Abstract
The antipsychotic drug, olanzapine, one of the most widely used drugs in clinical medicine, has a high rate of discontinuation due to inefficacy and/or adverse effects. We identified a SNP in the drug metabolizing enzyme cytochrome, P450 3A43 (CYP3A43; rs472660) that highly significantly predicted olanzapine clearance in the CATIE trial (p=5.9e−7). Moreover, at standard antipsychotic doses, 50% of individuals with the high clearance genotype (AA) have trough blood levels below the therapeutic range. Interestingly, a much higher proportion of African Americans carry the A allele compared to Caucasians (allele frequency 67% vs. 14%). After accounting for CYP3A43 genotype, race is no longer a significant predictor of olanzapine clearance. Olanzapine clearance was associated with measures of clinical response. Patients with greater clearance had higher symptom ratings and were more likely to discontinue treatment due to an inadequate response. Our data identify a genetic mechanism for variation in olanzapine response and demonstrate that blood level monitoring of olanzapine treatment is advisable.
Keywords: Genetics, Pharmacokinetics, Pharmacogenetics, Olanzapine, CYP450, CYP3A43
Introduction
The Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) was a systematic evaluation of the clinical response to antipsychotics in the treatment of Alzheimer’s disease and schizophrenia. In the schizophrenia trial (CATIE-SZ), olanzapine was the most effective antipsychotic studied; with the lowest rate of discontinuation for any cause (64% discontinued treatment before 18 months).1 There was a wide variability in response to olanzapine and the other antipsychotics in the trial and although the response to olanzapine was overall more positive, treatment with olanzapine was also associated with greater weight gain and alterations in glucose and lipid metabolism.1 One reason for the high rates of discontinuation may relate to the wide variability in the pharmacokinetics of these drugs, which often results in differences in the pharmacodynamics, both in the response to a drug and the incidence of adverse effects. Sex, smoking, and race have previously been shown to impact clearance of olanzapine,2 which likely contributes to the variable therapeutic response. This study aimed to identify genetic predictors of olanzapine clearance and ultimately response.
Patients and Methods
The study design details for the CATIE-SZ study have been published.3 The study was approved by an institutional review board at each site, and written informed consent was obtained from each patient or his or her legal guardian. Patients with schizophrenia were recruited from multiple US sites and patients in this analysis were randomized to receive olanzapine (7.5 to 30 mg/day taken a single dose or split into two doses). Demographic information was collected at study visits and race was self-reported. A subset of patients provided a sample to be used for genetic analysis (CATIE genetics distribution 7, n=235). The patients in this subset who had all available data pertinent to this study did not differ demographically from all subjects randomized to olanzapine in phase 1 of the CATIE trials (n=336, 33% female, 60% Caucasian, 36% African American). Likewise, baseline PANSS scores did not differ between this group and the entire sample (74.84 ± 19.45 vs. 76.13 ± 18.18, p=0.48).
Plasma samples were collected during the study visits and each patient provided between 1 and 6 samples for determination of olanzapine concentrations. Data were excluded for missing or incorrect dose, time of dose, sample, or time of sample. Plasma levels of olanzapine were determined using liquid chromatography tandem mass spectrometry, as previously described.2,4
Genotyping methods have been previously published.5 We used a candidate gene approach and selected single nucleotide polymorphisms (SNPs) that are in or near known olanzapine metabolizing enzymes (CYP1A2, CYP2D6, and FMO3)6 as well as CYP3A, which is involved in metabolism of approximately half of all drugs and has been reported to be altered during olanzapine treatment.7 All of the available SNPs in these genes on the Affymetrix 500K chip that was used for genotyping of these patients were tested. A total of 26 SNPs were tested in or near CYP1A2 (rs2472300, rs2069522, rs2069526, rs4646425, rs11631682), CYP2D6 (rs6002616, rs9306356), FMO3 (rs2213712, rs2859228, rs2859229, rs1492899, rs929087, rs10458360) and CYP3A4, 3A5, 3A7 or 3A43 (rs2527894, rs2527887, rs4215, rs2525557, rs6960542, rs4729562, rs651430, rs12535293, rs472660, rs17161981, rs17161983, rs2572023, rs2527927).
