Summary
Midzones, also called central spindles, are an array of antiparallel microtubules that form during animal cell cytokinesis between the separated chromosomes [1]. Midzones can be considered platforms that recruit specific proteins and orchestrate cytokinetic events, such as keeping sister nuclei apart, furrow ingression, and abscission [2–3]. Despite this important role, many aspects of midzone biology remain unknown, including the dynamic organization of midzone microtubules. Investigating midzone microtubule dynamics has been difficult in part because their plus-ends are interdigitated and buried in a dense matrix, making them difficult to observe. We employed monopolar cytokinesis to reveal that midzone plus-ends appear non-dynamic. We identified the chromokinesin KIF4 as a negative regulator of midzone plus-end dynamics, whose activity controls midzone length, but not stability. KIF4 is required to terminate midzone elongation in late anaphase. In the absence of KIF4, midzones elongate abnormally, and their overlap regions are unfocused. Electron dense material and midbodies are both absent from the elongated midzones, and actin filaments from the furrow cortex are not disassembled after ingression. KIF4 mediated midzone length regulation appears to occur by terminating midzone elongation at a specific time during cytokinesis, making midzones and mitotic spindles differ in their dynamics and length regulating mechanisms.
Results and discussion
KIF4 negatively regulates microtubule plus-end dynamics in monopolar cytokinesis
The dynamic behavior of midzone (also called central spindle) microtubules, and how their length is regulated, are poorly understood. Midzones are antiparallel arrays with plus-ends oriented towards the center [4], like the metaphase spindles they follow during cell division. It has thus been natural to assume these two are spatially organized and length regulated according to similar principles [5–8]. In this study, we investigated midzone dynamics and length regulation, and found they are in fact very different from that of mitotic spindles.
In normal midzones, the overlapping, antiparallel plus-ends are buried in a dense matrix that excludes antibodies against tubulin and stains darkly by thin section EM [9–10], making it difficult to observe their dynamics. We therefore employed monopolar cytokinesis in which the plus-ends of monopolar midzones are exposed in the cytoplasm [11]. Monopolar midzones resemble normal bipolar midzones in protein composition, competency to initiate furrows, and presence of electron dense material coating microtubule near plus-ends [10]. We arrested HeLa cells in monopolar mitosis with the Kinesin-5 inhibitor S-trityl-L-cysteine (STLC) [12] and forced them into cytokinesis with the Cdk1 inhibitor RO-3306 [13]. After an early Aurora-B dependent polarization step that breaks symmetry, midzone microtubules assemble and polarize towards a region of the plasma membrane that recruits furrow proteins. Midzone plus-ends first terminate in or near the cortex, but as the cortex recruits furrow proteins, changes shape and protrudes, the plus-end of midzone bundles are left isolated in the cytoplasm several microns from the cortex (Figure 1) [11].
Figure 1. KIF4 negatively regulates midzone plus-end dynamics during monopolar cytokinesis.
(A) Representative images of different stages in monopolar cytokinesis – polarization, cortex elongation/midzone termination, and furrow ingression. Cells were fixed and stained with antibodies as labeled.
(B) Summary of monopolar cytokinesis phenotypes after depleting cytokinesis proteins. Depletions of PRC1, MKLP1, RacGAP1, and Ect2 blocked the initial polarization. KIF4 depleted midzones polarized, but were not terminated during monopolar cytokinesis.
(C) KIF4 was required to terminate midzone plus-end dynamics in monopolar cytokinesis. HeLa cells stably expressing GFP-β-tubulin were imaged using spinning disk confocal microscope (Movie 1, 2). Imaging started at time zero. Monopolar midzones plus-ends were not polymerizing during furrow ingression in control (red arrow), but were growing towards the furrow region in KIF4 depleted cells (green arrow).
(D) KIF4 was required to properly localize proteins on monopolar midzones. KIF4 depleted monopolar midzone polymerized into the furrow-like cortex region, which was marked in dotted yellow line according to Anillin localization on the cortex. Furrow regions were boxed and magnified in the right. PRC1, MKLP1, and Aurora B localized differently in the absence of KIF4 during monopolar cytokinesis.
Bars = 5 μm. See also Figure S1.
