Abstract
Causative genetic variants have to date been identified for only a small proportion of familial colorectal cancer (CRC). While conditions such as Familial Adenomatous Polyposis and Lynch syndrome have well defined genetic causes, the search for variants underlying the remainder of familial CRC is plagued by genetic heterogeneity. The recent identification of families with a heritable predisposition to malignancies arising through the serrated pathway (familial serrated neoplasia or Jass syndrome) provides an opportunity to study a subset of familial CRC in which heterogeneity may be greatly reduced. A genome-wide linkage screen was performed on a large family displaying a dominantly-inherited predisposition to serrated neoplasia genotyped using the Affymetrix GeneChip Human Mapping 10 K SNP Array. Parametric and nonparametric analyses were performed and resulting regions of interest, as well as previously reported CRC susceptibility loci at 3q22, 7q31 and 9q22, were followed up by finemapping in 10 serrated neoplasia families. Genome-wide linkage analysis revealed regions of interest at 2p25.2-p25.1, 2q24.3-q37.1 and 8p21.2-q12.1. Finemapping linkage and haplotype analyses identified 2q32.2-q33.3 as the region most likely to harbour linkage, with heterogeneity logarithm of the odds (HLOD) 2.09 and nonparametric linkage (NPL) score 2.36 (P = 0.004). Five primary candidate genes (CFLAR, CASP10, CASP8, FZD7 and BMPR2) were sequenced and no segregating variants identified. There was no evidence of linkage to previously reported loci on chromosomes 3, 7 and 9.
Electronic supplementary material
The online version of this article (doi:10.1007/s10689-010-9408-8) contains supplementary material, which is available to authorized users.
Keywords: Serrated neoplasia, Familial cancer, Linkage
Introduction
The identification of a causative genetic variant in cancer-prone families such as those with Lynch syndrome is an important determinant of decreased mortality through more focussed cancer surveillance [1]. In addition, colorectal cancers (CRC) arising in Lynch syndrome show molecular and pathology features, such as immunohistochemical loss of DNA mismatch repair (MMR) gene expression and increased tumour infiltrating lymphocytes, which allow them to be distinguished from among unselected series. However, less than 3% of all CRC is accounted for by this well-characterised condition, in which the cancers develop from adenomatous polyps [2, 3].
Several studies examining linkage in non-syndromic CRC families have reported statistically significant associations at 3q22 [4–7] and 9q22 [8–11]. A more recent study found linkage to 7q31 in colorectal neoplasia families, and confirmed the region at 3q22 as a susceptibility locus for CRC [12]. To date no confirmed causative variants have been identified from these regions, though the reported associations remain robust.
The balance of families with CRC predisposition are likely to show genetic heterogeneity and variable penetrance, thereby limiting the power of genome-wide linkage approaches to define critical regions in the absence of stratification. The description of families with multiple cases of neoplasia in which serrated polyps are prominent has facilitated the exploration of a proportion of the remaining unexplained familial aggregation. Such families were initially reported from New Zealand by the late Professor Jass and colleagues in the mid-nineties [13, 14], and a further publication demonstrated the familial involvement of hyperplastic polyposis [15], associated with LOH at chromosome 1p. Families fulfilling the Amsterdam I criteria with mixed epithelial polyps and variable levels of microsatellite instability (MSI) in which Lynch syndrome could be excluded, and which showed some overlap with hyperplastic polyposis, were reported in 2005 [16]. Such families are characterised by a mixture of serrated and adenomatous polyps, and, in contrast to Lynch syndrome, somatic BRAF mutation is commonly observed in CRC. Only a minority of cancers demonstrate MMR deficiency, attributable to methylation of the MLH1 gene promoter. We refer to this condition of serrated neoplasia and adenomas clustering in families as Jass syndrome, after the pathologist who first described it [13, 14].
Studies on unselected series of CRCs have shown that molecular features of the serrated pathway, such as widespread CpG island methylation and somatic BRAF mutation, as well as the presence of serrated lesions with atypical histology (sessile serrated adenomas), are associated with a family history of CRC [17–19], lending further support to the idea that the observed familial aggregation of lesions arising through the serrated pathway is the result of an inherited predisposition. In addition, the presence of a sessile serrated adenoma is associated with polyp multiplicity [19, 20], and with conventional adenomas in patients who do not meet the criteria for hyperplastic polyposis [21]. In patients with hyperplastic polyposis, polyps with adenomatous elements increase the risk of CRC [22–24], and are the likely lesions of origin for at least some of the cancers occurring in this condition [25].
