Abstract
AIM: To automate breast cancer diagnosis and to study the inter-observer and intra-observer variations in the manual evaluations.
METHODS: Breast tissue specimens from sixty cases were stained separately for estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2/neu). All cases were assessed by manual grading as well as image analysis. The manual grading was performed by an experienced expert pathologist. To study inter-observer and intra-observer variations, we obtained readings from another pathologist as the second observer from a different laboratory who has a little less experience than the first observer. We also took a second reading from the second observer to study intra-observer variations. Image analysis was carried out using in-house developed software (TissueQuant). A comparison of the results from image analysis and manual scoring of ER, PR and HER-2/neu was also carried out.
RESULTS: The performance of the automated analysis in the case of ER, PR and HER-2/neu expressions was compared with the manual evaluations. The performance of the automated system was found to correlate well with the manual evaluations. The inter-observer variations were measured using Spearman correlation coefficient r and 95% confidence interval. In the case of ER expression, Spearman correlation r = 0.53, in the case of PR expression, r = 0.63, and in the case of HER-2/neu expression, r = 0.68. Similarly, intra-observer variations were also measured. In the case of ER, PR and HER-2/neu expressions, r = 0.46, 0.66 and 0.70, respectively.
CONCLUSION: The automation of breast cancer diagnosis from immunohistochemically stained specimens is very useful for providing objective and repeatable evaluations.
Keywords: Automation, Breast cancer diagnosis, Computer aided diagnosis, Image analysis, Immunohistochemical study
INTRODUCTION
Cancer of the breast is the second most common human neoplasm and accounts for approximately one quarter of all cancers in females after cervical carcinoma[1]. The accurate diagnosis of cancer plays a very important role in the treatment of patients with neoplastic breast disease. Immunohistochemical evaluation of hormone receptor expressions in tumor cell nuclei is an integral part of routine breast cancer diagnosis and provides important information for prognosis and choice of therapeutic approach.
Estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2/neu) over-expression as a predictor for herceptin therapy are crucially important in the biology of breast carcinoma. ER and PR expressions are the only predictive factors with proven usefulness in selecting patients who are likely to respond to adjuvant endocrine therapy. Patients lacking these receptors tend to have a shorter disease-free survival and earlier recurrence than those expressing these receptors[2]. In around 20%-30% of breast carcinoma, HER-2/neu is amplified and over-expressed. It is associated with an adverse prognosis independent of other prognostic factors in most cases and appears to be stronger in node-positive carcinoma. Immunohistochemical reactivity of tumor cells to ER, PR, and HER-2/neu helps the clinician to establish the mode of therapy and indicates the survival and recurrence rates of the tumor. The recurred tumor or metastatic tumor may not show the same immunoreactivity, and the unstable status of HER-2/neu in breast cancer is clinically significant[3]. Receptor status in recurred or metastasized breast cancer can be different from the original tumor. It was reported that ER status changed in around 33% of cases, and HER-2/neu status changed in around 10%.
In addition, ER-positive and ER-negative breast cancers have distinct disease-specific patterns[4]. A molecular classification of breast cancer is also performed based on their reactivity. The treatment protocol varies with the pattern of reactivity and is based on the molecular classification. Immunohistochemistry (IHC) is expected to play an increasingly important role in the clinical management of breast cancer[5].
The main challenges that pathologists are currently facing are productivity, accuracy and objective evaluation. Manual evaluation takes more time and resources. It is less accurate and is also highly subjective. Qualitatively, the immunohistochemically stained specimen can be evaluated visually as the presence of a specific color. However, to perform a quantitative evaluation, the number of stained cell nuclei and/or the amount of specimen that has been stained needs to be measured. For this purpose, computerized image analysis based methods are needed.
Traditionally, pathologists distinguish between positive and negative results based on visual judgment of the percentage of positive tumor cells, the cutoff being arbitrarily defined between 5% and 45%[6-9]. Some studies report the use of semiquantitative scores to assess nuclear staining intensity as a marker of the number of receptors per cell[10-13]. Diverse computerized image analysis systems have also been employed to provide more standardized data for quantification and were found to correlate well with semiquantitative scoring methods[14-16]. A large number of studies have reported the use of image analysis as a means of evaluating histological staining. Substantial efforts have been made to correlate the evaluations made by experienced pathologists with quantitative values[17-26]. Initial studies on the use of computerized image analysis were limited to evaluation of images based on gray levels[27,28]. Recently, studies have used the color spectrum of histological stains rather than gray levels for analysis of the images to discriminate cellular details[29,30]. The receptors can be accurately quantified by measuring the strength of expression[13]. Hence, automation of quantification could be very useful for the evaluation of histological staining for the diagnosis of breast cancer. However, their use in the routine diagnostic laboratory is limited due to the high cost and the complexity of the image analysis systems[17].
The goals of the present study were to establish the validity of the in-house developed image analysis system (TissueQuant version 1.0) for classification of the images for the diagnosis of breast cancer and to determine data variability due to investigator bias by calculating inter- and intra-observer variability in the case of manual evaluations. Each case was subjected to immunohistochemical evaluation along with the image analysis system and validation was performed by comparing visual and computer analysis of the same tissue fields.