To identify proteins that regulate monopolar midzone length, we depleted a list of cytokinetic proteins which are either midzones associated or cytokinesis regulators (Figure 1B). Depletion of PRC1, MKLP1, RacGAP1, or Ect2 blocked the early polarization step; neither midzones nor furrows formed. Depletion of other proteins, including CLASPs which regulate midzone dynamics in yeast [14], had no obvious effect. Uniquely, depletion of the chromokinesin KIF4 allowed polarization and midzone assembly, but midzones no longer terminated coherently in the cytoplasm. As shown in the timelapse movies of monopolar HeLa cells expressing GFP-β-tubulin (Figure 1C, Movie 1, 2) and GFP-EB3, a marker for growing microtubule plus-ends (Figure S1, Movie 3,4), midzones in control cells terminated in cytoplasm with plus-ends that appeared non-dynamic. However, KIF4 depleted midzone plus-ends continued to grow and entered the protruding furrow region. A furrow could still be assembled, but it was typically less robust. These data confirm that PRC1, MKLP1, RacGAP1, and Ect2 are essential for midzone assembly and the early polarization step. KIF4, in contrast, is required later, to inhibit midzone plus-end dynamics. KIF4 was previously shown to block plus-end dynamics in vitro as a pure protein [15–16]. Here we hypothesized that KIF4 acts directly to block polymerization dynamics at midzone plus-ends in vivo. Given that yeast does not have Kinesin-4 motors, this also suggests the regulation of midzone dynamics in animal cells is different to what has been found in yeast [14, 17].
We examined midzone protein localizations using immunofluorescence. As KIF4 relocalizing from chromosomes to midzone plus-ends in normal cytokinesis [18], KIF4 was observed on monopolar midzone plus-ends (Figure 1D). KIF4 depletion did not affect furrow specification but did perturb midzone proteins recruitments to plus-ends, evidenced by normally polarized furrow protein Anillin on the cortex and diffusely localized PRC1 and MKLP1 along microtubules in the furrow region (Figure 1D, yellow dotted line). Aurora B was delocalized from microtubules, though it still targeted to the furrowing cortex (Figure 1D).
KIF4 terminates midzone plus-end elongation in normal cytokinesis
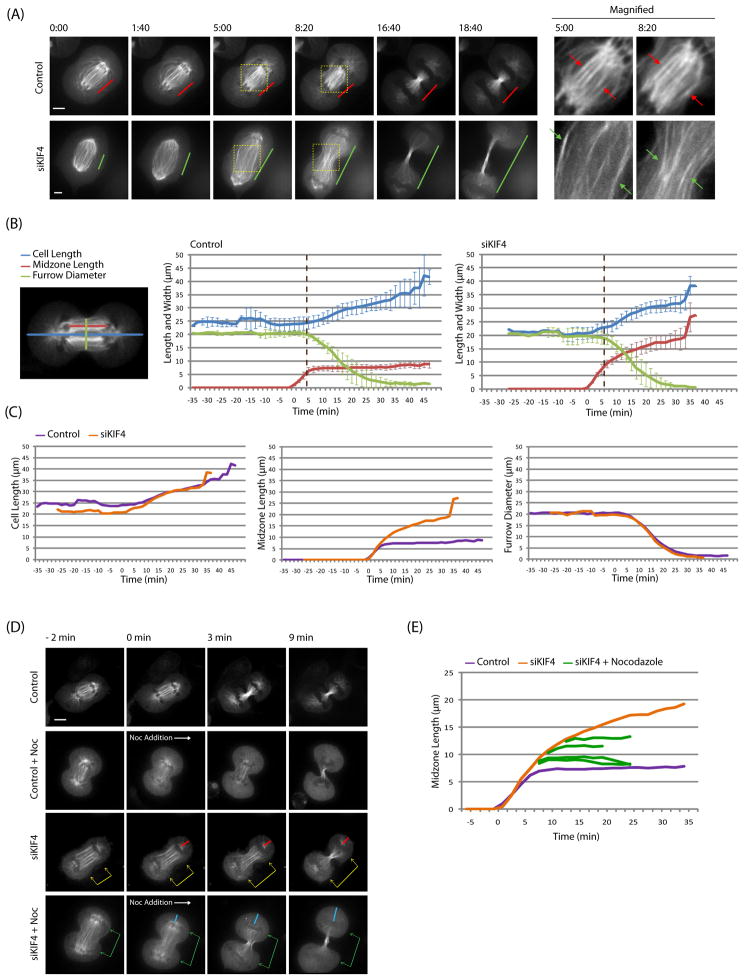
To test if KIF4 also terminates midzone elongation in normal bipolar cytokinesis, we used confocal timelapse microcopy of HeLa cells stably expressing GFP-β-tubulin (Figure 2A, Movie 5, 6). In control cells, midzones elongated rapidly until furrow initiation, and then held an approximately constant length for the rest of cytokinesis (Figure 2A, red lines). Antiparallel overlapping regions, visualized by higher GFP intensity at the center of midzones, were also constant in length and well aligned between individual midzone bundles (Figure 2A, red arrows). When KIF4 was depleted, midzones formed with normal kinetics, but continued to elongate throughout cytokinesis (Figure 2A, green lines). KIF4 depleted midzones were less organized, with antiparallel overlapping regions of variable lengths that were poorly aligned between bundles (Figure 2A, green arrows). Midzone length, cell length, and furrow diameter were measured over time and averaged for twenty controls and ten KIF4 depleted cells (Figure 2B, S2A). The main difference of KIF4 depletion is that midzones elongate continually, instead of stopping at fixed length when furrow ingression starts. In Figure 2C, we superimposed averaged plots of controls and siKIF4 to facilitate comparison. The velocity and extent of cell elongation and furrow ingression were not changed much in the absence of KIF4. We conclude that KIF4 terminates plus-end dynamics in midzones and keeps midzone length constant in normal bipolar cytokinesis, but it is not required for cell elongation of furrow ingression, as in its monopolar variant.
