Abstract
Many concerns have been raised about the potential allergenicity of novel, recombinant proteins into food crops. Guidelines, proposed by WHO/FAO and EFSA, include the use of bioinformatics screening to assess the risk of potential allergenicity or cross-reactivities of all proteins introduced, for example, to improve nutritional value or promote crop resistance. However, there are no universally accepted standards that can be used to encode data on the biology of allergens to facilitate using data from multiple databases in this screening. Therefore, we developed AllerML a markup language for allergens to assist in the automated exchange of information between databases and in the integration of the bioinformatics tools that are used to investigate allergenicity and cross-reactivity. As proof of concept, AllerML was implemented using the Structural Database of Allergenic Proteins (SDAP; http://fermi.utmb.edu/SDAP/) database. General implementation of AllerML will promote automatic flow of validated data that will aid in allergy research and regulatory analysis.
Keywords: Allergen database, allergen markup language, risk assessment, bioinformatics guidelines
1. Introduction
The incorporation of novel, recombinant proteins into food crops, whether for example to improve the nutritional value of the products or to promote resistance of the plants to pests, has raised concerns about their potential allergenicity(Ivanciuc et al., 2009c; Ladics and Selgrade, 2009; Selgrade et al., 2009; Thomas et al., 2009). Current methods to assess the potential allergenicity of transgenic proteins use a weight of evidence approach(Ladics et al., 2011), including serum tests(Goodman, 2008; Thomas et al., 2007), animal models(Bowman and Selgrade, 2008; Bowman and Selgrade, 2009; Fecek et al., 2010; Ganeshan et al., 2009; Gonipeta et al., 2010; Parvataneni et al., 2009; Yamamoto et al., 2009) and bioinformatics approaches(Gendel, 2009; Gendel and Jenkins, 2006; Goodman, 2006; Ivanciuc et al., 2009a; Ivanciuc et al., 2009b; Randhawa et al., 2011; Saha and Raghava, 2006; Schein et al., 2010; Silvanovich et al., 2009; Silvanovich et al., 2006). Allergenicity assessment guidelines proposed by the World Health Organisation(FAO/WHO, 2009) the European Food Safety Authority (EFSA)(EFSA, 2006) and the US Environmental Protection Agency(FIFRA, 2009) include the use of bioinformatics screening of the recombinant protein in allergen databases. As there are no universally accepted standards that can be used to encode data in these sources on the biology of allergens to facilitate the use of data from multiple databases in this screening, we propose here a specific markup language to define molecular, structural and clinical information for allergens. The language is based on the concept of ontologies that are in widespread use for the exchange of information in other areas of biology. The language should allow automatic, virtual screening of all major allergen databases, starting from the sequence of a novel protein. These tools will help to define (and later search for) structural and functional descriptors of allergenicity, and will enhance the sensitivity of bioinformatics search tools for regulatory purposes.
Ontologies (Hunter et al., 2003; Schuurman and Leszczynski, 2008) can be used to describe both specific pieces of data and the relationships between different data types. Most implementations of ontologies in biology use a form of markup language (ML), i.e., a flexible general-purpose text format, to encode, store, and exchange structured information. An ideal ML combines particular terminologies and relationships from many different sources into a common ontology, and provides a rigorous and extensible set of rules that may encode all information categories in a specific field. Once annotated in a markup language, data can be handled automatically and computationally processed. Markup languages have been developed for many areas of medicine, biology, and biochemistry. Examples include the BIOpolymer Markup Language (BIOML) which encodes information for biopolymers in a fashion that relates them to organisms and organelles (Fenyo, 1999), the Microarray Gene Expression Markup Language (MAGE-ML) for storing and displaying microarray data (Durinck et al., 2004), the Annotated Gel Markup Language (AGML) to describe annotated two-dimensional gel electrophoresis data (Stanislaus et al., 2004), and the Systems Biology Markup Language (SBML) to represent computational models of biochemical networks (Finney and Hucka, 2003; Hucka et al., 2003).
Here we show the use and a structure for a markup language to connect databases that contain information on allergenic proteins, such as those in Table 1 (Brusic et al., 2003; Gendel, 2004; Gendel and Jenkins, 2006; Goodman, 2006; Mari et al., 2006; Schein et al., 2006; Schein et al., 2007). In addition to the list of allergenic proteins provided by the International Union of Immunological Societies (IUIS, http://www.allergen.org) (Chapman et al., 2007) or AllAllergy (http://allallergy.net/), there are now many cross-indexed databases potentially useful for allergenicity assessment such as Allergome (http://www.allergome.org), Central Science Laboratory database (CSL, http://allergen.csl.gov.uk/), InformAll (http://foodallergens.ifr.ac.uk/), AllergenOnline (http://www.allergenonline.org), and Structural Database of Allergenic Proteins (SDAP; http://fermi.utmb.edu/SDAP/). As the number of identified allergens is continuously increasing, along with the secondary data describing these allergens, facilitating data exchange between these databases would allow automated communication, analysis and comparisons combining the information in these different resources (Gendel, 2009; Mari et al., 2009; Schein et al., 2010).
Table 1.
Allergen Databases and Servers that can Exchange Data through AllerML
| Web Site | URL |
|---|---|
| IUIS (International Union of Immunological Societies) | http://www.allergen.org |
| SDAP (Structural database of Allergenic proteins) and SDAP-Food | http://fermi.utmb.edu/SDAP |
| FARRP (Food Allergy Research and Resource Program) | http://www.allergenonline.org/ |
| Allergome | http://www.allergome.org |
| CSL (Central Science Laboratory, UK) | http://allergen.csl.gov.uk/ |
| InformAll | http://foodallergens.ifr.ac.uk/ |
| ADFS (Allergen Database for Food Safety) | http://allergen.nihs.go.jp/ADFS/ |
| All Allergy | http://allallergy.net/ |
| AllFam | http://www.meduniwien.ac.at/allergens/allfam/ |
| Allermatch | http://www.allermatch.org/ |
The Allergen Markup Language (AllerML) that we describe here is a first step in developing automated tools to access data on allergens in multiple databases. AllerML is based on the allergen nomenclature developed and maintained by the IUIS Allergen Nomenclature Sub-committee. This official allergen nomenclature is recognized by the WHO, and it applies the Linnean system to generate a systematic, unique and comprehensive nomenclature for allergenic proteins. AllerML consists of a hierarchical set of tags that describe the most important information normally contained in allergen databases, including common names, sources, sequence, structure, IgE and T-cell epitopes, and cross-reactivity. The allergen description in AllerML is augmented with a set of special tags that link allergen-specific databases to other general purpose biological data sets, such as the Pfam classification. In its current form, AllerML can be used to automate the dynamic exchange of information on allergens, to incorporate data on new allergenic proteins as they are identified, and to support computational and bioinformatics studies of allergenicity and clinically significant cross-reactivity. Wide implementation of AllerML will simplify automatic exchange of data between allergen databases, and improve data access for integrated computational and bioinformatics analysis.