MATERIALS AND METHODS
Specimens from sixty patients were subjected to immunohistochemistry separately with antibodies for ER, PR and HER-2/neu. The patient’s name, age, sex and clinical data were recorded. Out of a total of 60 cases studied 23 (38%) were younger than 50 years while the remaining 37 (62%) were older than 50 years of age. The youngest patient was 30 years and the oldest was 72 years old. The mean age of the patients was 52.5.
The specimens were received in 10% formalin and were sampled after fixation for less than 24 h. Care was taken not to over fix the tissue, as this would interfere with the receptor analysis. The specimens were examined grossly for ulceration, peau d’ orange, and retraction of nipple. The deeper resected margin was stained with India ink. Adequate numbers of sections were taken from the nipple and areola, the tumor proper, all the margins with and without tumor, adjacent breast parenchyma and other relevant areas.
Staining protocol
Hematoxylin and eosin (HE) was used as a routine stain to establish the histopathological diagnosis and for general study of the tissue; the markers ER, PR and Her-2/neu were assessed using immunohistochemistry; image analysis was performed using TissueQuant software for each immunohistochemically stained slide.
Immunohistochemistry staining for ER, PR and HER-2/neu was carried out by the polymer labeling 2-step method using the Super SensitiveTM Polymer- HRP IHC detection system (Biogenex).
Slide preparation
Slides were washed in soapy water and then washed three times with distilled water. Thereafter, they were rinsed in methanol and dried at room temperature. Poly-L-lysine solution was applied to the slides which were dried overnight at room temperature. Sections were treated as follows: (1) 5 μ thick sections were cut and mounted on the slides coated with poly-L-lysine. The sections were deparaffinized with 3 changes in xylene, 2 changes in methanol and then a decreasing concentration of isopropyl alcohol (i.e. 90%, 70%, 50% alcohol) and finally in distilled water for 5 min each; (2) The slides were then immersed in 3% hydrogen peroxide for 20 min to quench the endogenous peroxidase; (3) Antigen retrieval and unmasking was carried out by immersion in citrate buffer and incubation in a pressure cooker with 250 mL of water at 100°C for 15-20 min; (4) After cooling at room temperature, the slides were washed in buffer (0.1 mol/L Tris-HCl, 0.15 mol/L NaCl, pH 7.5) with the washing procedure carried out in a jar containing Tris buffer and immersing the slides for two 5 min periods (total of 10 min); (5) This was followed by incubation for 20 min in the Power block (buffered casein solution with sodium azide) to suppress non-specific binding of subsequent reagent; (6) The slides were then incubated in mouse primary antibody for 75 min at room temperature, after which they were washed in Tris buffer. For the ER study, mouse monoclonal antibody diluted with HK941-YAK in buffered glycine phosphate pH 7.1, 6% protein and 0.09% sodium azide was used. For the PR study, mouse monoclonal antibody in phosphate buffered saline pH 7.6 containing 1% bovine serum albumin (BSA) was used. For the HER-2/neu study, mouse monoclonal antibody from tissue culture supernatant diluted in phosphate buffered saline pH 7.6 containing 1% BSA and 0.09% sodium azide was used; (7) The slides were then immersed in Super Enhancer for 30 min, after which the washing procedure was repeated; (8) The slides were immersed again in Poly HRP (horseradish peroxidase) for 20 min, after which they were washed with Tris buffer followed by distilled water for 5 min each; (9) The slides were then treated with diaminobenzidine (DAB) chromogen for 5 min to develop the brown color; (10) Thorough washing in Tris buffer and distilled water was then performed; and (11) Counter staining with Meyer’s hematoxylin was performed for 1 min and then the slides were washed in tap water. The slides were dried and mounted with a cover slip and mounting media.
Immunohistochemistry scoring
ER and PR expressions: According to the International Breast Cancer Study Group, ER and PR were graded as: (1) None (grade 0): none of the tumor cells showed nuclear staining; (2) Low (grade 1): 1%-9% of cells showed nuclear positivity; and (3) High (grade 2): ≥ 10% of the cells showed nuclear positivity.
When considering the hormone receptor status, grade 1 and 0 were considered hormone receptor negative, and grade 2 was considered positive.
HER-2/neu expression: According to US FDA panel findings, (1) Grade 3: cell surface protein expression- positive: defined as uniform intense membrane staining of > 30% of invasive tumor cells; (2) Grade 2: cell surface protein expression- equivocal: defined as complete membrane staining that is either non uniform or weak in intensity but with obvious circumferential distribution in at least 10% of cells; and (3) Grade 0 or 1: cell surface protein expression- negative: no staining or weak, incomplete membrane staining in any proportion of the tumor cells.
When considering the HER-2/neu membrane status, grade 0, 1 and 2 were considered negative and grade 3 was considered positive.