Figure 2. KIF4 mediated plus-end termination regulates midzone length in bipolar cytokinesis.
(A) KIF4 regulated constant midzone lengths during normal bipolar cytokinesis. HeLa cells expressing GFP-β-tubulin were imaged using spinning disk confocal microscope (Movie 5, 6). Time zero was the point furrow ingression started. Midzone length was defined as the distance between segregated chromosomes. Note that midzone lengths were constant in control (red lines), but increased with time in KIF4 depleted cell (green lines) throughout furrow ingression. Antiparallel microtubule overlapping regions were boxed and magnified in the right. The overlaps were constant in length in controls (red arrows) but not in KIF4 depleted cells (green arrows).
(B, C) Correlations between cortex elongation, midzone elongation, and furrow ingression in normal and KIF4 depleted cells. Twenty controls and ten KIF4 depleted cells stably expressing GFP-β-tubulin were measured, averaged, plotted, and grouped differently to facilitate the comparisons. Definition of cell length, midzone length, and furrow diameter were shown. Time zero referred to the point chromosome segregation started (anaphase onset). Error bars in (B) were standard deviations and were omitted in (C) for clarity. Coincident with furrow initiation (dotted brown line), anaphase midzones elongation was terminated in controls, but not in KIF4 depleted cells. Velocities of midzones elongation in KIF4 depleted cells were similar before and after furrow ingression (3.83 and 3.47 μm/min). The velocities of cortex elongation and furrow ingression were similar between controls and KIF4 depleted cells.
(D) KIF4 depleted midzones were resistant to 10 μM nocodazole. HeLa cells expressing GFP-β-tubulin were imaged using spinning disk confocal microscope (Movie 8). Midzone length was defined as the distance between segregated chromosomes and nocodazole was added at time zero. KIF4 depleted midzone continuously elongated after furrow ingression (yellow arrows) which pushed chromosomes along with elongated cortex (red lines). Nocodazole stopped the continuous elongation of KIF4 depleted midzones (green arrows). The distance between chromosomes and the polar cortex increased while cortex elongation continued (blue lines).
(E) Nocodazole stopped post-furrow elongation of KIF4 depleted midzones. Midzone lengths of four KIF4 depleted cells treated with 10 μM nocodazole after furrow ingression were measured, plotted, and superimposed with the midzone length plot in (C) by their lengths before nocodazole exposure. KIF4 depleted midzones were constant in length after nocodazole treatment.
Bars = 5 μm. Also see Figure S2.
KIF4 mediated termination of plus-end dynamics is not essential for midzone stability
Midzones are more stable than mitotic spindles and astral microtubules [19], although it is still unclear how and when they are stabilized during cytokinesis. To test when stabilization occurs, we pulsed cells with microtubule assembly inhibitor nocodazole. Midzones gradually became insensitive to nocodazole after furrow initiation (Figure S2B, Movie 7). Unexpectedly, KIF4 depleted midzone microtubules were still resistant to nocodazole after furrow initiation. Nocodazole rapidly blocked the continued post-furrow elongation, causing the assembly to freeze at the length it had achieved at the time of drug addition (Figure 2D, comparing green and yellow arrows, Movie 8). The distance between chromosomes and polar cortex increased with time since cell elongation continued (Figure 2D, comparing blue to red lines). Lengths of four KIF4 depleted midzones with nocodazole added after furrow initiation were superimposed with Figure 2C for comparison (Figure 2E, green lines). Because KIF4 depleted midzones did not disassemble in the presence of nocodazole, we believe some other molecule(s) is responsible for stabilizing midzones. Midzone bundling protein PRC1 might stabilize microtubules towards depolymerization [20]. Indeed, when we depleted PRC1 and pulsed with nocodazole after furrow initiation, unbundled microtubules between chromosomes were depolymerized (data not shown). However, PRC1 depletion delocalizes other midzone proteins as well, including KIF4, centralspindlin and CENPE [21–22]. It is also possible that any or all of them could be responsible for midzone stabilization. Taken together, these data further confirmed that inhibition of microtubule plus-end growth by KIF4 blocks midzone growth, but also showed it is not essential for midzone stabilization.