2. Methods
2.1. Allergen Nomenclature and Data Sources
The IUIS was the primary source for allergen nomenclature and classification. Other databases that provide additional information are listed in Table 1.
2.2. Development of AllerML Tags
A set of XML tags were defined for the most common data types found in allergen databases, including tags for data that identify individual allergenic proteins, the source organisms, and sequences information. Relationships between the elements represented by these tags (such as “contained in” or “part of”) were also defined. Additional tags and relationships were defined for molecular and structural characteristics of allergenic proteins, including any known epitopes. To ensure that the sources for all data within a database are fully described, tags were created for both literature citations and to link allergen databases to larger repository databases such as GenBank and EMBL.
2.3. Automated generation of AllerML Tags by software tools
The AllerML tags proposed here encode all molecular information on allergens and IgE epitopes, as present in the major allergen databases. As an example for an AllerML implementation in practice we used the information collected in SDAP (Ivanciuc et al., 2002; Ivanciuc et al., 2003; Schein et al., 2007) and wrote a C program to translate the data listed in MySQL tables to AllerML schemes. The major core information on all allergen entries as AllerML documents can be downloaded from the SDAP web site (http://fermi.utmb.edu/SDAP/). For each allergen in SDAP, the AllerML record can be obtained from the link “Translate to AllerML” located immediately below the title line with the allergen name in the SDAP page corresponding to an allergen.
2.4. Illustration of AllerML Tags for SDAP entries
The database component in SDAP contains information regarding the allergen name, source, sequence, structure, IgE epitopes, and literature references. When supplementary information is available from the major protein databases (GenBank, SwissProt, UniProt, PIR, and PDB) and literature databases (PubMed), SDAP provides links for easy reference. A FASTA sequence search starting from an allergen selected from SDAP or from a user-provided sequence can identify all similar allergens in SDAP. Each SDAP allergen sequence is cross-linked to the protein motif database MotifMate (http://born.utmb.edu/motifmate/), which lists the corresponding physicochemical property (PCP) motifs (Ivanciuc et al., 2009a). To identify groups of similar allergens that might cross-react, SDAP provides the Pfam classification of all allergens. SDAP also contains reliable models or links to the experimentally determined structures for more than 80% of allergens included in SDAP (Oezguen et al., 2008). The AllerML implementation demonstrated in this report may be readily applied to all existing allergen databases, and extensions to new data fields may be easily defined.
2.5. Links to other Protein Databases
Other protein databases also contain relevant information about allergenic proteins, and we propose the following set of AllerML tags to link to the most relevant such services:
- EMBL (http://www.ebi.ac.uk/), tag <AllerML_EMBL>
- GenBank (http://www.ncbi.nlm.nih.gov), tag <AllerML_GenBank>
- DDBJ (DNA Data Bank of Japan, http://www.ddbj.nig.ac.jp/), tag <AllerML_DDBJ>
- HSSP (http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-page+LibInfo+-lib+HSSP), tag <AllerML_HSSP>
- PDB (http://www.pdb.org), tag <AllerML_PDB>
- IntAct (http://www.ebi.ac.uk/intact/site/index.jsf), tag <AllerML_IntAct>
- GO (http://www.geneontology.org/), tag <AllerML_GO>
- InterPro (http://www.ebi.ac.uk/interpro/), tag <AllerML_InterPro>
- Gene3D, tag <AllerML_Gene3D>
- Pfam (http://pfam.sanger.ac.uk/), tag <AllerML_Pfam>
- PRINTS (http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS), tag <AllerML_PRINTS>
- PROSITE (http://www.expasy.org/prosite/), tag <AllerML_PROSITE>
3. Results
3.1. AllerML General Sections
The core information contained in all major allergen databases is provided by IUIS, namely the allergen name, source taxonomy, gene and protein sequences, PDB structures, and literature references. In individual databases, this common information may be augmented with data collected from the literature and by processing sequence and structural data with bioinformatics tools. In the following sections we describe AllerML encoding of the common information categories, and provide examples of how this basic vocabulary can be extended to include information specific to individual databases.
3.2. Allergen Name and Taxonomy
The initial set of AllerML terms are listed in Table 2. These terms describe the fundamental data commonly contained in allergen databases. The central data item is the official IUIS designation which is defined in AllerML as a major attribute. The entire set of data related to an allergen is recorded under the tag <AllerML_Allergen> or <AllerML_Isoallergen>, depending on the IUIS classification of allergens. Other data such as a description of the biochemical function of the protein, gene and protein sequences, 3D structures, if available, epitopes, sequence motifs and relevant literature information are described using additional AllerML data designations that are related in a hierarchical fashion. The hierarchical structure between these building blocks in AllerML can be further extended by including data on protein family classification, additional information on the nature of the epitopes, or taxonomy information from the NCBI taxonomy database. Other data may be easily incorporated in AllerML by defining appropriate tags within the hierarchy.