Image analysis
Images were analyzed using the in-house developed software (TissueQuant). The facility for choosing the color representing the maximum density of hormone expression as a reference color is provided in the software. Using this facility, the color setting was used for analysis purposes. The software assigns scores to the various shades of the color represented by each pixel of the image, based on how close the shade is to the reference color. Using these values, the total hormone expression in the image is quantified. For this, the image is represented in the HSI color model. Gaussian weighting functions are used for scoring the shades. The widths of the Gaussian weighting functions are decided by the different ranges for the hue, saturation and intensity components. These values decide the range of shades of the color which should be considered as positive staining. These weighting functions provide the flexibility of fine adjustments of the color shades to be included.
Considering two parameters based on the color scores, a classification system was developed to classify a particular case as positive, negative and strongly positive. There were two parameters assigned-Mean and Mean-Max. Mean was the main criterion on which the decision was made. It represents the average hormone expression present in the image. Mean-Max was used as a helpful parameter for decision-making. This represents the maximum depth of the color shade present in the image. This was useful when the expression was concentrated in a small area. A grade was calculated based on the above two scores.
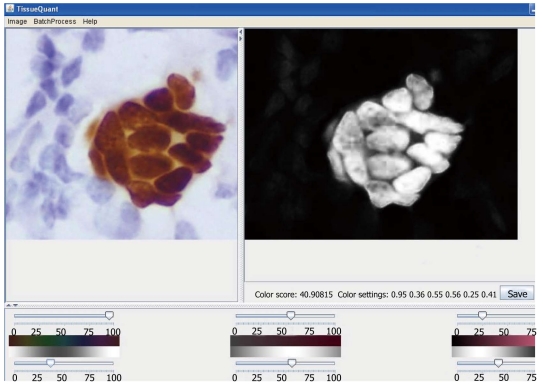
A screenshot of the software is shown in Figure 1. The user is provided with a facility to open the image and click on the region of the image with the reference pixel color. A set of sliders are provided to adjust the color parameters on the lower mid panel. The centers and widths of the Gaussian weighting functions can be adjusted with these sliders. The color parameters are set this way to calculate the color score for each pixel which is mapped from 0 to 255 for the purpose of display as a grayscale image in the right upper panel of the user interface. The color score and the color settings for the particular study are displayed just below the resulting image. This facilitates user interaction to select the appropriate color settings for the quantification. A “Save”button is also provided which facilitates saving of the resulting image. We could also save the color settings by specifying a name for the particular study. These color settings can be used for batch processing of image sets with the same color settings in an automated manner. To work on huge sets of images with the same color setting directly, the “Batch Analysis” option is used. The directory in which the images are saved, the color setting and the image type used for the batch analysis need to be specified by the user. All images in the specified directory are processed and the resulting images are stored in the ‘result-images’ directory, which is generated under the specified directory. The mean color score and the image name are stored in an MS Excel file in the same directory.
Figure 1.
Screenshot of the TissueQuant software. On the left panel of the main window the original image is opened. The right panel displays the color score representation of the image. In the lower panel are the sliders which could be adjusted to select the color shade of interest.
Manual method and image analysis
The specimens from all sixty cases were stained separately for ER, PR and HER-2/neu. All cases were assessed by manual grading as well as image analysis. The manual grading was carried out by an experienced expert pathologist. To study inter-observer and intra-observer variations, we obtained readings from another pathologist as the second observer from a different laboratory who has a little less experience than the first observer. We also took a second reading from the second observer after 30 d to study intra-observer variations.
For statistical analysis of the results, we used SPSS 11.5 for Windows and GraphPad Prism 4.03. The inter-observer and intra-observer variations were evaluated with SPSS software. The Spearman correlation coefficient was obtained using GraphPad Prism software. A ROC curve was drawn using Microsoft Excel software using the sensitivity and specificity of the algorithm for each of the expressions.
RESULTS
A comparison of the grading of the cases by the expert and the image analysis software TissueQuant for ER, PR and HER-2/neu expressions are shown in Table 1, 2, 3, 4, 5 and Figure 2.
Table 1.
Comparison of manual grading and image analysis of estrogen receptor
| Manual grading |
Image analysis |
Total | ||
| Grade 0 | Grade 1 | Grade 2 | ||
| Grade 0 | 30 | 6 | 6 | 42 |
| Grade 1 | 0 | 2 | 2 | 4 |
| Grade 2 | 0 | 0 | 14 | 14 |
| Total | 30 | 8 | 22 | 60 |
Table 2.
Comparison of manual grading and image analysis of progesterone receptor
| Manual grading |
Image analysis |
Total | ||
| Grade 0 | Grade 1 | Grade 2 | ||
| Grade 0 | 35 | 6 | 2 | 43 |
| Grade 1 | 0 | 2 | 5 | 7 |
| Grade 2 | 0 | 0 | 10 | 10 |
| Total | 35 | 8 | 17 | 60 |
Table 3.
Comparison of manual grading and image analysis of human epidermal growth factor receptor-2
| Manual grading |
Image analysis |
Total | |||
| Grade 0 | Grade 1 | Grade 2 | Grade 3 | ||
| Grade 0 | 6 | 0 | 1 | 0 | 7 |
| Grade 1 | 0 | 0 | 0 | 0 | 0 |
| Grade 2 | 0 | 0 | 2 | 5 | 7 |
| Grade 3 | 0 | 0 | 0 | 46 | 46 |
| Total | 6 | 0 | 3 | 51 | 60 |
Table 4.