We have investigated genomic regions associated with Jass syndrome by performing a genome-wide linkage screen in a single large family, followed by finemapping in a further 10 families, and present evidence for linkage to chromosome 2q32.2-q33.3. Through further finemapping analysis, we also provide evidence that previously reported CRC susceptibility loci at 3q22, 7q31 and 9q22 are unlikely to contribute to Jass syndrome.
Materials and methods
Families
The 11 families in this study, five of which have been described previously [16], were enrolled from high-risk genetics clinics in Australia as part of the Colon Cancer Family Registry, an international collaborative registry for the study of genetics and epidemiology of colorectal cancer [26]. All participants gave written informed consent to take part in research, and the project was performed under QIMR Human Research Ethics Committee Approval P912 (Genetics of Serrated Neoplasia).
Polyps were reviewed by a specialist gastro-intestinal pathologist (JRJ). MSI status of tumours was determined using a panel of 10 microsatellite markers (BAT-25, BAT-26, BAT-34C4, BAT-40, D5S346, D10S197, D17S250, D18S55, ACTC and MYCL) and standard techniques [16, 27]. BRAF V600E mutation status of tumours was analysed as previously described [28]. Lynch syndrome was excluded in all families as determined by: (1) proficient expression of the MMR proteins in tumours; (2) absence of pathogenic mutations or variants of uncertain clinical significance in the MMR genes after sequencing of the coding and splice site regions and MLPA analysis for large deletions or duplications; and (3) methylation analysis of the MLH1 gene promoter. No mutations were found in any patients. Three cancers showed loss of MLH1 protein expression, of which two had sufficient DNA available for methylation analysis and tested positive for methylation of the MLH1 promoter in their tumour tissue [29].
Criteria for inclusion of families were: at least 2 individuals with CRC, with one aged under 60 years; AND at least 2 individuals with polyps, with one aged under 60 years; AND at least two of the following characteristics associated with serrated neoplasia:
a mixture of hyperplastic and adenomatous polyps;
variable levels of MSI in cancers and/or polyps
the presence of BRAF V600E somatic mutation in one or more cancers; and
at least one individual with multiple hyperplastic polyps under age 60.
Characteristics of each family are listed in Table 1.
Table 1.
Phenotypic characteristics of families
| Family | Individuals with CRC | CRC ages | Polyp ages (and numbers) | MSI-V | BRAF CRC | Hyperplastic and adenomatous polyps | Individuals with multiple hyperplastic polyps under 60 (ages) | Evidence of linkage to Region 1 |
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 45, 53, 74 | 63 (1), 53 (3), 51 (2), 57 (2), 46 (1) | Yes | Yes | Yes | 1 (57) | Yes |
| 2aa | 3 | 25a, 31a, 32 | 29 (1), 53 (1) | No | No | Yes | 0 | Yes |
| 2ba | 8 | 25a, 31a, 54, 72, 53, 65, 63, 52 | 57 (10), 53 (2), 63 (1) | No | Yes | Yes | 1 (57) | No |
| 3 | 4 | 52, 65, 67, 70 | 71(1), 66 (1), 65 (7), 55 (11), 60 (2), 49 (1) | No | No | Yes | 1 (55) | No |
| 4 | 4 | 37, 44, 58, 62 | 37 (2), 57 (11), 51 (4), 58 (5) | No | No | Yes | 3 (51, 57, 58) | No |
| 5 | 4 | 31, 47, 52, 59 | 49 (1), 38 (1), 45 (2), 50 (7), 28 (1) | Yes | No | Yes | 1 (50) | Yes |
| 6 | 3 | 43, 53, 74 | 53 (6), 43 (4), 52 (4) | Yes | Yes | Yes | 2 (52, 53) | Yes |
| 7 | 4 | 31, 39, 50, 61 | 49 (2), 31 (2) | No | No | Yes | 1 (49) | No |
| 8 | 3 | 27, 57, 60 | 52 (1) | Yes | Yes | Yes | 0 | Yes |
| 9 | 3 | 54, 59, 76 | 54 (2), 75 (1), 50 (1) | Yes | No | Yes | 0 | No |
| 10 | 2 | 55, 62 | 58 (2), 65 (6) | Yes | No | Yes | 1 (58) | Yes |
| 11 | 4 | 48, 53, 74, 85 | 85 (2), 48 (5), 50 (3), 51 (17), 53 (6) | Yes | Yes | Yes | 2 (48, 51) | Yes |
MSI-V variable levels of MSI between lesions
aFamilies 2a and 2b are branches of the same family which were analysed separately due to independent segregation of affected status. Two individuals with CRC were included as part of both families
Genome-wide linkage screen
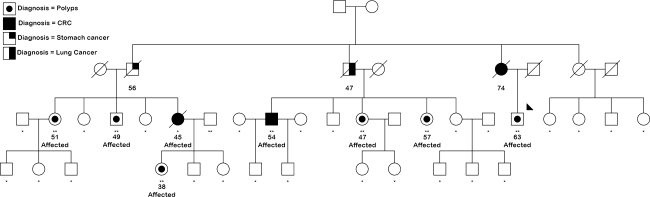