Table 2.
AllerML Tags (only the start tag is shown for each section)
| Tag | Description |
|---|---|
| <AllerML_Allergen> | Allergen section |
| <AllerML_Allergen_Name> | Allergen name |
| <AllerML_Allergen_SDAP_ID> | Allergen SDAP ID |
| <AllerML_Isoallergen> | Isoallergen section |
| <AllerML_Isoallergen_Name> | Isoallergen name |
| <AllerML_Isoallergen_SDAP_ID> | Isoallergen SDAP ID |
| <AllerML_Allergen_Type> | Allergen type: IUIS or non-IUIS |
| <AllerML_Organism> | Allergen organism |
| <AllerML_Systematic_Name> | Systematic name |
| <AllerML_Taxonomy_ID> | Taxonomy ID |
| <AllerML_Common_Name> | Common name |
| <AllerML_Taxonomy> | Taxonomy |
| <AllerML_Comment> | Comment |
| <AllerML_Protein> | Protein section |
| <AllerML_Protein_Source> | Protein source: UniProt, GenBank, PubMed, or DOI |
| <AllerML_UniProt> | UniProt section |
| <AllerML_UniProt_ID> | UniProt ID |
| <AllerML_UniProt_Accession> | UniProt accession number |
| <AllerML_GenBank> | GenBank section |
| <AllerML_GenBank_Locus> | GenBank locus |
| <AllerML_GenBank_Accession> | GenBank accession number |
| <AllerML_GenBank_Version> | GenBank version |
| <AllerML_GenBank_GI> | GenBank GI |
| <AllerML_Protein_Length> | Protein length |
| <AllerML_Protein_Sequence> | Protein sequence |
| <AllerML_PDBML> | PDBML section |
| <AllerML_Epitopes> | Epitopes section |
| <AllerML_Epitope_Set> | Section for an epitope set, normally from a single publication |
| <AllerML_Epitope_Type> | Epitope type: IgE, T-cell |
| <AllerML_Epitope_Position> | Epitope position |
| <AllerML_Epitope_Sequence> | Epitope sequence |
| <AllerML_Epitope_Comment> | Epitope comment |
| <AllerML_MotifMate_Motifs> | Section for a collection of MotifMate motifs |
| <AllerML_MotifMate_Motif> | MotifMate motif |
| <AllerML_MotifMate_Motif_Position> | MotifMate motif position |
| <AllerML_MotifMate_Motif_Sequence> | MotifMate motif sequence |
| <AllerML_Cross-references> | Cross-references to other databases |
| <AllerML_EMBL> | EMBL (http://www.ebi.ac.uk/) |
| <AllerML_GenBank> | GenBank (http://www.ncbi.nlm.nih.gov) |
| <AllerML_DDBJ> | DDBJ (DNA Data Bank of Japan, http://www.ddbj.nig.ac.jp/) |
| <AllerML_HSSP> | HSSP (http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-page+LibInfo+-lib+HSSP) |
| <AllerML_PDB> | PDB (http://www.pdb.org) |
| <AllerML_IntAct> | IntAct (http://www.ebi.ac.uk/intact/site/index.jsf) |
| <AllerML_GO> | GO (http://www.geneontology.org/) |
| <AllerML_InterPro> | InterPro (http://www.ebi.ac.uk/interpro/) |
| <AllerML_Gene3D> | Gene3D (http://gene3d.biochem.ucl.ac.uk/Gene3D/) |
| <AllerML_Pfam> | Pfam (http://pfam.sanger.ac.uk/) |
| <AllerML_PRINTS> | PRINTS (http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS) |
| <AllerML_PROSITE> | PROSITE (http://www.expasy.org/prosite/) |
| <AllerML_References> | Set of references |
| <AllerML_Reference> | Reference |
| <AllerML_Reference_PubMed> | PubMed ID |
| <AllerML_Reference_Authors> | Set of authors |
| <AllerML_Author> | Author |
| <AllerML_Reference_Journal> | Journal |
| <AllerML_Reference_Title> | Title |
| <AllerML_Reference_Year> | Year |
| <AllerML_Reference_Volume> | Volume |
| <AllerML_Reference_Pages> | Pages |
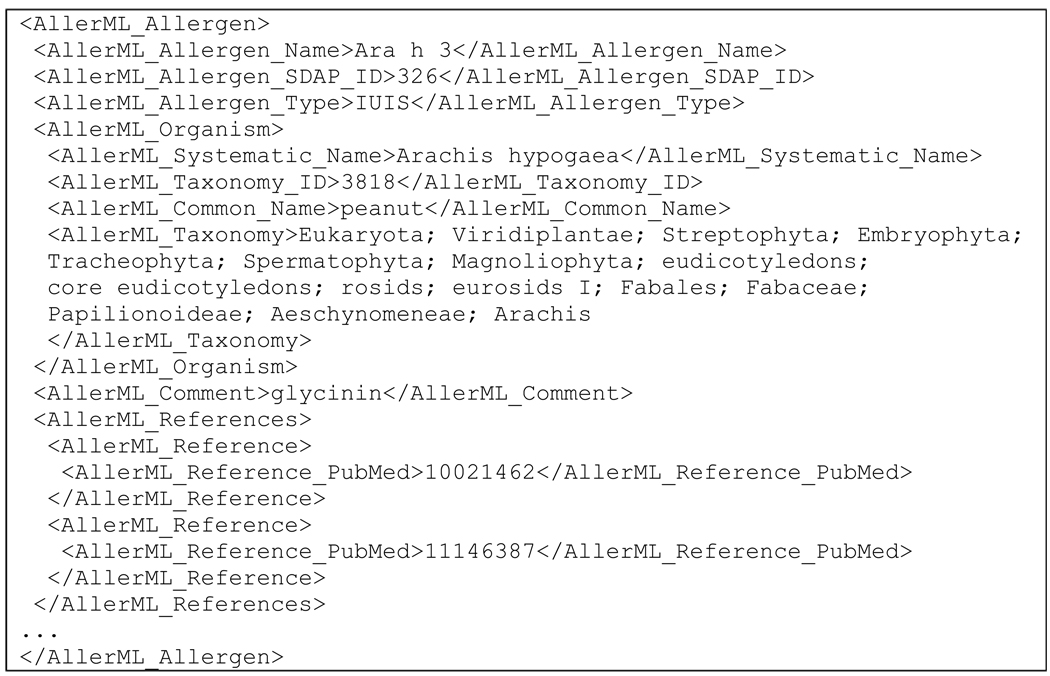
An example of the implementation of AllerML for the peanut allergen Ara h 3 is shown in Scheme 1. The first part of the record indicates the IUIS allergen name and type (IUIS or non-IUIS). Other unique identifiers from each allergen database can be included (if available) using appropriate tags. The SDAP ID is given here as an example. The second part of the record contains source organism information including the accession number from the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/). Comments may be included in any AllerML section under the tag <AllerML_Comment>, and may contain HTML tags used to format the text for display or to include other HTML elements such as tables or hyperlinks.
Scheme 1.
Core information of Ara h 3 as an AllerML document
3.3 Cross-references to Other Web Databases
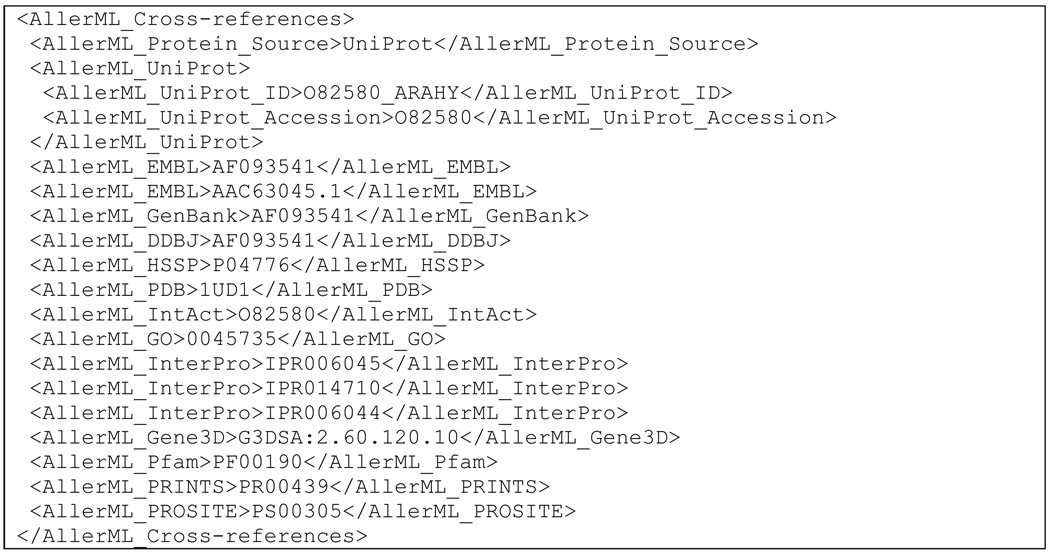
AllerML offers also a mechanism to identify and link to allergen gene and protein sequences or 3D structure information in other biology databases. Cross-references to other databases are included in the section <AllerML_Cross-references>, as shown for Ara h 3 in Scheme 2, based on the UniProt sequence O82580. Sequences in UniProt (http://www.uniprot.org/) are referenced by their ID and accession number, with the tags <AllerML_UniProt_ID> and <AllerML_UniProt_Accession>, respectively. Similar tags can be used to link to EMBL, PDB, Pfam, PROSITE, and other databases. The cross-linking with Pfam is of particular relevance, as all major allergens belong to a relatively small number of protein families (Ivanciuc et al., 2009a; Radauer et al., 2008). Allergens from the same Pfam class are significantly similar in amino acid sequence and have the same 3D fold. Members of the same PFAM often show clinically relevant cross-reactivity (Schein et al., 2010). Currently, the Pfam classification of allergens may be accessed from AllFam (Radauer et al., 2008), Allergome (Truffer et al., 2006), and SDAP (Ivanciuc et al., 2009a).