Nonlinear mixed-effects modeling was used to determine the effects of genotype on olanzapine clearance using NONMEM (version 5, level 1.1; GloboMax, Ellicott City, MD) and the first order conditional estimation method with interaction. The baseline population pharmacokinetic model has been previously published.2 A one-compartment pharmacokinetic model with additive error best describes the data. In a previous analysis of these data, we identified sex, race, and smoking status as independent significant contributors to olanzapine clearance,2 therefore the model used to test each genetic variant included sex, race, and smoking status as covariates. Details on the pharmacokinetic modeling have been previously published.2 Smoking status was given by patient self-report as either active smoker or non-smoker (including past smokers). Concomitant medications that had an incidence of approximately 1% or greater were individually tested as discrete covariates in the original characterization of the population pharmacokinetics of olanzapine, and none had a significant effect.2 Each SNP was tested separately as a categorical covariate to determine its effect on the olanzapine population clearance.
To determine the effect of olanzapine concentrations on clinical response measures, we performed a series of linear regressions between olanzapine clearance estimated using the final model with all significant covariates including CYP3A43 genotype and several measures of clinical response to olanzapine for patients in phase 1 or 1A. We chose PANSS ratings as a measure of symptom severity, time to discontinuation for all cause, as well as reason for discontinuation as measures of clinical response to olanzapine. There were a total of 153 patients that were randomized to olanzapine for phase 1 or 1A for which pharmacokinetic data, genetic data, and any clinical response data were available. Four patients were excluded after blood level analysis indicated the use of an antipsychotic drug outside of the study.
The likelihood ratio test was used to determine the effect of each SNP on the model. The likelihood ratio test is based on the property that the ratio of the NONMEM objective function values (−2 log-likelihood) is asymptotically chi-square distributed. The objective function value is the sum of squared deviations between the predictions and the observations. An objective function decrease of 3.84 units was considered significant (χ2, df=1, p=0.05). That is to say that the SNP had a significant impact on olanzapine clearance if the objective function value decreased at least 3.84 points.
Post-processing of NONMEM outputs and all plots were generated using GraphPad Prism 5 (version 5.02, GraphPad Software, San Diego, CA). Linear regression was performed to determine the magnitude of contribution to the variability of clearance for the significant covariates (sex and smoking status) as well as genotype effects. In order to correct for multiple comparisons for the 26 SNPs tested, we set an a priori threshold based on the conservative Bonferroni correction of p=0.002 for the effect of genotype on olanzapine clearance. Data are reported as mean ± standard deviation. Data in the plots were estimated using the final model, which included the covariates (sex and smoking status) and genotype.
To address the possibility of population stratification, which could result in finding false positive associations in this subset of patients, we completed a multidimensional scaling (MDS) analysis. We performed the MDS analysis based on identify by state (IBS) of the genome-wide SNP genotypes within each racial group and the first two MDS components were extracted to be used as covariates in drug clearance analysis. The IBS and MDS analyses were completed using PLINK8 and R package (http://www.r-project.org/).
Results
Patient demographics are listed in table 1. There was a 9-fold difference in clearance across the population (range 6.941 to 60.09 L/h) with a mean clearance of 25.83 ± 9.920 L/h. Similar to the original pharmacokinetic analysis,2 men had 25% higher clearance than women (27.31 ± 9.975 vs. 21.77 ± 8.613 L/h), smokers had 53% higher clearance compared to nonsmokers (29.54 ± 9.361 vs. 19.26 ± 7.103 L/h), and African Americans had a 17% higher clearance than Caucasians (28.74 ± 10.08 vs. 24.49 ± 9.584 L/h).
Table 1.
Patient demographics by CYP3A43 genotype.
| All patients (n=235) | GG genotype (n=124) | GA genotype (n=76) | AA genotype (n=35) | |
|---|---|---|---|---|
| Sex | 63 F (27%) | 31 F (25%) | 18 F (24%) | 14 F (40%) |
| Smokers | 150 (64%) | 76 (61%) | 51 (67%) | 23 (66%) |
We identified a single SNP in cytochrome P450 3A43 (CYP3A43; rs472660) that highly significantly predicted olanzapine clearance. The addition of CYP3A43 genotype to the pharmacokinetic model, which already included sex, smoking status, and race, resulted in a 10.261 point decrease in objective function value (χ2, df=1, p=0.0014; table 2). Interestingly, when CYP3A43 genotype was entered into the model, race was no longer a significant predictor of olanzapine clearance. When race was removed from the model, the objective function did not change, again indicating that race no longer contributed to the estimate of olanzapine clearance. The difference between a base model (including only smoking and sex) and the addition of genotype to the base model is a highly significant change in −15.75 points (p=7.2e−5), and the addition of genotype explained an additional 5% of the variance not explained by the race effect (table 2). As shown in table 3, 95% of GG carriers are Caucasian (compared to only 5% African American), while the opposite, 89% of AA carriers are African American (compared to only 11% Caucasian). These allele frequencies are similar to those in the HapMap project.9
Table 2.