Results of the classification of estrogen receptor, progesterone receptor and human epidermal growth factor receptor-2 expression images
| True positive | True negative | False positive | False negative | Sensitivity (%) | Specificity (%) | PPV1 (%) | NPV2 (%) | |
| ER expression | 14 | 38 | 8 | 0 | 100 | 82.6 | 63.6 | 100 |
| PR expression | 10 | 43 | 7 | 0 | 100 | 86.0 | 58.8 | 100 |
| HER-2/neu expression | 46 | 9 | 5 | 0 | 100 | 64.3 | 90.2 | 100 |
PPV: Positive predictive value;
NPV: Negative predictive value. It can be seen that the sensitivity has been maintained at 100%. The specificity of the evaluations for ER and PR is good. The specificity of HER-2/neu expression is comparatively less than the ER and PR expression evaluations. ER: Estrogen receptor; PR: Progesterone receptor; HER-2/neu: Human epidermal growth factor receptor-2.
Table 5.
Spearman correlation coefficient (95% confidence interval) of the different evaluations
| Expert vs automated analysis1 | Expert vs pathologist2 | Pathologist reading 1 vs pathologist reading 23 | |
| ER expression | 0.73 (0.59 to 0.83) | 0.53 (0.32 to 0.69) | 0.46 (0.24 to 0.64) |
| PR expression | 0.82 (0.73 to 0.90) | 0.63 (0.45 to 0.76) | 0.66 (0.49 to 0.78) |
| HER-2/neu expression | 0.92 (0.88 to 0.96) | 0.68 (0.52 to 0.80) | 0.70 (0.54 to 0.81) |
Represents the correlation of the automated analysis with the expert’s evaluation;
Represents the correlation between two pathologists;
Represents the correlation between readings by the same pathologists taken at different time periods. It can be seen that the Spearman correlation coefficients for evaluations by automated analysis are better for all receptor expressions.
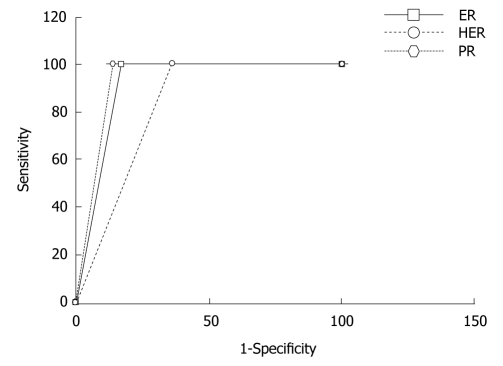
Figure 2.
ROC curves for the automated evaluation of estrogen receptor, progesterone receptor and human epidermal growth factor receptor-2 expression images using TissueQuant. For all three expressions the sensitivity of 100% is maintained, the specificity for PR expression is best at, 86%, for ER the specificity is 82.3% and for HER-2/neu expression the specificity is least at, 64.3%. ER: Estrogen receptor; PR: Progesterone receptor; HER-2/neu: Human epidermal growth factor receptor-2.
ER
All cases were assessed by manual grading as well as image analysis. Thirty cases with grade 0 on manual grading showed a similar grade on image analysis. Out of 12 cases with a manual grading of 0, 6 showed grade 1 and 6 showed grade 2 on image analysis. Two cases with manual grade 1 showed grade 2 on image analysis. All cases with manual grade 2 showed a similar grade on image analysis. These findings were statistically highly significant (Table 1).
The ER expression evaluation performed by the expert was compared with the evaluation by the second pathologist. Evaluation by both pathologists in 24 cases of grade 0 out of 42 cases, 2 cases of grade 1 out of 4 cases and 8 cases of grade 2 out of 14 cases matched correctly. The ER expression evaluation was repeated by the second pathologist and was compared to the first evaluation. The evaluation in both readings matched in 19 cases of grade 0 out of 26 cases, 12 cases of grade 1 out of 21 cases and 6 cases of grade 2 out of 13 cases.
PR
PR status was also assessed by manual grading as well as image analysis. Thirty five cases with grade 0 on manual grading showed a similar grade on image analysis. Out of 8 cases with a manual grading of 0, 6 showed grade 1 and 2 showed grade 2 on image analysis. Five cases with manual grade 1 showed grade 2 on image analysis. All cases with manual grade 2 showed a similar grade on image analysis. These findings were statistically highly significant (Table 2).
The PR expression evaluation performed by the expert was compared with the evaluation by the second pathologist. Evaluation by both pathologists in 32 cases of grade 0 out of 43 cases, 6 cases of grade 1 out of 7 cases and 6 cases of grade 2 out of 10 cases matched correctly.
The PR expression evaluation was repeated by the second pathologist and was compared to the first evaluation. The evaluation in both readings matched in 28 cases of grade 0 out of 34 cases, 8 cases of grade 1 out of 18 cases and 3 cases of grade 2 out of 8 cases.