Scheme 2.
Cross-references section of Ara h 3
3.4. Protein Sequence
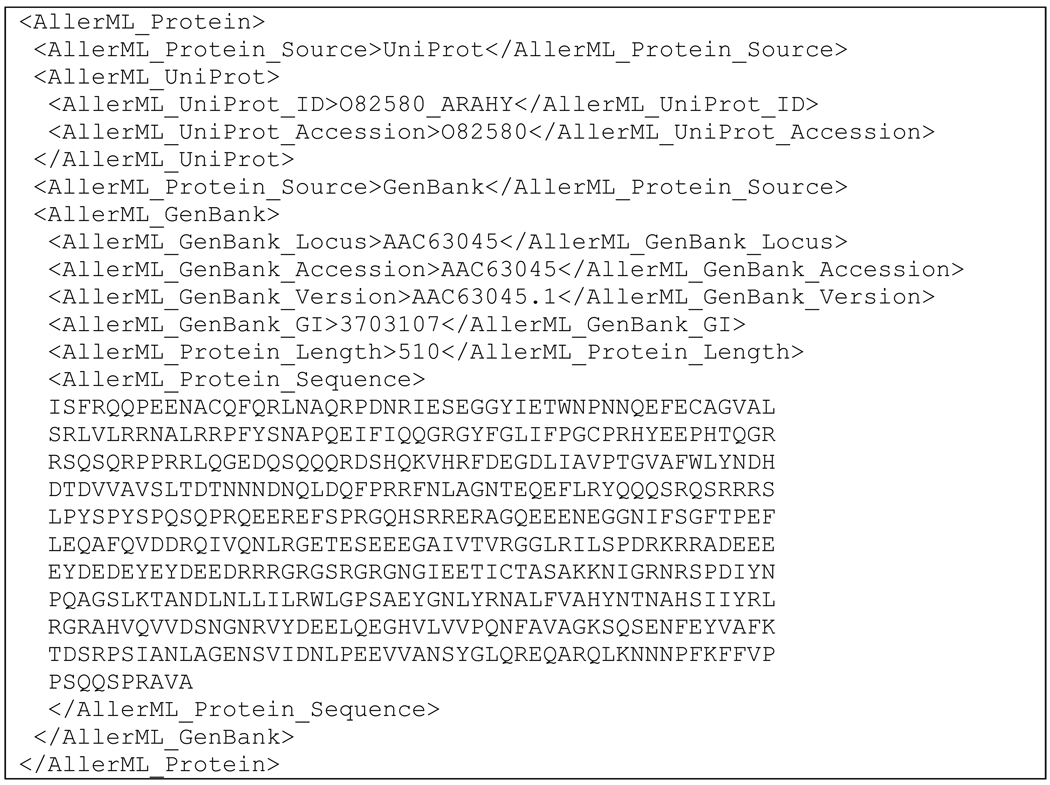
Protein sequences may be described in AllerML by indicating their source and accession numbers, by showing the source, sequence, and reference (for protein sequences retrieved from literature), or by specifying the source, accession numbers, and sequence. The sequences of Ara h 3 are shown in Scheme 3 in the AllerML format. The GenBank protein sequence is characterized by its locus, accession number, version, and GI, together with the corresponding sequence. Genes may be encoded with the same four GenBank accession tags, namely <AllerML_GenBank_Locus>, <AllerML_GenBank_Accession>, <AllerML_GenBank_Version>, and <AllerML_GenBank_GI>, respectively, whereas the gene sequence is collected under the tag <AllerML_Gene_Sequence>.
Scheme 3.
Protein section of Ara h 3
3.5. Protein 3D Structure
The representation of 3D structures of allergens in AllerML is based on the ML format adopted by the Protein Data Bank (PDB). The Protein Data Bank Markup Language (PDBML) developed by Westbrook et al. (Westbrook et al., 2005) is used to represent all PDB structures in XML (http://pdbml.rcsb.org/), based on a translation of the mmCIF files into PDBML driven by the PDB Exchange Data Dictionary. Allergen structure information is designated in AllerML using the start tag <AllerML_PDBML> and end tag </AllerML_PDBML>. Similar ML encoding can be used to represent computational allergen models. This scheme can be applied to all experimentally determined 3D structures or 3D models of allergens (Oezguen et al., 2008; Schein et al., 2010). The ML representation of the protein structure can be extended to depict regions (e.g., for conformational epitopes) or domains in the 3D structures of allergens.
3.6. Allergen Epitopes
Accurate data on IgE epitopes of allergens, although currently only rarely available in public databases, are critical data for predicting cross-reactivity among related allergens. The majority of experimental information on allergen epitopes was obtained by investigating linear IgE epitopes. These are usually determined by synthesizing overlapping peptides from the allergen sequence and then determining the peptide interaction with IgE from sera obtained from patients that react to a particular allergen. Such linear IgE epitopes have been reported for several proteins, such as the peanut allergens Ara h 1 (Shin et al., 1998), Ara h 2 (Stanley et al., 1997), and Ara h 3 (Rabjohn et al., 1999), the shrimp allergen Pen a 1 (Ayuso et al., 2002), and the mountain cedar allergens Jun a 1 (Midoro-Horiuti et al., 2003; Midoro-Horiuti et al., 2006) and Jun a 3 (Soman et al., 2000).
We propose here an AllerML set of tags for linear epitopes (Scheme 4), that can also be used to encode conformational epitopes. For an allergen, a set of epitopes collected from a single publication are collected under the tag <AllerML_Epitope_Set>, whereas all sets of epitopes are collected under the tag <AllerML_Epitopes>. For each epitope set, the epitope type is indicated only once, and may be IgE or T-cell. Then, for each epitope, AllerML specifies the epitope position, sequence, and an optional comment. The section concludes with a tag indicating the reference of the epitope data. Supplementary information for each epitope is collected under a comment tag, <AllerML_Epitope_Comment>, which marks a text field that may contain HTML tags to format the text. Conformational epitopes are encoded by recording a list with all amino acids that form the epitope, and then adding after each amino acid its position number in the protein sequence.
Scheme 4.