Blocking plus-end growth is the main function of KIF4 in midzones during cytokinesis
KIF4 is known to be required for focusing of PRC1 to a tight mid-line in the center of the midzone [22–23]. We confirmed it and showed similar behavior for other midzone proteins (Figure S3A). We further noticed that these proteins relocalized differently on KIF4 depleted midzones (Figure 3A, S3B). In the absence of KIF4, Aurora B was absent from microtubules but remained on the furrow cortex; CENPE and PRC1 were broadly diffuse on midzones albeit the latter was slightly enriched in center overlaps; Plk1 and MKLP1 localized on midzones but were less diffuse than CENPE. Our results showed this diffuse localization was not PRC1 specific. All tested midzone proteins were more or less diffuse on KIF4 depleted midzones. Thus, KIF4 is required for focusing of midzone proteins, even though only PRC1 and Aurora B among them are known KIF4 interacting proteins [22–24].
Figure 3. KIF4 mediated midzone termination focuses midzone proteins on midzones.
(A) Midzone proteins relocalized differently on KIF4 depleted midzones. Cells were fixed and co-stained as labeled. CENPE was used as a reference for protein localization comparison. In the absence of KIF4, Aurora B preferentially relocalized to the furrow cortex. Plk1 and MKLP1 to be less diffuse, and PRC1 diffuse all along midzone microtubules as CENPE did.
(B) Experiment design to replace endogenous KIF4 with mouse KIF4 using human KIF4 specific siRNA.
(C) Summary of expressed mouse GFP-KIF4 (mKIF4).
(D) GFP-mKIF4-FL and GFP-mKIF4-tailless focused and colocalized with PRC1 on endogenous KIF4 depleted midzones. GFP-mKIF4-headless did not focus on midzones and could not be rescued by endogenous KIF4. PRC1 focusing was rescued by GFP-mKIF4-FL, tailless, but not headless.
Bars = 5 μm. Also see Figure S3.
KIF4 could function in midzone assembly as a plus-end polymerization blocker, a transport motor, or both. It has a typical kinesin family organization with a N-terminal motor domain, a central coiled-coil stalk and a C-terminal cargo binding tail domain [25]. KIF4 motor domain alone is sufficient to block microtubule plus-end dynamics in vitro [15], while it requires both its stalk and tail domains to associate with PRC1 [23]. To characterize if PRC1 diffuse localization is a consequence of dynamic and therefore mislocalized midzone plus-ends, or the absence of direct KIF4 interaction, endogenous KIF4 was depleted by human KIF4-specific siRNA and replaced with GFP-fused mouse KIF4 (mKIF4): FL (full-length), tailless (without tail domain), and headless (without motor domain) (Figure 3B, C). As shown in Figure 3D and S3C, GFP-mKIF4-FL rescued both midzone termination and PRC1 focusing in the absence of endogenous KIF4. Albeit with slightly lower efficiency to localize on microtubules, GFP-mKIF4-tailless rescued midzone length regulation. However, PRC1 was also recruited and focused. Consistent with human KIF4 [23], our results showed PRC1 co-immunoprecipitated with GFP-mKIF4-FL, but not GFP-mKIF4-tailless lacking the C-terminal tail domain (data not shown). Therefore KIF4’s ability to focus PRC1 does not depend on a direct transport function. GFP-mKIF4-headless kept its ability to associate with chromosomes, but was unable to terminate midzones. PRC1 and GFP-mKIF4-headless were non-colocalizedly diffuse on elongated midzones. We conclude that the most important function of KIF4 in midzone assembly is to block plus-end growth at the right time. This promotes normal midzone architecture, which is sufficient to focus KIF4 and PRC1 at the mid-line, even without a direct interaction between them.