HER-2/neu
For HER-2/neu status, 6 cases with manual grade 0 showed a similar grade on image analysis. One case with a manual grade of 0 showed grade 2 on image analysis. None of the cases showed grade 1 on manual as well as image analysis. Five cases with manual grade 2 showed grade 3 on image analysis. All 46 cases with manual grade 3 showed a similar grade on image analysis. These findings were highly significant (Table 3).
HER-2/neu expression evaluation performed by the expert was compared with the evaluation by the second pathologist. Evaluation by both pathologists in 6 cases of grade 0 out of 7 cases, 3 cases of grade 2 out of 7 cases and 29 cases of grade 3 out of 46 cases matched correctly.
HER-2/neu expression evaluation was repeated by the second pathologist and was compared to the first evaluation. The evaluation in both readings matched in 6 cases of grade 0 out of 7 cases, 2 cases of grade 2 out of 7 cases and in all 46 cases of grade 3.
DISCUSSION
Various types of solutions are available to quantify staining intensity and range from inexpensive, general purpose software to specific, expensive software. Some of the image analysis systems used for such studies are SAMBA, Image Pro Plus, Metaview, Lucia software, and BioQuant Nova Prime. Charpin et al used Metaview software for staining intensity quantification. This is a general purpose image processing software[31]. Suitable threshold values for the Red (R), Green (G) and Blue (B) components are selected to choose the stained area. The amount of positively stained area gives the measure of staining. Charpin et al[32] used the SAMBA 4000 image analysis system for quantification of hormone receptor expression. For each marker’s positive cell surface, integrated and mean optical densities and IOD histograms were compared. Pauschinget et al[33] and Soukupova et al[34] made use of Lucia software for stain quantification, which uses a measure of optical density. Diaz Encarnacion et al[31] and Niendorf et al[35] used a threshold and area measurement approach. Hatanaka et al[20] used WinROOF with macroinstructions for analyzing each captured area. Lehr et al[17] used Adobe Photoshop-based image analysis to quantify hormone receptor expression in breast cancer. The feature selection was done with the Magic Wand tool which could reliably select all immunostained nuclei. The nuclear immunostaining index was calculated as the difference between nuclear and background immunostaining intensity. Vrekoussis et al[36] reported the use of freeware ImageJ for the analysis of immunohistochemically stained sections of breast cancer. McCabe et al[37] and Chung et al[38] carried out quantitative analysis of hormone receptor expressions in breast cancer using the AQUA system. BioQuant Nova Prime is an advanced image analysis tool designed for biomedical research. Ariol SL 50 is an automated microscope slide analysis tool, which acquires monochrome images through three bright field filters, using cell masking templates and applies area analysis. Image Pro Plus (Media Cybernetics) and EMPIX Imaging solutions are also being used for stain quantification. Sharangpani et al[39] developed a semi-automatic system to quantify estrogen and progesterone receptor immunoreactivity in human breast cancer. All these applications work on the basis of threshold and area measurement or change in optical density. It is not always possible to select appropriate R, G and B thresholds to suitably select shades of a particular color. Hence, the approach of threshold and area measurement is inadequate. In addition, when a tissue section is studied for a particular substance, other components present in the section may also take up the stain, expressing different colors. In such cases, the change in optical density is not a suitable indicator to measure the amount of the substance under study. However, the in-house developed software, TissueQuant, overcomes all these drawbacks by facilitating discrimination between colors and also between depths of color. Thus, it provides a fully automated solution for more efficient quantification of staining intensity.
Automation of image analysis holds promise for improving inter- and intra-observer reproducibility which is the main problem with manual analysis. However, the lack of standards in system performance makes automation less reliable. Automation could also face problems with variations in illumination while imaging, variations in staining intensities, and section thickness. These could be solved with automated sectioning and staining systems. Standardized guidelines for transmission of baseline colors are very important since the evaluation is based on intensity. The clinical utility of automated analysis depends on strict adherence to quality assurance of the systems. Discrepancies in evaluation could be really serious; hence, automated evaluation does not eliminate the role of the pathologist. In our study, the focus was to identify all possible positive cases to ensure there were no false negative results. Once this is done, the expert pathologist can prioritize the slides to confirm the evaluation of the automated system.
The benefit of automation and computer-aided diagnosis has been demonstrated here. It was observed that there was a very good correlation between image analysis and expert opinion in evaluating the ER, PR and HER-2/neu expression images. The system was designed to avoid any false negative findings, hence, the specificity was compromised to obtain 100% sensitivity. This can be seen from the ROC curve in Figure 2. Table 5 shows the inter-observer variations and the intra-observer variations. It can be seen that the correlation between two readings by the same observer was slightly higher than the correlation of the readings by the two pathologists in the PR and HER-2/neu expression images. However, in the case of the ER expression images, intra-observer variation was greater than inter-observer variation. It was observed that in the majority of cases of mismatch, the grade 0 cases were mostly evaluated as grade 1 rather than grade 2, and similarly cases of grade 3 were mostly evaluated as grade 2 rather than grade 1. The automated analysis correlated best with the expert’s opinion in all three cases and this was significantly higher than the correlation between the two different observers and between two readings by the same observer.