Epitope section of Ara h 3
3.7. Reference Section
Information describing data sources is coded in the Reference section identified with the <AllerML_References> tag. This section may be embedded within other sections, such as Allergen Name and Taxonomy, Protein Sequence, or Allergen Epitope. The main source of data will generally be journal articles, but information may be obtained from sources such as patents or direct submission (in the case of NCBI sequences), or government documents. Each reference starts with the tag <AllerML_Reference>, and contains enough information to uniquely identify the reference. The minimum amount of information to indicate a reference may be a database accession number or a patent number. Usually, the source of information for journal articles is PubMed, and any such article may be encoded by the PubMedID indicated in the tag <AllerML_Reference_PubMed>. A complete record of a journal article contains tags for authors, journal title, article title, year, volume, and pages. The Reference section may include also subsections, such as Abstract, Keywords, Accession number in other literature databases, or Comments.
3.8. AllerML Special Sections
The AllerML implementation proposed in this report may be readily applied to all existing allergen databases, and extensions to new data fields may be easily defined. As each allergen database and bioinformatics server has specific data and applications, AllerML can be extended with appropriate sets of tags. In the following sections we use examples from Allergome (Mari and Scala, 2006; Mari et al., 2006) and SDAP (Ivanciuc et al., 2002; Ivanciuc et al., 2003; Schein et al., 2007) to demonstrate how AllerML may be easily applied to encode particular categories of data.
3.8.1. Allergen Specific Motifs
AllerML may be easily extended to encode the INSCH motifs from the Allergome database. These motifs are MEME-type motifs developed by Marti et al. (Marti et al., 2007). The INSCH motif length was set to 50 to match the average size of a protein domain. The current collection of INSCH motifs is based on a set of 2189 sequences from the Allergome database (January 10, 2009), and consists of 97 motifs identified in 1885 sequences. No motif was found in the remaining 304 sequences. For each allergen that contains a motif, AllerML has an INSCH motif section marked by the tag <AllerML_INSCH_Motif>, followed by the motif ID tag <AllerML_INSCH_Motif_ID>, and the corresponding sequence <AllerML_INSCH_Motif_Sequence>. The AllerML encoding for the INSCH motif of Ara h 3 is shown in Scheme 5.
Scheme 5.
AllerML encoding for the INSCH motif section of Ara h 3.
A similar scheme can be applied to encode physicochemical property (PCP) motifs that were derived from a PFAM classification of all allergens in SDAP and archived in the database MotifMate (http://born.utmb.edu/motifmate/) (Ivanciuc et al., 2009a). PCP motifs are defined as protein regions where the side chains may not be identical but show conservation of physicochemical properties, such as hydrophobicity, size or alpha-helical propensity, among allergens of the same protein family (Venkatarajan and Braun, 2001). Previously, PCP motifs were shown to correlate with functionally important regions and to provide relevant fingerprints that can identify distantly related proteins (Ivanciuc et al., 2004; Mathura et al., 2003; Schein et al., 2010; Schein et al., 2005). The AllerML encoding for several MotifMate motifs of Ara h 3 is shown in Scheme 6. The MotifMate section is indicated by the tag <AllerML_MotifMate_Motifs>, whereas the information for each motif is collected under the tag <AllerML_MotifMate_Motif>. For linear IgE epitopes, AllerML indicates the motif position within the protein and the sequence.
Scheme 6.
AllerML encoding for the MotifMate motifs section of Ara h 3.
3.8.2. IgE Cross-reactive Peptides
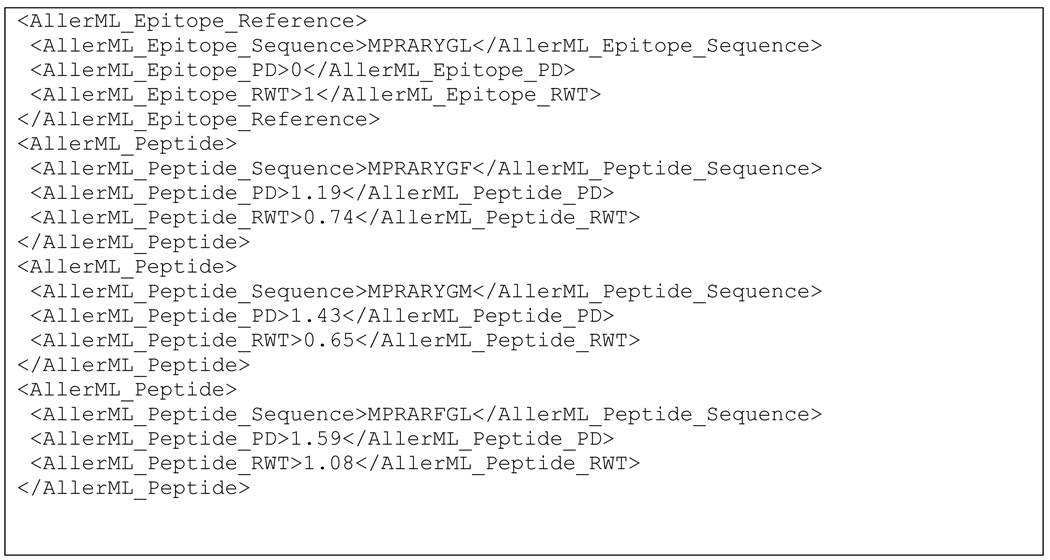
AllerML can also encode quantitative information regarding allergen cross-reactivity, such as the sequence similarity index PD and the experimentally measured binding affinity of peptides to IgE. The PD index was recently validated as a quantitative predictor for IgE cross-reactivity (Ivanciuc et al., 2009b) using sets of peptides related to three known linear IgE epitopes of Jun a 1 (Midoro-Horiuti et al., 2003; Midoro-Horiuti et al., 2006). These experimental results are translated into AllerML by recording for each peptide the sequence, the PD value, and the experimental measure of IgE binding. IgE binding is expressed as the intensity ratio RWT = SIP/SIE, where SIP is the spot intensity of the test peptide and SIE is the spot intensity of the corresponding Jun a 1 epitope in the same column on the membrane. To demonstrate the encoding of a peptide library we show a selection of the results obtained for peptides related to the epitope 3 from Jun a 1 (Scheme 7).
Scheme 7.
AllerML encoding of quantitative data regarding allergen cross-reactivity with peptides for Jun a 1
This approach may be also adapted to store the experimental results obtained during the identification of linear IgE epitopes with a library of overlapping peptides (Ayuso et al., 2002; Midoro-Horiuti et al., 2003; Midoro-Horiuti et al., 2006; Rabjohn et al., 1999; Shin et al., 1998; Stanley et al., 1997). Storing experimental results of epitope detection in a computer readable format facilitates their direct use in computational models that may enhance our ability to predict IgE epitopes. Additional tags should be defined to encode the experimental cross-reactivity data for individual patients.
3.9. AllerML Implementation
All tags currently defined in AllerML are summarized in Table 2 showing for each section the start tag <AllerML_Tag>. To demonstrate the application of the proposed allergen markup language in practice, we implemented AllerML for the core information of all allergens in SDAP. The AllerML translation for each allergen in SDAP is publicly available, to provide transparent access to the database. Broad AllerML implementation in other databases would provide standardized access to the common body of information on allergens, namely nomenclature, taxonomy, protein sequences, 3D structures, data on IgE and T-cell epitopes, and cross-references with literature, protein, and genomic databases. Besides these general sections, particular implementations of AllerML may encode database-specific information, such as protein motifs or peptide cross-reactivity data.