KIF4 mediated control of midzone length is required to focus the furrow
We next determined the consequence for furrow morphogenesis of elongated midzones with diffusely localized midzone proteins. Anillin and RhoA localized uniformly to broadened regions of the cortex in KIF4 depleted cells (Figure 4A, S4A). Furrow length (along the axis of cell division) was measured using Anillin as a marker. Cell length was used as a reference for different stages of cytokinesis. In early cytokinesis, furrow lengths were similar between control and KIF4 depleted cells. As cytokinesis progressed, furrow regions focused in controls but broadened in KIF4 depleted cells (Figure 4B, S4B). Furrow initiation and ingression are redundantly regulated by astral microtubules and midzones during cytokinesis [26]. Our results indicated a novel function of midzones which cannot be replaced by astral microtubules. Cells require midzones to focus the cleavage furrow from an initial broad range to a highly focused region during ingression.
Figure 4. KIF4 mediated midzone termination focuses furrow proteins and disassembles actin filaments on the cortex.
(A) KIF4 was required to focus furrow on the cortex during cytokinesis. Cells at different stages were fixed and stained with Anillin as a furrow marker. Furrow regions were focusing with time in controls, but not in KIF4 depleted cells. Bars = 5 μm.
(B) Lengths of cell and furrow region in (A) were measured and plotted. Definitions of cell length and furrow region were shown. Data were fitted with exponential trend lines. Furrow regions were focusing while cell length were increasing in controls (purple dots and line), but not in KIF4 depleted cells (orange dots and line). Cell numbers in control = 25 and in siKIF4 = 50.
(C) Thin section EM of furrow regions in cytokinesis. KIF4 depletion caused an elongated intracellular bridge between two daughter cells. Microtubules (green arrows) were less bundled and in lower density in KIF4 depleted cells. Electron dense materials, which coated central overlaps of midzones in controls (brown arrows), was missing in KIF4 depleted cells. The whole-cell views were shown as insets. Boxed regions were magnified in the right.
(D) Thin section EM of the furrow cortex. Long straight actin filaments (red arrows) were observed underneath the cortex in the unfocused furrow region of KIF4 depleted cells. Electron dense materials (white arrows) were observed coating on actin filaments but not on microtubules (green arrow).
Also see Figure S4.
Thin section electron microcopy (EM) revealed defects in the broad furrow region in KIF4 depleted cells. After furrow ingression, normal midzone microtubules (Figure 4Ca, green arrows) were bundled and coated with electron dense materials at central antiparallel overlaps to compose the midbody (Figure 4Ca, brown arrows). In KIF4 depleted cells, midzone microtubules were in lower density and significantly less bundled (Figure 4Cb, green arrows). Electron dense materials were not strongly accumulated and midbody assembly was defective. Normally, cortical actin filaments in furrows disassemble after furrow ingression. In KIF4 depleted midzones, we observed long, straight filaments with the appearance of F-actin (Figure 4Db, S4C, red arrows) with interspersed electon dense materials (Figure 4D, S4C, white arrows) underneath the cortex. Similar long linear fibrous structures were observed in phalloidin staining (Figure S4D), confirming they are actin filaments. Thus, KIF4 is required for accurate specification of midzone and midbody microtubule architecture, and for disassembly of furrow actin.
Spatiotemporal control of KIF4
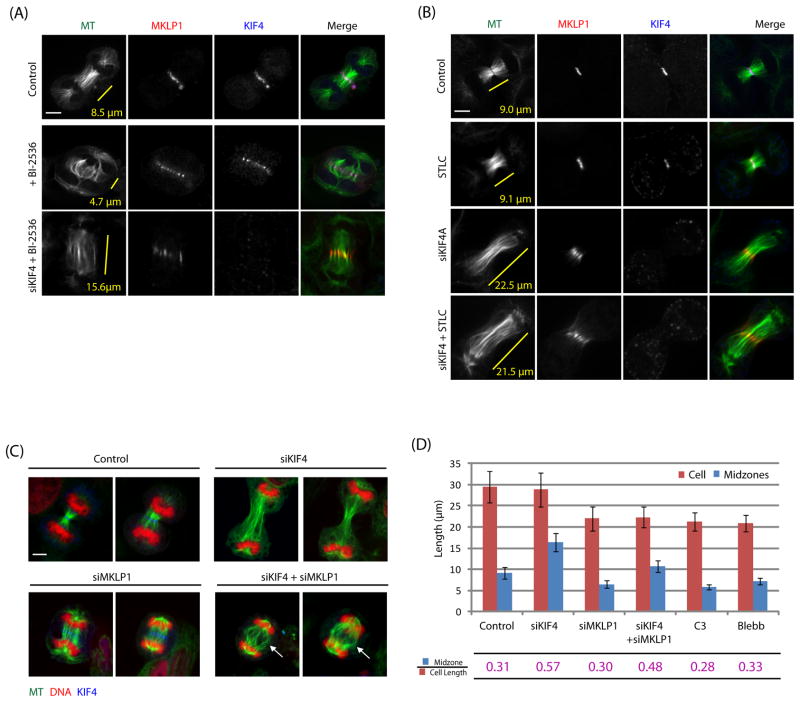