The 10 K Xba 142 GeneChip Human Mapping Array (Affymetrix Inc., Santa Clara, CA, USA) was used to genotype seven affected individuals and one unaffected spouse from Family 1 (Fig. 1), a multi-case CRC family with multiple young-onset, BRAF-mutated, MSI-variable (MSI-V) CRCs, and multiple individuals with multiple serrated polyps or those with atypical histology including sessile serrated adenomas, serrated adenomas, and mixed polyps [16, 25]. Sample call rates were all greater than 98.5%, with a total of 990 no calls, and 1280 SNPs (1.3%) were uninformative in the pedigree. Sex was verified by observing heterozygosity rates of X-linked markers. PedCheck [30] was used to detect Mendelian errors, which were manually corrected via the removal of inconsistent genotypes.
Fig. 1.
Family 1. Solid symbols, individuals with colorectal cancer. Dotted symbols, individuals with large or atypical serrated polyps. Individuals marked with two asterisks were genotyped for the genome-wide linkage scan and for finemapping; those with a single asterisk were typed for finemapping only. Affected individuals are marked as such. A single individual in generation 2 with CRC was also considered as affected. Numbers indicate age at first onset of cancer or polyps
Both parametric and nonparametric (NPL) linkage analyses were performed using GeneHunter version 2.1 [31] via a stepwise 900 SNP sliding window, implemented in the Alohomora GUI interface [32]. For parametric analysis, an autosomal dominant mode of inheritance was specified, with 60% penetrance, 10% phenocopy rate and a disease allele frequency of 0.001. Families segregating serrated neoplasia show clustering of CRC, including those with somatic BRAF mutation, and polyps of adenomatous, serrated and mixed histology. Individuals with CRC and/or large, multiple or atypical serrated polyps, or any serrated or adenomatous polyp diagnosed before age 60, were classed as affected. As the prevalence of adenomas in the population increases with age [33], a conservative approach was taken to the classification of affected individuals in the over 60 age group, requiring adenomas to be 5 mm or greater in size, or have villous components, and excluding individuals with diminutive rectal-only lesions. Haplotypes were constructed in GeneHunter and viewed graphically using HaploPainter version 029.5 [34].
Finemapping
Twelve microsatellite markers on chromosome 2p, 22 markers on 2q, 18 markers on chromosome 3, 10 markers on chromosome 7, 12 markers on chromosome 8 and 13 markers on chromosome 9 were typed in the 11 families. Markers and primer sequences are listed in Supplementary Table 1. Markers were amplified using True Allele PCR Premix (Applied Biosystems, Foster City, CA, USA) or GoTaq Colorless Master Mix (Promega, Madison, WI, USA) according to the manufacturer’s instructions. PCR products were labelled with FAM, HEX or NED fluorophores, either by labelling one primer in each pair or via an M13 tag and labelled M13 primer, and separated using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Data were analysed using GeneMarker version 1.70 or 1.75 (SoftGenetics, PA, USA). Mendelian error checking was performed using Progeny Lab 6 (Progeny Software, South Bend, IN, USA). Non-Mendelian error checking and parametric and nonparametric linkage analyses on full pedigrees were performed using SimWalk 2.91 [35], implemented under the easyLINKAGE Plus interface version 5.05 [36]. Parametric linkage analysis was performed under a dominant model with 60% penetrance, 10% phenocopy rate and disease allele frequency of 0.001. Affected-only parametric analysis was performed using GeneHunter version 2.1, also under the easyLINKAGE interface, under a dominant model with 99% penetrance, 10% phenocopy rate and disease allele frequency of 0.001. For affected-only analysis, all individuals with unknown or unaffected status were classed as unknown. Marker map locations were based on the deCODE map [37]; positions for markers not on the deCODE map were interpolated from the nearest adjacent deCODE markers using the NCBI Map Viewer Build 36.3 (http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9606). Haplotypes were constructed by SimWalk and visualised using HaploPainter.