This paper presents a technique for automation of the diagnosis of breast cancer from immunohistochemically stained biopsy specimens. Our goal was to provide 100% sensitivity and the software was successfully used to efficiently classify the cases. It was also demonstrated that the manual evaluation introduced a lot of variation, whereas the automated analysis provided an objective evaluation and was repeatable. Such automation could facilitate fast and efficient diagnosis of breast cancer and eliminate human errors, to a large extent. However, the results reported here could be further improved with the use of neural networks or other such classification models.
COMMENTS
Background
Cancer of the breast is the second most common human neoplasm and accounts for approximately one quarter of all cancers in females after cervical carcinoma. Immunohistochemical evaluation of hormone receptor expressions in tumor cell nuclei is an integral part of routine breast cancer diagnosis and provides important information for prognosis and choice of therapeutic approach. The main challenges that pathologists are currently facing are productivity, accuracy and objective evaluation. Pure visual estimates of immunohistochemically stained biopsy specimens provide very crude results with poor inter-observer and intra-observer reproducibility. For this purpose, computerized image analysis based methods are needed.
Research frontiers
Many studies have been carried out using computerized image analysis for the automation of evaluation. The use of various software packages such as ImageJ, the AQUA system, Image Pro Plus, and Adobe Photoshop has been reported. Another area which has seen very good advancement is the high throughput technology called tissue microarray (TMA) which generates a huge number of images in a fully automated and standardized manner which also makes it best suited for automation of evaluation.
Innovations and breakthroughs
In this article, the authors introduce the in-house developed software, TissueQuant, for automation of the evaluation of images of immunohistochemically stained biopsy specimens. The method they proposed provides a fully automated solution to these evaluations. The algorithm was designed to obtain 100% sensitivity. The inter-observer and intra-observer variations are reported. In addition, the correlation of the automated analysis with the expert’s evaluation is also reported. It can be seen that the automated evaluation correlated well with the expert and overcame the problem of inter-observer and intra-observer variation.
Applications
The proposed method can be used to automate evaluations of images generated with the Tissue MicroArray technique so as to handle high throughput. The same technique can also be modified to evaluate any image where staining intensity needs to be assessed for decision-making.
Terminology
Estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2/neu) are specific hormone receptors which are expressed when stained with the respective immunohistochemical stains.
Peer review
Keerthana Prasad and colleagues demonstrated that the automation of breast cancer diagnosis from immunohistochemically stained specimen is a useful tool to provide objective and repeatable evaluations.
Footnotes
Peer reviewer: Songsak Petmitr, PhD, Associate Professor, Department of Tropical Nutrition and Food Science, Mahidol University, 420/6 Ratchawithi Road, Bangkok 10400, Thailand
S- Editor Cheng JX L- Editor Webster JR E- Editor Ma WH
References
- 1.Ellis IO, Shnitt SJ, Sastre-Garau X, Bussolati G, Tavassoli FA, Eusebi V. “Invasive Breast Carcinoma”. In: Tavassoli FA, Devilee P, editors. Pathology and genetics- tumors of the breast and female genital organs World Health Organization classification of tumors. Lyon: IARC Press; 2006. pp. 9–110. [Google Scholar]
- 2.Almasri NM, Al Hamad M. Immunohistochemical evaluation of human epidermal growth factor receptor 2 and estrogen and progesterone receptors in breast carcinoma in Jordan. Breast Cancer Res. 2005;7:R598–R604. doi: 10.1186/bcr1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilking U, Karlsson E, Skoog L, Hatschek T, Lidbrink E, Elmberger G, Johansson H, Lindström L, Bergh J. HER2 status in a population-derived breast cancer cohort: discordances during tumor progression. Breast Cancer Res Treat. 2010:Jul 14. doi: 10.1007/s10549-010-1029-2. [DOI] [PubMed] [Google Scholar]