What controls KIF4 activity to determine an appropriate midzone length? Inhibiting Plk1 kinase activity during anaphase was shown to block midzone elongation [27]. We hypothesized that Plk1 negatively regulates KIF4 mediated midzone termination. To test this hypothesis, we asked if depleting KIF4 could override the block of midzone elongation caused by Plk1 inhibition. Cells were treated with Plk1 inhibitor BI-2536 for 60 min to assure that cells entered cytokinesis with drug. In cells that entered cytokinesis before their bipolar spindles collapsed into monopoles, midzones were significantly shorter than controls as reported [27]. MKLP1 and KIF4 were both focused at the center of these short midzones, suggesting microtubule plus-ends were coherently positioned as in normal midzones, but just terminated sooner (Figure 5A). When KIF4 was depleted, midzone elongation was no longer blocked by BI-2536. Midzones became longer than controls and in drug alone, and MKLP1 localized diffusely (Figure 5A). These data suggest that midzone elongation is regulated by a double negative pathway: Plk1 inhibits KIF4 which inhibits midzone elongation. However, there is no consensus Plk1 site identified on KIF4. We suspect that KIF4 might be indirectly regulated by Plk1 for midzone termination.
Figure 5. KIF4 mediated termination of midzone elongation regulates midzone length in cytokinesis.
(A) KIF4 depletion overrode BI-2536-caused block of midzone elongation. Midzone length was shorter than controls under BI-2536 treatment, but became longer than controls if KIF4 was further depleted. Midzone length was measured by the distance between chromosomes.
(B) Kinesin-5 inhibitor STLC did not block midzone elongation. KIF4 depleted midzones were longer than controls and in similar lengths with or without STLC treatment.
(C–D) Centralspindlin was not involved in midzone elongation. Compared to controls, MKLP1 depletion caused shorter cell length and midzones, but maintained a similar midzone over cell length ratio, unless KIF4 was absent. RhoA inhibitor C3 and Myosin II inhibitor blebbistatin (Blebb), which block the cortex elongation, also caused comparable ratios. Error bars were standard deviations. Cell numbers in control = 41, in siKIF4 = 41, in siMLP1 = 41, in siKIF4/siMKLP1 = 26, in C3 = 41, and in Blebb = 68.
In yeast, midzone elongation is driven by the Kinesin-5 homologs Cin8 sliding antiparallel microtubules [28]. Kinesin-5 inhibitor STLC, which does not block cytokinesis entry in cells that have already assembled bipolar spindles, had no effect on midzone elongation (Figure 5B), suggesting Kinesin-5 is not involved. Centralspindlin, a complex containing MKLP1 (Kinesin-6) and RacGAP1, crosslinks antiparallel microtubules in cytokinesis [29]. To test if it drives midzone elongation, cells were treated with either or both KIF4 and MKLP1 siRNAs. As expected, KIF4 depletion caused longer midzones which pushed chromosomes to distal ends of the cell (Figure 5C). Depleting MKLP1 blocks cortex elongation [30]. If MKLP1 were co-depleted, midzones still elongated, and buckled when they reached the limit of shorter cell length. This inhibition of cell elongation physically prevented KIF4 depleted midzones from achieving their maximal length, but they were still significantly longer than controls. These data suggest neither Kinesin-5 nor centralspindlin is required for midzone length increase during cytokinesis. However, the buckled midzones in MKLP1/KIF4 double depleted cells suggested that the force driving midzone elongation is generated internally, and antiparallel sliding is the obvious candidate. Thus we did not exclude the possibility that other kinesins might be involved, for example, Kif15 which can keep centrosomes apart during metaphase when Kinesin-5 is inhibited [31].
Midzone length is coupled with cell length during cytokinesis
Although cells depleted of MKLP1 alone were shorter in both cell and midzone lengths, we noticed the ratio of midzones over cell length was similar to controls (Figure 5D). Direct blocking cell elongation but leaving midzone function intact by RhoA inhibitor C3 toxin or the Myosin II inhibitor blebbistatin also caused comparable ratios. These suggest that cells are able to coordinate midzone lengths with their own cell lengths; unless the KIF4 mediated midzone termination is compromised. It also excluded the possible involvements of RhoA pathway from the cortex in midzone length regulation. HeLa cells seem to scale midzone lengths to cell lengths over the rather small length range we were able to probe (Figure 5D). Similar scaling, but over a larger length range, was observed in Caenorhabdtis elegans early embryos [32]. This suggests midzone length scaling might be a universal mechanism in both embryo and somatic cells.