Pharmacokinetic model statistics
| Model Parameters | Objective function | Change from base model | Change from previous model | p-value based on change from base model | Total R2 |
|---|---|---|---|---|---|
| Smoking and sex | 5814.858 | NA | NA | NA | 0.3327 |
| Smoking, sex, race | 5809.365 | −5.493 | −5.493 | 0.019 | 0.3627 |
| Smoking, sex, race, gene | 5799.104 | −15.754 | −10.261 | 7.2e−5 | 0.4122 |
| Smoking, sex, gene | 5799.104 | −15.754 | 0 | 7.2e−5 | 0.4122 |
Table 3.
CYP3A43 genotype frequency.
| GG | GA | AA | |
|---|---|---|---|
| Caucasian (n=161) | 118 (73%) | 39 (24%) | 4 (2%) |
| African American (n=74) | 6 (8%) | 37 (50%) | 31 (42%) |
CYP3A43 genotype was also a significant predictor of olanzapine clearance within each race, indicating that the overall effect is not an artifact of racial stratification. A linear regression in the 74 African Americans revealed that CYP3A43 genotype predicted more than 10% of olanzapine clearance in these patients (r2=0.1055, F=8.495, DFn=1, DFd=72, p=0.0047). A similar analysis in the 161 Caucasians found that CYP3A43 genotype predicted almost 5% of olanzapine clearance in this group (r2=0.04596, F=7.660, DFn=1, DFd=159, p=0.0063). The overall effect of genotype in both races is that AA carriers have 37% higher clearance than GG carriers, which translates into a 48% lower trough plasma concentration (25 ng/mL compared to 37 ng/mL).
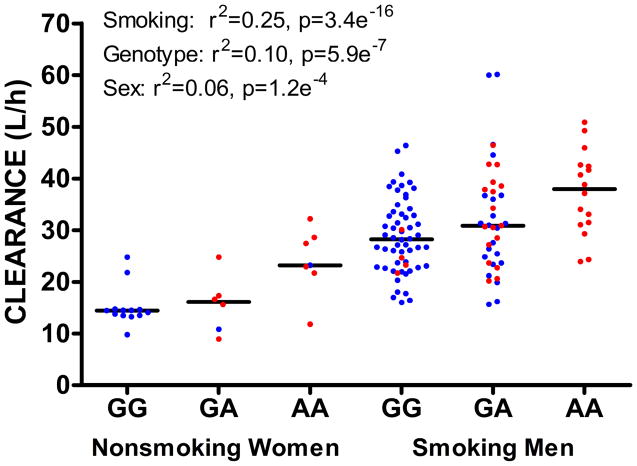
Linear regression analysis in all subjects revealed that patients with the AA genotype had significantly higher olanzapine clearance (31.60 ± 8.990 L/h) compared with GA (27.64 ± 10.70 L/h) and GG genotypes (23.09 ± 8.716 L/h; F= 26.39, DFn=1, DFd=233, p=5.89e−7, figure 1). A linear regression estimated that smoking accounts for 25%, sex accounts for 6%, and CYP3A43 genotype accounts for 10% of the variability in olanzapine clearance. The combined effect of all three covariates is that smoking men with AA genotype (n=16) have 2.5 times higher clearance of olanzapine than nonsmoking women who have the GG genotype (n=13), 37.37 ± 8.175 compared to 15.14 ± 3.870 L/h (figure 1). None of the other 25 SNPs tested were significantly associated with olanzapine clearance. In addition, pharmacokinetic analysis of the clearance data from another drug in the CATIE trial, ziprasidone, in which race was not a predictor,10 revealed no effect of rs472660 genotype (p=0.176), implicating a relatively specific effect of this SNP on olanzapine.
Figure 1.
The combined effect of smoking, sex, and CYP3A43 genotype on olanzapine clearance. Caucasians are in blue. African Americans are in red. Horizontal lines represent median.