- 4.Osborne JR, Port E, Gonen M, Doane A, Yeung H, Gerald W, Cook JB, Larson S, “18F-FDG PET of locally invasive breast cancer and association of estrogen receptor status with standardized uptake value: microarray and immunohistochemical analysis”. J Nucl Med. 2010;51:543–550. doi: 10.2967/jnumed.108.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan DJ, Gallagher WM. Prognostic ability of a panel of immunohistochemistry markers - retailoring of an 'old solution'. Breast Cancer Res. 2008;10:102. doi: 10.1186/bcr1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raymond WA, Leong AS. Oestrogen receptor staining of paraffin-embedded breast carcinomas following short fixation in formalin: a comparison with cytosolic and frozen section receptor analyses. J Pathol. 1990;160:295–303. doi: 10.1002/path.1711600405. [DOI] [PubMed] [Google Scholar]
- 7.Molino A, Micciolo R, Turazza M, Bonetti F, Piubello Q, Corgnati A, Sperotto L, Martignoni G, Bonetti A, Nortilli R. Estrogen receptors in 699 primary breast cancers: a comparison of immunohistochemical and biochemical methods. Breast Cancer Res Treat. 1995;34:221–228. doi: 10.1007/BF00689713. [DOI] [PubMed] [Google Scholar]
- 8.Veronese SM, Barbareschi M, Morelli L, Aldovini D, Mauri FA, Caffo O, Gambacorta M, Dalla Palma P, “Predictive value of ER1D5 antibody immunostaining in breast cancer”. Appl Immunohistochemistry. 1995;3:85–90. [Google Scholar]
- 9.Pertschuk LP, Feldman JG, Kim YD, Braithwaite L, Schneider F, Braverman AS, Axiotis C. Estrogen receptor immunocytochemistry in paraffin embedded tissues with ER1D5 predicts breast cancer endocrine response more accurately than H222Sp gamma in frozen sections or cytosol-based ligand-binding assays. Cancer. 1996;77:2514–2519. doi: 10.1002/(SICI)1097-0142(19960615)77:12<2514::AID-CNCR14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 10.Brey EM, Lalani Z, Johnston C, Wong M, McIntire LV, Duke PJ, Patrick CW Jr. Automated selection of DAB-labeled tissue for immunohistochemical quantification. J Histochem Cytochem. 2003;51:575–584. doi: 10.1177/002215540305100503. [DOI] [PubMed] [Google Scholar]
- 11.Leal S, Diniz C, Sá C, Gonçalves J, Soares AS, Rocha-Pereira C, Fresco P. Semiautomated computer-assisted image analysis to quantify 3,3'-diaminobenzidine tetrahydrochloride-immunostained small tissues. Anal Biochem. 2006;357:137–143. doi: 10.1016/j.ab.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Pham NA, Morrison A, Schwock J, Aviel-Ronen S, Iakovlev V, Tsao MS, Ho J, Hedley DW. Quantitative image analysis of immunohistochemical stains using a CMYK color model. Diagn Pathol. 2007;2:8. doi: 10.1186/1746-1596-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matkowskyj KA, Cox R, Jensen RT, Benya RV. Quantitative immunohistochemistry by measuring cumulative signal strength accurately measures receptor number. J Histochem Cytochem. 2003;51:205–214. doi: 10.1177/002215540305100209. [DOI] [PubMed] [Google Scholar]
- 14.Aziz DC. Quantitation of estrogen and progesterone receptors by immunocytochemical and image analyses. Am J Clin Pathol. 1992;98:105–111. doi: 10.1093/ajcp/98.1.105. [DOI] [PubMed] [Google Scholar]
- 15.Auger M, Katz RL, Johnston DA, Sneige N, Ordonez NG, Fritsche H. Quantitation of immunocytochemical estrogen and progesterone receptor content in fine needle aspirates of breast carcinoma using the SAMBA 4000 image analysis system. Anal Quant Cytol Histol. 1993;15:274–280. [PubMed] [Google Scholar]
- 16.Santeusanio G, Mauriello A, Schiaroli S, Anemona L, Spagnoli LG, Scambia G, Oberholzer M. Densitometric and morphometric study of immunocytochemical estrogen receptors detection in breast carcinomas. Pathol Res Pract. 1992;188:478–483. doi: 10.1016/S0344-0338(11)80041-5. [DOI] [PubMed] [Google Scholar]
- 17.Lehr HA, Mankoff DA, Corwin D, Santeusanio G, Gown AM. Application of photoshop-based image analysis to quantification of hormone receptor expression in breast cancer. J Histochem Cytochem. 1997;45:1559–1565. doi: 10.1177/002215549704501112. [DOI] [PubMed] [Google Scholar]
- 18.Ruifrok AC, Katz RL, Johnston DA. Comparison of quantification of histochemical staining by hue-saturation-intensity (HSI) transformation and color-deconvolution. Appl Immunohistochem Mol Morphol. 2003;11:85–91. doi: 10.1097/00129039-200303000-00014. [DOI] [PubMed] [Google Scholar]
- 19.Mofidi R, Walsh R, Ridgway PF, Crotty T, McDermott EW, Keaveny TV, Duffy MJ, Hill AD, O'Higgins N. Objective measurement of breast cancer oestrogen receptor status through digital image analysis. Eur J Surg Oncol. 2003;29:20–24. doi: 10.1053/ejso.2002.1373. [DOI] [PubMed] [Google Scholar]
- 20.Hatanaka Y, Hashizume K, Nitta K, Kato T, Itoh I, Tani Y. Cytometrical image analysis for immunohistochemical hormone receptor status in breast carcinomas. Pathol Int. 2003;53:693–699. doi: 10.1046/j.1440-1827.2003.01547.x. [DOI] [PubMed] [Google Scholar]