KIF4 blocks plus-end dynamics of midzone microtubules
Our experiments reveal that KIF4 is required to block midzones from growing continually during cytokinesis by inhibiting plus-end polymerization. This finding provides an in vivo complement to elegant pure protein experiments where KIF4 was shown to inhibit plus-ends dynamics [15] and to limit plus-end growth in reconstituted antiparallel bindles, together with PRC1 [16]. Lack of dynamics may suggest microtubule plus-ends are effectively capped. Microtubule capping was firstly proposed a decade ago [33]. It is not clear from the pure protein data that KIF4 acts as a classic capper, like CapZ for actin. Whatever the precise mechanism, our study provides evidence that KIF4 negatively regulates midzone plus-end dynamics in vivo in mammalian cell.
Our finding that KIF4 lacking its cargo binding domain can largely rescue all these functions suggests that its main role in cytokinesis is to inhibit plus-end growth, not transport cargoes. KIF4 seems to gain its growth-limiting activity a few minutes into anaphase, perhaps when its inhibition by Plk1 is relieved by an unknown mechanism. When we treated KIF4 depleted midzones with nocodazole, they rapidly stopped elongating, but did not depolymerize either, suggesting that plus-end assembly and disassembly might be regulated separately. KIF4 seems to act as a break on growth, and another mechanism plays the role on shrinkage, at least in nocodazole. However, since XKLP1, the KIF4 homolog in Xenopus laevis, is able to inhibit both growing and shrinking on microtubule plus-ends in an in vitro pure protein reconstitution assay [15–16], we do not exclude the possibility that the catastrophe of midzone microtubules is redundantly inhibited by KIF4 capper and another mechanism, such as PRC1-mediated bundling.
Midzone length regulation during cytokinesis
Midzone dynamics are perhaps best understood in yeast. However, neither budding nor fission yeasts have KIF4 homologue in their genomes. In these cells, midzone plus-ends elongate continually, with antiparallel microtubules sliding past each other [14, 17] driven by Kinesin-5 [28]. Cls1p/Peg1p (CLASP) stabilizes fission yeast midzones through preventing microtubule disassembly in the overlapping regions but still allowing plus-end growth [14]. From our data, this type of regulation does not apply in HeLa cells. CLASP depletions in HeLa cells have no defect on midzones in monopolar cytokinesis (Figure 1B), and potential antiparallel sliding kinesins (Kinesin-5 and MKLP1) are not required for midzone elongation (Figure 5). Animal cells might regulate their midzone lengths differently, with evolutionally newly acquired components, such as KIF4.
Current models for midzone length regulation in animal cells tend to assume that midzones exhibit dynamic instability of plus-ends [5–8], as mitotic spindles in metaphase. Length regulation of metaphase spindles is posited that microtubules growth and shrink by dynamic instability, and that length of the bipolar assembly is determined by the balance of these activities combined with antiparallel sliding (Figure 6 left). Our data suggest a quite different model (Figure 6 right). We propose that shortly after they grow, midzone plus-ends are coherently terminated by KIF4, and appear not to undergo polymerization or dynamic instability thereafter. KIF4 at plus-ends blocks polymerization but the mechanism is currently unknown – it may act by distorting the lattice [15]. KIF4 mediated midzone termination appears to be regulated in a timely manner coincident with furrow ingression (Figure 2B). Termination does not, however, depend on furrow ingression, since it continues when ingression is blocked with a RhoA or myosin II poison (Figure 5D). How the timing of termination is governed is not clear. It might involve Plk1, since its inhibition rapidly blocks midzone elongation [27].
Figure 6. Model of KIF4 mediated midzone plus-end termination regulates midzone length in cytokinesis.
Different to mitotic spindles regulating its length through the balance between dynamic plus-ends growing and shrinking, and motor mediated microtubule sliding, midzones regulate its length through a KIF4 mediated termination of anaphase midzone elongation. Negatively regulated by PLk1, KIF4 terminates midzone elongation by inhibiting microtubule plus-end polymerization. A microtubule (MT) stabilizer, which inhibits plus-end depolymerization, provides midzone stability.