In the CATIE olanzapine dataset, there were 150 subjects treated with 15 to 25 mg/day (once or twice a day; median dose 20 mg/day), with 474 measurements of olanzapine plasma concentrations. The mean olanzapine clearance was 25.63 ± 9.934 (range 6.940 to 50.87) L/h, which resulted in a mean steady state concentration of 36.67 ± 24.71 (range 0.09 to 178.5) ng/mL. Based on these data, the mean individual predicted trough concentration would be 31.74 ± 19.20 (range 7.760 to 132.0 ng/mL, which is a 17 fold difference). If we compare groups based solely on genotype, GG carriers have a mean individual predicted trough concentration of 34.84 ± 18.99, GA carriers 31.17 ± 21.07, and AA carriers 23.47 ± 13.59 ng/mL. Moreover, 50% of AA carriers (13 of 26), 33% of GA carriers (15 of 46) and 18% of GG carriers (14 of 78) have individual predicted trough concentrations less than 20 ng/mL, a blood level thought to represent the lower therapeutic limit.
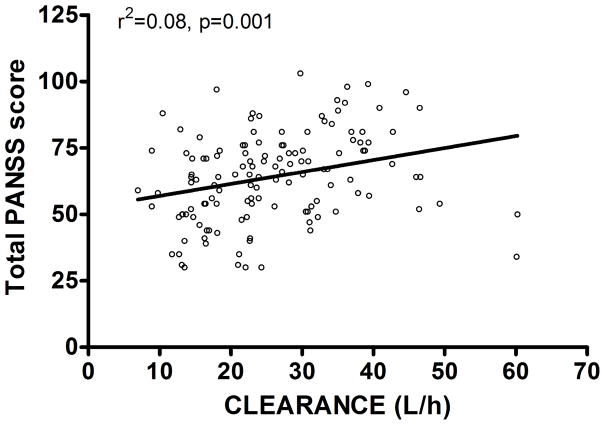
The olanzapine clearance model based on genotype was also significantly associated with measures of clinical response. Patients with a higher clearance (i.e. lower concentrations of olanzapine), had a higher total PANSS score at the end of phase 1/1A (n=131, F=11.39, r2=0.0811, p=0.0010; figure 2), meaning that they had more symptoms, particularly positive symptoms (F=11.48, r2=0.0817, p=0.0009) and general psychopathology ratings (F=11.24, r2=0.0801, p=0.0011) rather than negative symptoms (F=2.774, r2=0.0211, p=0.098), consistent with the fact that antipsychotic drugs are more efficacious in ameliorating positive psychotic symptoms than negative symptoms. While olanzapine clearance did not predict time to discontinuation for all cause in phase 1/1A (n=149, F=1.620, r2=0.011, p=0.205), it did significantly predict the reason for discontinuation. Patients who discontinued olanzapine due to unacceptable side effects (n=34) had the lowest clearance (24.41 ± 8.900 L/h); patients who discontinued due to an inadequate response (n=19) had the highest clearance (30.97 ± 9.192 L/h); and those who discontinued for other reasons (n=15) had an intermediate clearance (29.91 ± 6.687 L/h; ANOVA F=4.361, p=0.0167). Patients with unacceptable side effects had 27% lower clearance than those who had an inadequate response (Bonferroni corrected p=0.031). This means that patients who had a higher olanzapine clearance/lower olanzapine concentrations were more likely to discontinue due to an inadequate therapeutic effect (F=3.981, r2=0.0569, p=0.050), and those with lower olanzapine clearance/higher olanzapine concentrations were more likely to discontinue due to unacceptable side effects (F=8.705, r2=0.117, p=0.0044).
Figure 2.
Olanzapine clearance predicts total PANSS score at end of phase 1/1A.
A multidimensional scaling analysis of the whole genome data revealed no evidence of stratification as an explanation of these genotype associations with clearance both within and across racial groups (supplemental figure 1). While there were some individuals in the Caucasian group that were outside the main cluster, the proportion of outlier individuals was similar for each genotype. In addition, we performed an analysis controlling for sex, smoking and first two dimensions of the MDS data, and the effect of genotype remained in both Caucasians (p=0.0131) and African Americans (p=0.0018) using a general linear model.