4. Discussion
The markup language AllerML facilitates the automated storage, import, export, comparison and analysis of biological data catalogued in databases that contain information on allergenic proteins. Our goal in developing AllerML was not to create a single database, but rather to permit easier integration of related data across different databases. Each of the existing databases has its own target field of application, protocols for information collection, selectivity in the type of data displayed, and computational tools for data processing and interpretation, making it quite unlikely that one particular database would be accepted as a common standard that satisfies the diverse needs for all allergen researchers. To facilitate the evaluation of protein allergenicity or cross-reactivity it is necessary to consolidate information from diverse sources into a comprehensive database that can offer validated data for bioinformatics models. The AllerML markup language presented here (Schemes 1–7) is designed to provide a path for easy access to data from these web servers for the user. AllerML may be also used as a convenient tool to provide input for novel bioinformatics servers.
AllerML contains a group of general tags, that are common to all allergen databases, and a group of specific tags to codify information particular to a database or bioinformatics server. The general information on allergens comes mainly from IUIS, and includes allergen name and taxonomy, gene and protein sequences, PDB structures, epitopes, and literature references. The corresponding AllerML tags may be used to store, retrieve, request, and download such information in an automatic way, which may speed-up the process of database update and may eliminate typing errors. The specific tags from AllerML are characteristic for each database. We anticipate that interested researchers and groups will define new AllerML tags to encode allergen properties of interest to them, or data currently present in their databases or bioinformatics servers. AllerML is flexible and easy to extend, and starting from the common group of tags it is easy to diversify and adapt AllerML for various specialized applications, either clinical, laboratory, or computational.
There are several specific advantages in exchanging information between allergen databases with AllerML. First, different types of literature and database information stored for a given allergen can be rapidly assembled from multiple sources. For example, files for allergens that are clinically cross-reactive, according to biological data stored in Allergome, can be combined with data on their sequences from Uniprot, structures from PDB or 3D models from SDAP. This allows one to rapidly determine common features of the allergens that could account for the cross-reactivity, and may indicate what other allergens should be listed as potentially cross-reactive. Secondly, it will streamline the collection of data for novel computational tools for allergenicity and cross-reactivity prediction. For example, by combining data on allergen sequences (Scheme 3), known IgE epitopes (Scheme 4), INSCH motifs (Scheme 5), and MotifMate motifs (Scheme 6), one may readily obtain the input data for a new method of identifying allergens. Finally, the ontology is flexible enough to permit data exchange between other databases that are not specifically for allergenic proteins, such as the Immune Epitope Database and Analysis Resource (IEDB; http://immuneepitope.org), which contains an extensive list of T- and B-cell epitopes assembled from large scale screening (Zhang et al., 2008). Other extensions of AllerML could include data on allergenic medical symptoms or biomarkers from new technologies, such as proteomics (Chapman et al., 2007; Guo et al., 2008; Reisdorph et al., 2009) or microarray approaches (Bublin et al., 2011; Hiller et al., 2002; Hochwallner et al., 2010; Mari et al., 2010; Shreffler et al., 2005).
5. Concluding remarks
A common data structure of archiving allergens across several databases greatly simplifies the development of standard computational procedures for estimating the risk of allergenic response to novel proteins in genetically engineered organisms. Even though a common IUIS nomenclature exists for allergens, the data structure and content differs vastly between different databases. So far, the progress in allergen analysis was driven by the accumulation of data on allergens, such as sequences, structures, and IgE epitopes. AllerML provides the tools for automated comparisons and analysis of allergens catalogued in generic and specialized databases by providing a uniform representation of biological and biochemical data. We emphasize that the current AllerML is a first draft of such a flexible data structure that is a good basis for a common language for the automatic exchange of data on allergens. If broadly adopted, it would ease the establishment of a standardized set of accepted allergens for use in regulatory bioinformatics guidelines for food safety. We also envision that an automatic flow of information between allergen databases and servers will help to improve algorithms that predict allergenicity and cross-reactivity.
Acknowledgements
This work was supported by grants from the National Institute of Health (R01 AI 064913; WB and CHS), the U.S. Environmental Protection Agency STAR Research Assistance Agreement (No. RD 83482301 to WB), an NIH/EPA STAR joint program award (RE-83406601-0 to CHS), and a contract from the U.S. Food and Drug Administration (HHSF22320011I). The article has not been formally reviewed by the EPA, and the views expressed in this document are solely those of the authors.
Footnotes
Conflict of Interest Statement:
None of the authors have a conflict of interest.
References
- Ayuso R, Lehrer SB, Reese G. Identification of continuous, allergenic regions of the major shrimp allergen Pen a 1 (tropomyosin) Int. Arch. Allergy Immunol. 2002;127:27–37. doi: 10.1159/000048166. [DOI] [PubMed] [Google Scholar]
- Bowman CC, Selgrade MK. Failure to induce oral tolerance in mice is predictive of dietary allergenic potency among foods with sensitizing capacity. 2008;106:435–443. doi: 10.1093/toxsci/kfn200. [DOI] [PubMed] [Google Scholar]
- Bowman CC, Selgrade MK. Utility of rodent models for evaluating protein allergenicity. Regul. Toxicol. Pharmacol. 2009;54:S58–S61. doi: 10.1016/j.yrtph.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Brusic V, Millot M, Petrovsky N, Gendel SM, Gigonzac O, Stelman SJ. Allergen databases. Allergy. 2003;58:1093–1100. doi: 10.1034/j.1398-9995.2003.00248.x. [DOI] [PubMed] [Google Scholar]
- Bublin M, Dennstedt S, Buchegger M, Antonietta Ciardiello M, Bernardi ML, Tuppo L, Harwanegg C, Hafner C, Ebner C, Ballmer-Weber BK, Knulst A, Hoffmann-Sommergruber K, Radauer C, Mari A, Breiteneder H. The performance of a component-based allergen microarray for the diagnosis of kiwifruit allergy. Clin. Exp. Allergy. 2011;41:129–136. doi: 10.1111/j.1365-2222.2010.03619.x. [DOI] [PubMed] [Google Scholar]