- 21.Kostopoulos S, Cavouras D, Daskalakis A, Bougioukos P, Georgiadis P, Kagadis GC, Kalatzis I, Ravazoula P, Nikiforidis G. Colour-texture based image analysis method for assessing the hormone receptors status in breast tissue sections. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:4985–4988. doi: 10.1109/IEMBS.2007.4353459. [DOI] [PubMed] [Google Scholar]
- 22.Yeh IT, Mies C. Application of immunohistochemistry to breast lesions. Arch Pathol Lab Med. 2008;132:349–358. doi: 10.5858/2008-132-349-AOITBL. [DOI] [PubMed] [Google Scholar]
- 23.Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS) Pathol Res Pract. 1993;189:862–866. doi: 10.1016/S0344-0338(11)81095-2. [DOI] [PubMed] [Google Scholar]
- 24.Detre S, Saclani Jotti G, Dowsett M. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goedkoop AY, de Rie MA, Teunissen MB, Picavet DI, van der Hall PO, Bos JD, Tak PP, Kraan MC. Digital image analysis for the evaluation of the inflammatory infiltrate in psoriasis. Arch Dermatol Res. 2005;297:51–59. doi: 10.1007/s00403-005-0578-4. [DOI] [PubMed] [Google Scholar]
- 26.Kayser G, Radziszowski D, Bzdyl P, Sommer R, Kayser K. Theory and implementation of an electronic, automated measurement system for images obtained from immunohistochemically stained slides. Anal Quant Cytol Histol. 2006;28:27–38. [PubMed] [Google Scholar]
- 27.Humm JL, Macklis RM, Yang Y, Bump K, Chin LM. Image analysis for the study of radionuclide distribution in tissue sections. J Nucl Med. 1994;35:1217–1225. [PubMed] [Google Scholar]
- 28.Raisman-Vozari R, Hirsch E, Javoy-Agid F, Vassort C, Savasta M, Feuerstein C, Thibault J, Agid Y. Quantitative autoradiography of tyrosine hydroxylase immunoreactivity in the rat brain. J Neurochem. 1991;57:1212–1222. doi: 10.1111/j.1471-4159.1991.tb08282.x. [DOI] [PubMed] [Google Scholar]
- 29.Lamaziere JM, Lavallee J, Zunino C, Larrue J. Semiquantitative study of the distribution of two cellular antigens by computer-directed color analysis. Lab Invest. 1993;68:248–252. [PubMed] [Google Scholar]
- 30.Kim D, Gregory CW, Smith GJ, Mohler JL. Immunohistochemical quantitation of androgen receptor expression using color video image analysis. Cytometry. 1999;35:2–10. doi: 10.1002/(sici)1097-0320(19990101)35:1<2::aid-cyto2>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 31.Diaz Encarnacion MM, Griffin MD, Slezak JM, Bergstralh EJ, Stegall MD, Velosa JA, Grande JP. Correlation of quantitative digital image analysis with the glomerular filtration rate in chronic allograft nephropathy. Am J Transplant. 2004;4:248–256. doi: 10.1046/j.1600-6143.2003.00311.x. [DOI] [PubMed] [Google Scholar]
- 32.Charpin C, Andrac L, Habib MC, Vacheret H, Xerri L, Devictor B, Lavaut MN, Toga M. Immunodetection in fine-needle aspirates and multiparametric (SAMBA) image analysis. Receptors (monoclonal antiestrogen and antiprogesterone) and growth fraction (monoclonal Ki67) evaluation in breast carcinomas. Cancer. 1989;63:863–872. doi: 10.1002/1097-0142(19890301)63:5<863::aid-cncr2820630512>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 33.Pauschinger M, Knopf D, Petschauer S, Doerner A, Poller W, Schwimmbeck PL, Kühl U, Schultheiss HP. Dilated cardiomyopathy is associated with significant changes in collagen type I/III ratio. Circulation. 1999;99:2750–2756. doi: 10.1161/01.cir.99.21.2750. [DOI] [PubMed] [Google Scholar]
- 34.Soukupova J, Albrechtova J. Image analysis - tool for quantification of histochemical detections of phenolic compounds, lignin and peroxidases in needles of Norway spruce. Biologica plantarium. 2003;46:595–601. [Google Scholar]
- 35.Niendorf A, Rath M, Wolf K, Peters S, Arps H, Beisiegel U, Dietel M. Morphological detection and quantification of lipoprotein(a) deposition in atheromatous lesions of human aorta and coronary arteries. Virchows Arch A Pathol Anat Histopathol. 1990;417:105–111. doi: 10.1007/BF02190527. [DOI] [PubMed] [Google Scholar]
- 36.Vrekoussis T, Chaniotis V, Navrozoglou I, Dousias V, Pavlakis K, Stathopoulos EN, Zoras O. Image analysis of breast cancer immunohistochemistry-stained sections using ImageJ: an RGB-based model. Anticancer Res. 2009;29:4995–4998. [PubMed] [Google Scholar]
- 37.McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 38.Chung GG, Zerkowski MP, Ghosh S, Camp RL, Rimm DL. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest. 2007;87:662–669. doi: 10.1038/labinvest.3700543. [DOI] [PubMed] [Google Scholar]
- 39.Sharangpani GM, Joshi AS, Porter K, Deshpande AS, Keyhani S, Naik GA, Gholap AS, Barsky SH. Semi-automated imaging system to quantitate estrogen and progesterone receptor immunoreactivity in human breast cancer. J Microsc. 2007;226:244–255. doi: 10.1111/j.1365-2818.2007.01772.x. [DOI] [PubMed] [Google Scholar]