Discussion
Approximately half of all drugs are metabolized by the CYP3A family of enzymes,11 which spans 220 kb on chromosome 7q21.1 and consists of four genes, CYP3A4, CYP3A5, CYP3A7, and the most recently identified CYP3A43.12–15 The amino acid sequence of CYP3A43 is 75% identical to CYP3A4 and CYP3A5,13 which results in some overlap in substrate specificity. The SNP in CYP3A43 showing association with olanzapine clearance is intronic and has not been established as a functional variant. However, as illustrated in supplemental figure 2, in the Caucasian sample, it is only in minor linkage disequilibrium (LD) with other CYP3A43 SNPs in HapMap, suggesting that it is either functional or in LD with a previously untyped CYP3A43 variant in this population. In the Yurubi sample there is more significant LD with SNPs (rs667660, rs585071, rs620020, rs2740566) in the 5′ domain of the gene which were not typed in the dataset, nor were any SNPs in LD with these SNPs (supplemental figure 2; SNP version 2.1, www.broadinstitute.org/mpg/snap). Further investigation of genetic variation in CYP3A43 is needed to identify the functional variants likely monitored by this SNP.
CYP3A43 is expressed at much lower levels in human liver compared to CYP3A416 and therefore has previously been thought to be less important in drug metabolism, although analogous to CYP3A5, it may contribute to a large fraction of total hepatic CYP3A enzyme activity due to genetic variation.17 Interestingly, CYP3A43 has higher levels of extrahepatic expression with highest levels in the brain,13, 18 which could result in even larger differences in olanzapine levels at the site of its therapeutic action. In fact, one study showed that differences in CYP3A43 expression resulted in differences in CYP3A43-specific metabolism of alprazolam to alpha-hydroxy alprazolam in human brain.18
While rs472660 was the only SNP tested to have a significant effect on olanzapine clearance, there are likely SNPs in other metabolizing enzymes that contribute to variability in olanzapine clearance, and future studies should test known and identify novel functional polymorphism in CYP450s and other metabolizing enzymes. For example, there is evidence that a genetic variant in UDP-glucuronosyltransferase 1A4 (UGT1A4), a phase 2 metabolizing enzyme that catalyzes the conjugation of a glucuronosylgroup, has a significant effect on olanzapine clearance.19
While further research into CYP3A genetics is needed, these data suggest that genetic variation in CYP3A43 contributes to the wide variability in olanzapine pharmacokinetics and therefore likely results in differences in clinical response. While genotype alone did not predict clinical outcome, olanzapine clearance estimated using CYP3A43 genotype significantly predicted PANSS score, and was also associated with reason for discontinuation. These results indicate that olanzapine pharmacokinetics can have a direct impact on clinical response and therefore it may be important to take CYP3A43 genotype and other contributors to pharmacokinetic variability into consideration when dosing olanzapine.
Dose-response curves20 and international dosing recommendations of olanzapine21 agree that effective doses of olanzapine are in the minimum range of 15 to 20 mg/day, with target plasma concentrations of 20–50 ng/mL22, 23 Based on our clearance data, 50% of AA carriers have individual predicted trough concentrations less than 20 ng/mL. These results suggest that while CYP450 genetic testing may ultimately have utility in clinical monitoring of olanzapine therapy, in the absence of more extensive genetic analyses, at the minimum, monitoring blood levels of olanzapine is advisable.
Supplementary Material
Acknowledgments
This research was supported (in part) by the Division of Intramural Research Programs, National Institute of Mental Health, National Institutes of Health. The principal investigators of the CATIE (Clinical Antipsychotic Trials of Intervention Effectiveness) trial were Jeffrey A. Lieberman, MD, T. Scott Stroup, MD, MPH and Joseph P. McEvoy, MD. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Genotyping was funded by Eli Lilly and Company. We thank Alette Wessels for sharing the ziprasidone population pharmacokinetic model.
Footnotes
None of the authors have a conflict of interest.