Sequencing of exons and exon–intron boundaries of candidate genes was performed using BigDye Terminator chemistry on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Where possible, variants were confirmed by sequencing in both forward and reverse directions.
Results
Genome-wide linkage screen
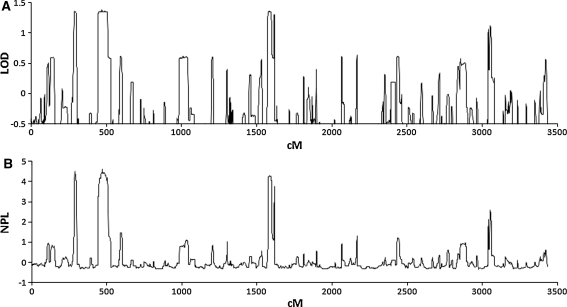
As there is currently little information on Jass syndrome in the literature, the parameters used for linkage analysis were estimated from the 11 families involved in the study. The pattern of affected individuals was consistent with a dominant mode of inheritance with 60% penetrance, and four affected spouses out of 37 resulted in an estimated phenocopy rate of 10%. Using these parameters, genome-wide linkage analysis on Family 1 revealed regions of interest at 2p25.2-p25.1 (logarithm of the odds (LOD) 1.36, nonparametric linkage (NPL) score 4.50 (nominal P = 0.004)), 2q24.3-q37.1 (LOD 1.38, NPL 4.62 (nominal P = 0.003)), and 8p21.2-q12.1 (LOD 1.36, NPL 4.28 (nominal P = 0.007)) (Fig. 2), although none of these attained genome-wide statistical significance.
Fig. 2.
Genome-wide linkage screen on Family 1. a Parametric and b Nonparametric analysis
Finemapping
Finemapping using microsatellite markers spanning each of the three regions of interest was carried out in Family 1 and a further 10 families (Supplementary Table 2). Family 2 showed independent segregation of affected status in two branches, each of which met the criteria for inclusion, and was therefore treated as two separate families for analysis, 2a and 2b. Five individuals overlapped between the two branches and were therefore included in the analysis as part of both Family 2a and Family 2b. In an effort to account for some of the uncertainties inherent in a newly described disease (such as penetrance, age at onset and the possibility of a proportion of affected individuals being asymptomatic), three separate analyses were performed: parametric analysis with full pedigrees, 10% phenocopy rate and 60% penetrance; nonparametric analysis; and affected-only parametric analysis with 10% phenocopy rate and 99% penetrance, in which unaffected individuals were retained only if they were parents of affected individuals, or were genotyped children of affected individuals for whom DNA was unavailable; these individuals were assigned unknown affected status.
LOD scores on chromosomes 2p and 8 were negative throughout. Evidence for linkage to these regions was limited, with no HLOD score greater than 0.6 and NPL reaching significance at the 5% level at a single location only (marker D2S262 at 30 cM on chromosome 2p, NPL 1.36 (P = 0.044)) (Table 2).
Table 2.
