [PubMed] [Google Scholar]
- 40.Hart A. Behavior. WormBook; 2006. pp. 1–67. [Google Scholar]
- 41.Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crocker A, Sehgal A. Genetic analysis of sleep. Genes Dev. 2010;24:1220–1235. doi: 10.1101/gad.1913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 44.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 46.Jensen CH, Meyer M, Schroder HD, Kliem A, Zimmer J, Teisner B. Neurons in the monoaminergic nuclei of the rat and human central nervous system express FA1/dlk. Neuroreport. 2001;12:3959–3963. doi: 10.1097/00001756-200112210-00021. [DOI] [PubMed] [Google Scholar]
- 47.Monti JM, Monti D. The involvement of dopamine in the modulation of sleep and waking. Sleep Med Rev. 2007;11:113–133. doi: 10.1016/j.smrv.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 48.White J, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans Soc London. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 49.Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes & Development. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- 50.Rohlfing AK, Miteva Y, Hannenhalli S, Lamitina T. Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS ONE. 2010;5:e9010. doi: 10.1371/journal.pone.0009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Greenwald I. LIN-14 Inhibition of LIN-12 Contributes to Precision and Timing of C. elegans Vulval Fate Patterning. Current biology: CB. 2010 doi: 10.1016/j.cub.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomon A, Mian Y, Ortega-Cava C, Liu VW, Gurumurthy CB, Naramura M, Band V, Band H. Upregulation of the let-7 microRNA with precocious development in lin-12/Notch hypermorphic Caenorhabditis elegans mutants. Dev Biol. 2008;316:191–199. doi: 10.1016/j.ydbio.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loria PM, Hodgkin J, Hobert O. A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans. J Neurosci. 2004;24:2191–2201. doi: 10.1523/JNEUROSCI.5462-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.