References
- 1.Lieberman JA, Stroup TS, McEvoy JP, et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. The New England Journal of Medicine. 2005;353(12):1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 2.Bigos KL, Pollock BG, Coley KC, et al. Sex, race, and smoking impact olanzapine exposure. Journal of Clinical Pharmacology. 2008;48(2):157–165. doi: 10.1177/0091270007310385. [DOI] [PubMed] [Google Scholar]
- 3.Stroup TS, McEvoy JP, Swartz MS, et al. The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophrenia Bulletin. 2003;29(1):15–31. doi: 10.1093/oxfordjournals.schbul.a006986. [DOI] [PubMed] [Google Scholar]
- 4.Aravagiri M, Marder SR. Determination of olanzapine in plasma by liquid chromatography/electrospray tandem mass spectrometry and its application to plasma level monitoring in schizophrenic patients. AAPS PharmSci. 2002;4(4) Abstract W5016. [Google Scholar]
- 5.Sullivan PF, Lin D, Tzeng JY, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13(6):570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callaghan JT, Bergstrom RF, Ptak LR, Beasley CM. Olanzapine: pharmacokinetic and pharmacodynamic profile. Clinical Pharmacokinetics. 1999;37(3):177–193. doi: 10.2165/00003088-199937030-00001. [DOI] [PubMed] [Google Scholar]
- 7.Ring BJ, Binkley SN, Vandenbranden M, Wrighton SA. In vitro interaction of the antipsychotic agent olanzapine with human cytochromes P450 CYP2C9, CYP2C19, CYP2D6 and CYP3A. Br J Clin Pharmacol. 1996;41(3):181–186. doi: 10.1111/j.1365-2125.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 8.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium TIH. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 10.Wessels AM, Bies RR, Pollock BG, et al. Population pharmacokinetic modeling of ziprasidone in patients with schizophrenia from the CATIE study. Journal of Clinical Pharmacology. doi: 10.1177/0091270010387604. In press. [DOI] [PubMed] [Google Scholar]
- 11.Evans WE, Relling MV. Pharmacogenomics: Translating Functional Genomics into Rational Therapeutics. Science. 1999;286(5439):487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 12.Gellner K, Eiselt R, Hustert E, et al. Genomic organization of the human CYP3A locus: identification of a new, inducible CYP3A gene. Pharmacogenetics. 2001;11(2):111–121. doi: 10.1097/00008571-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Domanski TL, Finta C, Halpert JR, Zaphiropoulos PG. cDNA Cloning and Initial Characterization of CYP3A43, a Novel Human Cytochrome P450. Molecular Pharmacology. 2001;59(2):386–392. doi: 10.1124/mol.59.2.386. [DOI] [PubMed] [Google Scholar]
- 14.Westlind A, Malmebo S, Johansson I, et al. Cloning and Tissue Distribution of a Novel Human Cytochrome P450 of the CYP3A Subfamily, CYP3A43. Biochemical and Biophysical Research Communications. 2001;281(5):1349–1355. doi: 10.1006/bbrc.2001.4505. [DOI] [PubMed] [Google Scholar]
- 15.Thompson EE, Kuttab-Boulos H, Yang L, Roe BA, Di Rienzo A. Sequence diversity and haplotype structure at the human CYP3A cluster. Pharmacogenomics J. 2005;6(2):105–114. doi: 10.1038/sj.tpj.6500347. [DOI] [PubMed] [Google Scholar]
- 16.Westlind-Johnsson A, Malmebo S, Johansson A, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metabolism and Disposition. 2003;31(6):755–761. doi: 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 17.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal V, Kommaddi RP, Valli K, et al. Drug Metabolism in Human Brain: High Levels of Cytochrome P4503A43 in Brain and Metabolism of Anti-Anxiety Drug Alprazolam to Its Active Metabolite. PLoS ONE. 2008;3(6):e2337. doi: 10.1371/journal.pone.0002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghotbi R, Mannheimer B, Aklillu E, et al. Carriers of the UGT1A4 142T>G gene variant are predisposed to reduced olanzapine exposure—an impact similar to male gender or smoking in schizophrenic patients. European Journal of Clinical Pharmacology. 2010;66(5):465–474. doi: 10.1007/s00228-009-0783-8. [DOI] [PubMed] [Google Scholar]
- 20.Davis J, Chen N. Dose response and dose equivalence of antipsychotics. Journal of Clinical Psychopharmacology. 2004;24(2):192–208. doi: 10.1097/01.jcp.0000117422.05703.ae. [DOI] [PubMed] [Google Scholar]
- 21.Gardner D, Murphy A, O’Donnell H, Centorrino F, Baldessarini R. International consensus study of antipsychotic dosing. American Journal of Psychiatry. 2010;167(6):686–693. doi: 10.1176/appi.ajp.2009.09060802. [DOI] [PubMed] [Google Scholar]
- 22.Mauri MC, Steinhilber CPC, Marino R, et al. Clinical outcome and olanzapine plasma levels in acute schizophrenia. European Psychiatry. 2005;20:55–60. doi: 10.1016/j.eurpsy.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Mauri MC, Volonteri LS, Colasanti A, Fiorentini A, De Gaspari IF, Bareggi SR. Clinical pharmacokinetics of atypical antipsychotics: a critical review of the relationship between plasma concentrations and clinical response. Clinical Pharmacokinetics. 2007;46(5):359–388. doi: 10.2165/00003088-200746050-00001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.