Finemapping results
| Chr | cM | Parametric (full pedigrees) | Nonparametric | Affected only | |||||
|---|---|---|---|---|---|---|---|---|---|
| LOD | HLOD | α | NPL | P | LOD | HLOD | α | ||
| Regions identified from genome-wide screen on Family 1 | |||||||||
| 2q (Region 1) | 193–199 | 0.48 | 0.48 | 0.95 | 2.36 | 0.004 | 1.12 | 2.09 | 0.54 |
| 2q (Region 2) | 224–231 | 0.79 | 0.96 | 0.60 | 1.75 | 0.018 | 0.82 | 1.73 | 0.54 |
| 2p | 2–34 | −1.06 | 0.04 | 0.15 | 1.36 | 0.04 | −1.23 | 0.60 | 0.38 |
| 8 | 39–89 | −1.56 | 0.00 | 0.00 | 0.86 | 0.14 | −1.73 | 0.41 | 0.17 |
| Chr | cM | Parametric (full pedigrees) | Nonparametric | Affected only | |||
|---|---|---|---|---|---|---|---|
| LOD | HLOD | NPL | P | LOD | HLOD | ||
| Previously published CRC susceptibility loci | |||||||
| 3 | 126–190 | −1.22 | 0.18 | 0.57 | 0.27 | −5.47 | 0.09 |
| 7 | 111–129 | −2.76 | 0.00 | 0.83 | 0.15 | −3.32 | 0.05 |
| 9 | 66–121 | −1.86 | 0.00 | 0.37 | 0.42 | −5.16 | 0.00 |
α proportion of linked families
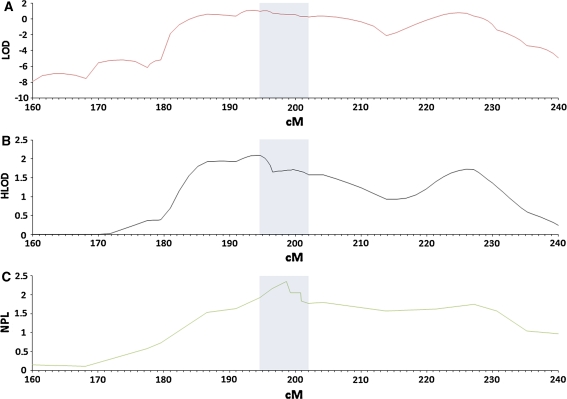
Parametric analysis on full pedigrees at 2q revealed LOD scores of 0.48 at 197 cM between markers D2S117 and D2S309, and 0.79 at 224 cM between markers D2S163 and D2S401, with corresponding HLOD scores of 0.48 and 0.96, respectively (Table 2). Nonparametric analysis revealed a single peak at 199 cM at D2S309 with NPL 2.36 (P = 0.004), while the strongest evidence for linkage in the affected-only analysis was at 193 cM between markers D2S118 and D2S115, with LOD 1.12 and HLOD 2.09, with a secondary peak at 225 cM between markers D2S163 and D2S133, with LOD 0.82 and HLOD 1.73 (Table 2; Fig. 3). Taking all three analyses into account, the strongest evidence for linkage was between markers D2S118 and D2S2309, with limited evidence for linkage between markers D2S163 and D2S133; these loci are defined respectively as Region 1 and Region 2. Affected-only linkage analysis accounting for heterogeneity gave estimates for the proportion of linked families (α) as 0.54 for both Region 1 and Region 2 (Table 2). Results for individual families are summarised in Supplementary Table 2.
Fig. 3.
Finemapping at chromosome 2q in 11 Jass syndrome families. Horizontal axis position along chromosome 2 in cM. a LOD score from affected-only analysis; b HLOD from affected-only analysis accounting for heterogeneity; c Nonparametric analysis. Shading indicates location of haplotype sharing at Region 1
Haplotype analysis at 2q and sequencing of candidate genes
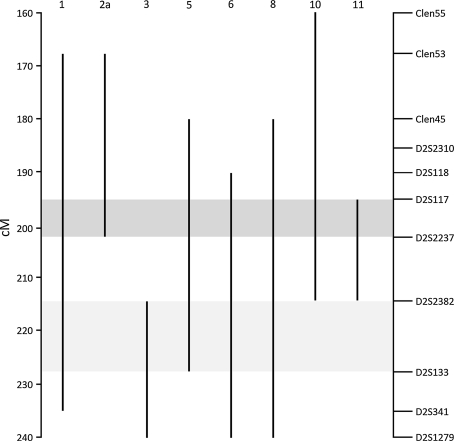
Eight families revealed a haplotype segregating with affected status. Although there was no evidence of a common haplotype shared between families, a consensus region covering approximately 12 Mb, bordered at the centromeric end by D2S117 and at the telomeric end by D2S2358 and showing considerable overlap with Region 1, segregated with affected status in 48/53 affected individuals in seven families (Figs. 3, 4, Supplementary Table 2). Of these, six families demonstrated variable MSI in cancers and/or polyps (Table 1). In contrast, only one of the five families not showing linkage to this region had variable MSI. The consensus region contains 60 genes, from which five (CFLAR, CASP10, CASP8, FZD7 and BMPR2) were selected based on previous evidence in the literature for a role in CRC or other cancers for sequencing of exons and exon–intron boundaries. At least two affected members of each family were screened, and where a variant showed possible segregation with affected status the rest of the family was also sequenced. Several variants were found, including two CASP8 polymorphisms, namely a 6-nucleotide promoter insertion/deletion (rs3834129) and the D302H SNP (rs1045485), with previously reported associations with colorectal and other cancers [38–40]. Neither these nor any other identified variants segregated with affected status.