Closing remarks
To summarize this work, we first used monopolar cytokinesis to demonstrate that midzone microtubules appear not following dynamic instability. We further identified KIF4 as uniquely required to negatively regulate midzone plus-end dynamics. We then returned to normal bipolar cytokinesis showing this KIF4 mediated negative regulation of plus-ends is essential to terminate anaphase midzone elongation at the onset of furrow ingression, though it is not required to render midzones stable to a nocodazole challenge. This KIF4 mediated midzone termination was also required to focus both midzone proteins and the furrow. These functions could be largely supplied by KIF4 lacking its cargo binding domain, suggesting KIF4 acts mainly as a plus-end dynamics regulator, not a transport motor, during cytokinesis. These data prompted a model for midzone length regulation that is conceptually different from those proposed for yeast midzones in anaphase, and metaphase spindles. In light of our data, current models of midzone length control should be revisited and significantly revised. In particular, models designed to account for mitotic spindle length, where plus-ends are dynamic, will fail for midzones, where KIF4 terminates plus-end dynamics. It is unusual to find a population of microtubules in an animal cell that are not undergoing dynamic instability. Perhaps KIF4 itself, or other proteins that block plus-end dynamics at defined times and places, have broader roles in morphogenesis of the cytoskeleton.
Experimental procedures
Detailed material and methods are described in the supplementary information.
Supplementary Material
Acknowledgments
We thank Nathanael Gray (HMS), James Orth (HMS), Paul Chang (MIT), and Nobutaka Hirokawa (University of Tokyo) for their reagents, and HMS Nikon Imaging Center for microscopy assistances. We thank David Burgess (Boston College) for insightful comments, and Aaron Groen, Edwin Tan, and Ani Nguyen for critical reading. This study was supported by NIH grant GM 23928 to TJM.
Footnotes
Supplemental information includes experimental procedures, 4 figures and 8 movies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bringmann H. Cytokinesis and the spindle midzone. Cell Cycle. 2005;4:1709–1712. doi: 10.4161/cc.4.12.2215. [DOI] [PubMed] [Google Scholar]
- 4.Euteneuer U, McIntosh JR. Polarity of midbody and phragmoplast microtubules. J Cell Biol. 1980;87:509–515. doi: 10.1083/jcb.87.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci U S A. 2004;101:15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goshima G, Scholey JM. Control of Mitotic Spindle Length. Annu Rev Cell Dev Biol. 2010 doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- 8.Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 9.Saxton WM, McIntosh JR. Interzone microtubule behavior in late anaphase and telophase spindles. J Cell Biol. 1987;105:875–886. doi: 10.1083/jcb.105.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoufias DA, DeBonis S, Saoudi Y, Lebeau L, Crevel I, Cross R, Wade RH, Hackney D, Kozielski F. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem. 2006;281:17559–17569. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- 13.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- 16.Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 17.Fridman V, Gerson-Gurwitz A, Movshovich N, Kupiec M, Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10:387–393. doi: 10.1038/embor.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmon ED, Goode D, Maugel TK, Bonar DB. Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J Cell Biol. 1976;69:443–454. doi: 10.1083/jcb.69.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mollinari C, Kleman JP, Saoudi Y, Jablonski SA, Perard J, Yen TJ, Margolis RL. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol Biol Cell. 2005;16:1043–1055. doi: 10.1091/mbc.E04-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci U S A. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozlu N, Monigatti F, Renard BY, Field CM, Steen H, Mitchison TJ, Steen JJ. Binding partner switching on microtubules and aurora-B in the mitosis to cytokinesis transition. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900308-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazumdar M, Misteli T. Chromokinesins: multitalented players in mitosis. Trends Cell Biol. 2005;15:349–355. doi: 10.1016/j.tcb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Bringmann H. Mechanical and genetic separation of aster- and midzone-positioned cytokinesis. Biochem Soc Trans. 2008;36:381–383. doi: 10.1042/BST0360381. [DOI] [PubMed] [Google Scholar]
- 27.Brennan IM, Peters U, Kapoor TM, Straight AF. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS One. 2007;2:e409. doi: 10.1371/journal.pone.0000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Mishima M. Cytokinesis: a view from centralspindlin. Tanpakushitsu Kakusan Koso. 2006;51:206–215. [PubMed] [Google Scholar]
- 30.Hickson GR, Echard A, O’Farrell PH. Rho-kinase controls cell shape changes during cytokinesis. Curr Biol. 2006;16:359–370. doi: 10.1016/j.cub.2005.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Hara Y, Kimura A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr Biol. 2009;19:1549–1554. doi: 10.1016/j.cub.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 33.Infante AS, Stein MS, Zhai Y, Borisy GG, Gundersen GG. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J Cell Sci. 2000;113(Pt 22):3907–3919. doi: 10.1242/jcs.113.22.3907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.