- Chapman MD, Pomés A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J. Allergy Clin. Immunol. 2007;119:414–420. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Durinck S, Allemeersch J, Carey VJ, Moreau Y, De Moor B. Importing MAGE-ML format microarray data into BioConductor. Bioinformatics. 2004;20:3641–3642. doi: 10.1093/bioinformatics/bth396. [DOI] [PubMed] [Google Scholar]
- EFSA. EFSA Journal. Parma, Italy: European Food Safety Authority; 2006. Guidance document for the risk assessment of genetically modified plants and derived food and feed. [Google Scholar]
- FAO/WHO. Foods derived from modern biotechnology. Rome: World Health Organization; 2009. Codex Alimentarius. [Google Scholar]
- Fecek RJ, Marcondes Rezende M, Busch R, Hassing I, Pieters R, Cuff CF. Enteric reovirus infection stimulates peanut-specific IgG2a responses in a mouse food allergy model. 2010;215:941–948. doi: 10.1016/j.imbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenyo D. The biopolymer markup language. Bioinformatics. 1999;15:339–340. doi: 10.1093/bioinformatics/15.4.339. [DOI] [PubMed] [Google Scholar]
- FIFRA. Data required to register plant-incorporated protectants. Arlington, VA: US Environmental Protection Agency; 2009. [Google Scholar]
- Finney A, Hucka M. Systems biology markup language: Level 2 and beyond. Biochem. Soc. Trans. 2003;31:1472–1473. doi: 10.1042/bst0311472. [DOI] [PubMed] [Google Scholar]
- Ganeshan K, Neilsen CV, Hadsaitong A, Schleimer RP, Luo X, Bryce PJ. Impairing oral tolerance promotes allergy and anaphylaxis: a new murine food allergy model. 2009;123:231–238. doi: 10.1016/j.jaci.2008.10.011. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendel SM. Bioinformatics and food allergens. J. AOAC Int. 2004;87:1417–1422. [PubMed] [Google Scholar]
- Gendel SM. Allergen databases and allergen semantics. Regul. Toxicol. Pharmacol. 2009;54:S7–S10. doi: 10.1016/j.yrtph.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Gendel SM, Jenkins JA. Allergen sequence databases. Mol. Nutr. Food Res. 2006;50:633–637. doi: 10.1002/mnfr.200500271. [DOI] [PubMed] [Google Scholar]
- Gonipeta B, Parvataneni S, Paruchuri P, Gangur V. Long-term characteristics of hazelnut allergy in an adjuvant-free mouse model. 2010;152:219–225. doi: 10.1159/000283028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RE. Practical and predictive bioinformatics methods for the identification of potentially cross-reactive protein matches. Mol. Nutr. Food Res. 2006;50:655–660. doi: 10.1002/mnfr.200500277. [DOI] [PubMed] [Google Scholar]
- Goodman RE. Performing IgE serum testing due to bioinformatics matches in the allergenicity assessment of GM crops. 2008;46 Suppl 10:S24–S34. doi: 10.1016/j.fct.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Guo BZ, Liang XQ, Chung SY, Holbrook CC, Maleki SJ. Proteomic analysis of peanut seed storage proteins and genetic variation in a potential peanut allergen. Protein Pept. Lett. 2008;15:567–577. doi: 10.2174/092986608784966877. [DOI] [PubMed] [Google Scholar]
- Hiller R, Laffer S, Harwanegg C, Huber M, Schmidt WM, Twardosz A, Barletta B, Becker WM, Blaser K, Breiteneder H, Chapman M, Crameri R, Duchêne M, Ferreira F, Fiebig H, Hoffmann-Sommergruber K, King TP, Kleber-Janke T, Kurup VP, Lehrer SB, Lidholm J, Müller U, Pini C, Reese G, Scheiner O, Scheynius A, Shen HD, Spitzauer S, Suck R, Swoboda I, Thomas W, Tinghino R, Van Hage-Hamsten M, Virtanen T, Kraft D, Muller MW, Valenta R. Microarrayed allergen molecules: diagnostic gatekeepers for allergy treatment. FASEB J. 2002;16:414–416. doi: 10.1096/fj.01-0711fje. [DOI] [PubMed] [Google Scholar]
- Hochwallner H, Schulmeister U, Swoboda I, Balic N, Geller B, Nystrand M, Harlin A, Thalhamer J, Scheiblhofer S, Niggemann B, Quirce S, Ebner C, Mari A, Pauli G, Herz U, van Tol EA, Valenta R, Spitzauer S. Microarray and allergenic activity assessment of milk allergens. Clin. Exp. Allergy. 2010;40:1809–1818. doi: 10.1111/j.1365-2222.2010.03602.x. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr J-H, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novère N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Hunter A, Kaufman MH, McKay A, Baldock R, Simmen MW, Bard JBL. An ontology of human developmental anatomy. J. Anat. 2003;203:347–355. doi: 10.1046/j.1469-7580.2003.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O, Garcia T, Torres M, Schein CH, Braun W. Characteristic motifs for families of allergenic proteins. Mol. Immunol. 2009a;46:559–568. doi: 10.1016/j.molimm.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O, Midoro-Horiuti T, Schein CH, Xie L, Hillman GR, Goldblum RM, Braun W. The property distance index PD predicts peptides that cross-react with IgE antibodies. Mol. Immunol. 2009b;46:873–883. doi: 10.1016/j.molimm.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O, Oezguen N, Mathura VS, Schein CH, Xu Y, Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions of proteins. Curr. Med. Chem. 2004;11:583–593. doi: 10.2174/0929867043455819. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Schein CH, Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. 2002;18:1358–1364. doi: 10.1093/bioinformatics/18.10.1358. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Schein CH, Braun W. SDAP: Database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O, Schein CH, Garcia T, Oezguen N, Negi SS, Braun W. Structural analysis of linear and conformational epitopes of allergens. Regul. Toxicol. Pharmacol. 2009c;54:S11–S19. doi: 10.1016/j.yrtph.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladics GS, Cressman RF, Herouet-Guicheney C, Herman RA, Privalle L, Song P, Ward JM, McClain S. Bioinformatics and the allergy assessment of agricultural biotechnology products: Industry practices and recommendations. Regul. Toxicol. Pharmacol. 2011 doi: 10.1016/j.yrtph.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Ladics GS, Selgrade MK. Identifying food proteins with allergenic potential: Evolution of approaches to safety assessment and research to provide additional tools. Regul. Toxicol. Pharmacol. 2009;54:S2–S6. doi: 10.1016/j.yrtph.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Mari A, Alessandri C, Bernardi ML, Ferrara R, Scala E, Zennaro D. Microarrayed Allergen Molecules for the Diagnosis of Allergic Diseases. Curr. Allergy Asthma Rep. 2010;10:357–364. doi: 10.1007/s11882-010-0132-0. [DOI] [PubMed] [Google Scholar]
- Mari A, Rasi C, Palazzo P, Scala E. Allergen databases: Current status and perspectives. Curr. Allergy Asthma Rep. 2009;9:376–383. doi: 10.1007/s11882-009-0055-9. [DOI] [PubMed] [Google Scholar]
- Mari A, Scala E. Allergome: a unifying platform. Arb Paul Ehrlich Inst Bundesamt Sera Impfstoffe Frankf A M. 2006;95:29–39. [PubMed] [Google Scholar]