Fig. 4.
Haplotype analysis at 2q. Vertical lines show chromosomal segments segregating with affected status within each family. Dark and light shading show areas of segregation at Region 1 and Region 2 respectively
Exclusion of reported CRC susceptibility loci
The contribution of recently reported CRC susceptibility loci on chromosomes 3, 7 and 9 to Jass syndrome was tested using microsatellite markers selected to cover the reported regions, and parametric, nonparametric and affected-only analyses performed as above. No evidence was found to support linkage (Table 2).
Discussion
Since the discoveries of the genes responsible for Familial Adenomatous Polyposis, Lynch syndrome, and the hamartomatous polyposes, little progress has been made in the identification of genetic causes for the remaining unexplained majority of familial CRC. One of the reasons for this lack of success is likely to be extensive genetic heterogeneity, which has proved to be a difficult obstacle to overcome for investigators undertaking linkage studies on collections of CRC families. Other than exclusion of known syndromes, approaches aimed at limiting the heterogeneity within a sample set have included enrichment for families with young-onset cases [8, 41], exclusion of multiple polyposis phenotypes [8], exclusion or inclusion of adenomas and hyperplastic polyps [41, 42], and stratification by occurrence of extracolonic cancers [41]. While these approaches certainly limit phenotypic heterogeneity, the extent to which they reduce genetic heterogeneity is unknown.
The recent identification of a familial predisposition to colonic malignancies arising through the serrated pathway [13, 14, 16, 43] has presented an opportunity to study a subset of familial CRC in which genetic heterogeneity is more limited. Serrated neoplasia families show distinct features, such as variable levels of MSI and frequent somatic BRAF mutation in tumours and a mixture of serrated and adenomatous polyps, that allow them to be distinguished from the remainder of familial CRC. Such features characterised families with evidence of linkage to 2q. Incomplete penetrance, small family size, under-reporting due to a proportion of individuals being asymptomatic, and practical difficulties in obtaining tumour and polyp samples for all patients, however, mean that all features will not be observed in all families. In an effort to account for this, a set of criteria was developed for this study that allowed for some flexibility while still requiring families to show a clustering of CRC and polyps in conjunction with characteristics that define Jass syndrome. In this attempt to include phenotypically homogeneous families, the possibility remains that a minority of these families may have a genetic predisposition different to that responsible for the majority of Jass syndrome. While this leaves open the possibility of some families being incorrectly included, these are likely to be a minority.
A linkage strategy was selected that would maximise the possibility of identifying linkage, while minimising the effects of any heterogeneity that may remain within the selected families. To this end, the family considered the most likely to allow the identification of a strong linkage signal was subjected to a genome-wide linkage screen, followed by finemapping of regions of interest in the remaining 10 families. Loci at 2p25.2-p25.1, 2q24.3-q37.1 and 8p21.2-q12.1 were considered worthy of further investigation, with 2q24.3-q37.1 showing the strongest evidence of linkage. This region has also been identified in three previous genome-wide linkage studies [4, 6, 12] and one association study [40].