- Mari A, Scala E, Palazzo P, Ridolfi S, Zennaro D, Carabella G. Bioinformatics applied to allergy: Allergen databases, from collecting sequence information to data integration. The Allergome platform as a model. Cell. Immunol. 2006;244:97–100. doi: 10.1016/j.cellimm.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Marti P, Truffer R, Stadler MB, Keller-Gautschi E, Crameri R, Mari A, Schmid-Grendelmeier P, Miescher SM, Stadler BM, Vogel M. Allergen motifs and the prediction of allergenicity. Immunol. Lett. 2007;109:47–55. doi: 10.1016/j.imlet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Mathura VS, Schein CH, Braun W. Identifying property based sequence motifs in protein families and superfamilies: Application to DNase-1 related endonucleases. Bioinformatics. 2003;19:1381–1390. doi: 10.1093/bioinformatics/btg164. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Mathura V, Schein CH, Braun W, Yu SN, Watanabe M, Lee JC, Brooks EG, Goldblum RM. Major linear IgE epitopes of mountain cedar pollen allergen Jun a 1 map to the pectate lyase catalytic site. Mol. Immunol. 2003;40:555–562. doi: 10.1016/s0161-5890(03)00168-8. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Schein CH, Mathura V, Werner B, Czerwinski EW, Togawa A, Kondo Y, Oka T, Watanabe M, Goldblum RM. Structural basis for epitope sharing between group 1 allergens of cedar pollen. Mol. Immunol. 2006;43:509–518. doi: 10.1016/j.molimm.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, Braun W. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol. Immunol. 2008;45:3740–3747. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvataneni S, Gonipeta B, Tempelman RJ, Gangur V. Development of an adjuvant-free cashew nut allergy mouse model. 2009;149:299–304. doi: 10.1159/000205575. [DOI] [PubMed] [Google Scholar]
- Rabjohn P, Helm EM, Stanley JS, West CM, Sampson HA, Burks AW, Bannon GA. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Invest. 1999;103:535–542. doi: 10.1172/JCI5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 2008;121:847–852. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Randhawa GJ, Singh M, Grover M. Bioinformatic analysis for allergenicity assessment of Bacillus thuringiensis Cry proteins expressed in insect-resistant food crops. 2011;49:356–362. doi: 10.1016/j.fct.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Reisdorph NA, Reisdorph R, Bowler R, Broccardo C. Proteomics methods and applications for the practicing clinician. Ann. Allergy Asthma Immunol. 2009;102:523–529. doi: 10.1016/S1081-1206(10)60128-7. [DOI] [PubMed] [Google Scholar]
- Saha S, Raghava GPS. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Ivanciuc O, Braun W. Structural Database of Allergenic Proteins (SDAP) In: Maleki SJ, Burks AW, Helm RM, editors. Food Allergy. Washington, D.C.: ASM Press; 2006. pp. 257–283. [Google Scholar]
- Schein CH, Ivanciuc O, Braun W. Bioinformatics approaches to classifying allergens and predicting cross-reactivity. Immunol. Allerg. Clin. North Am. 2007;27:1–27. doi: 10.1016/j.iac.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Ivanciuc O, Midoro-Horiuti T, Goldblum RM, Braun W. An allergen portrait gallery: Representative structures and an overview of IgE binding surfaces. Bioinform. Biol. Insights. 2010;4:113–125. doi: 10.4137/BBI.S5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Zhou B, Oezguen N, Mathura VS, Braun W. Molego-based definition of the architecture and specificity of metal-binding sites. Proteins. 2005;58:200–210. doi: 10.1002/prot.20253. [DOI] [PubMed] [Google Scholar]
- Schuurman N, Leszczynski A. Ontologies for bioinformatics. Bioinformatics Biol. Insights. 2008;2:187–200. doi: 10.4137/bbi.s451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selgrade MK, Bowman CC, Ladics GS, Privalle L, Laessig SA. Safety assessment of biotechnology products for potential risk of food allergy: Implications of new research. Toxicol. Sci. 2009;110:31–39. doi: 10.1093/toxsci/kfp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Compadre CM, Maleki SJ, Kopper RA, Sampson H, Huang SK, Burks AW, Bannon GA. Biochemical and structural analysis of the IgE binding sites on Ara h1, an abundant and highly allergenic peanut protein. J. Biol. Chem. 1998;273:13753–13759. doi: 10.1074/jbc.273.22.13753. [DOI] [PubMed] [Google Scholar]
- Shreffler WG, Lencer DA, Bardina L, Sampson HA. IgE and IgG4 epitope mapping by microarray immunoassay reveals the diversity of immune response to the peanut allergen, Ara h 2. J. Allergy Clin. Immunol. 2005;116:893–899. doi: 10.1016/j.jaci.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Silvanovich A, Bannon G, McClain S. The use of E-scores to determine the quality of protein alignments. 2009;54:S26–S31. doi: 10.1016/j.yrtph.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Silvanovich A, Nemeth MA, Song P, Herman R, Tagliani L, Bannon GA. The value of short amino acid sequence matches for prediction of protein allergenicity. Toxicol. Sci. 2006;90:252–258. doi: 10.1093/toxsci/kfj068. [DOI] [PubMed] [Google Scholar]
- Soman KV, Midoro-Horiuti T, Ferreon JC, Goldblum RM, Brooks EG, Kurosky A, Braun W, Schein CH. Homology modeling and characterization of IgE binding epitopes of mountain cedar allergen Jun a 3. Biophys. J. 2000;79:1601–1609. doi: 10.1016/S0006-3495(00)76410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislaus R, Jiang LH, Swartz M, Arthur J, Almeida JS. An XML standard for the dissemination of annotated 2D gel electrophoresis data complemented with mass spectrometry results. BMC Bioinformatics. 2004;5:9. doi: 10.1186/1471-2105-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, Helm RM, West CM, Bannon GA. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch. Biochem. Biophys. 1997;342:244–253. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- Thomas K, Bannon G, Herouet-Guicheney C, Ladics G, Lee L, Lee SI, Privalle L, Ballmer-Weber B, Vieths S. The utility of an international sera bank for use in evaluating the potential human allergenicity of novel proteins. 2007;97:27–31. doi: 10.1093/toxsci/kfm020. [DOI] [PubMed] [Google Scholar]
- Thomas K, MacIntosh S, Bannon G, Herouet-Guicheney C, Holsapple M, Ladics G, McClain S, Vieths S, Woolhiser M, Privalle L. Scientific advancement of novel protein allergenicity evaluation: An overview of work from the HESI Protein Allergenicity Technical Committee (2000–2008) Food Chem. Toxicol. 2009;47:1041–1050. doi: 10.1016/j.fct.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Truffer R, Stadler MB, Vogel M, Mari A, Stadler BM. Computational resources: a regulatory need, a tool for research. Arb Paul Ehrlich Inst Bundesamt Sera Impfstoffe Frankf A M. 2006;95:11–15. [PubMed] [Google Scholar]
- Venkatarajan MS, Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical-chemical properties. J. Mol. Model. 2001;7:445–453. [Google Scholar]
- Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Fujiwara K, Yoshida M, Kageyama-Yahara N, Kuramoto H, Shibahara N, Kadowaki M. Therapeutic effect of kakkonto in a mouse model of food allergy with gastrointestinal symptoms. 2009;148:175–185. doi: 10.1159/000161578. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, Lund O, Lundegaard C, Nielsen M, Ponomarenko J, Sette A, Zhu Z, Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]