The consensus region at 2q32.2-q33.3 identified through finemapping linkage and haplotype analyses contains five genes that stand out as potential candidates and as a result were prioritised for sequencing. CFLAR, CASP10 and CASP8 are located in close proximity to each other at 2q33.1 and are all involved in the death receptor-induced apoptotic pathway [44]. CASP8 somatic mutation has been reported in CRC [45], and a CASP8 promoter polymorphism has been reported to be associated with susceptibility to multiple cancers including CRC [40], although the association has not been replicated in other populations [46, 47]. CFLAR has attracted attention as a therapeutic target as well as a potential oncogene due to its role in the TRAIL pathway [48]. FZD7 is a member of the Frizzled family which plays an important role in Wnt signalling [49], central to the development and progression of CRC [50], and has recently been shown to activate the Wnt pathway in CRC cell lines [51]. BMPR2 inactivation leads to epithelial cell proliferation and mixed polyp formation in the mouse colon [52] and reduced protein expression associated with somatic mutation is observed in human CRC and cell lines [53]. Although sequencing of these genes did not reveal any variants segregating with disease, only coding regions and exon–intron boundaries were sequenced, leaving noncoding regions, which may harbour regulatory elements and potential splicing variants, largely unexplored. With current re-sequencing technology and exome capture, the investigation of the remaining genes and regulatory regions becomes more feasible.
Several studies have reported evidence of linkage among a proportion of non-syndromic CRC families, with 3q22, 7q31 and 9q22 prominent [4–6, 8–10, 12]. As well as suggesting that they are unlikely to contribute to Jass syndrome, the failure to find evidence of linkage to these loci in the present study suggests that further characterisation of the 3q22, 7q31 and 9q22 regions should aim to exclude families demonstrating molecular and/or histological evidence of serrated neoplasia.
Approximately half of the families studied showed evidence of linkage to 2q32.2-q33.3. The balance of the families did not demonstrate linkage, suggesting that other as yet unidentified loci may contribute to families with serrated neoplasia. Continuing efforts to identify and characterise serrated neoplasia families will serve the dual purpose of allowing a greater understanding of the phenotypic presentation of the syndrome, and facilitating further genetic studies which will allow confirmation and refinement of the linked region, paving the way for more comprehensive analysis of the locus, and potentially the identification of the underlying genetic defect.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from the Cancer Council Queensland, the National Health and Medical Research Council, and the National Cancer Institute under RFA CA-95-011 (Australasian Colorectal Cancer Family Registry Centre UO1 CA097735), and through cooperative agreements with members of the Colon Cancer Family Registry and Principal Investigators, and 1R01CA123010 (Genetics of Serrated Neoplasia). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. JY is a Cancer Council Queensland Senior Research Fellow. The authors also thank the Jeremy Jass Memorial Tissue Bank for supply of polyps and cancers for the study.
Conflict of interest
Authors declare no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CRC
Colorectal cancer
- HLOD
Heterogeneity logarithm of the odds
- LOD
Logarithm of the odds
- MMR
Mismatch repair
- MSI
Microsatellite instability
- MSI-V
Microsatellite instability-variable
- NPL
Nonparametric linkage
References
- 1.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Clendenning M, Sotamaa K, Prior T, Westman JA, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iino H, Simms L, Young J, Arnold J, Winship IM, Webb SI, Furlong KL, Leggett B, Jass JR. DNA microsatellite instability and mismatch repair protein loss in adenomas presenting in hereditary non-polyposis colorectal cancer. Gut. 2000;47:37–42. doi: 10.1136/gut.47.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA, et al. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:10. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemp Z, Carvajal-Carmona L, Spain S, Barclay E, Gorman M, Martin L, Jaeger E, Brooks N, Bishop DT, Thomas H, et al. Evidence for a colorectal cancer susceptibility locus on chromosome 3q21-q24 from a high-density SNP genome-wide linkage scan. Hum Mol Genet. 2006;15:2903–2910. doi: 10.1093/hmg/ddl231. [DOI] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, Carvajal-Carmona L, Sellick GS, Kemp Z, Webb E, Spain S, Sullivan K, Barclay E, Lubbe S, Jaeger E, et al. Deciphering the genetics of hereditary non-syndromic colorectal cancer. Eur J Hum Genet. 2008;16:1477–1486. doi: 10.1038/ejhg.2008.129. [DOI] [PubMed] [Google Scholar]
- 6.Picelli S, Vandrovcova J, Jones S, Djureinovic T, Skoglund J, Zhou XL, Velculescu VE, Vogelstein B, Lindblom A. Genome-wide linkage scan for colorectal cancer susceptibility genes supports linkage to chromosome 3q. Bmc Cancer. 2008;8:7. doi: 10.1186/1471-2407-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Middeldorp A, Jagmohan-Changur SC, van der Klift HM, van Puijenbroek M, Houwing-Duistermaat JJ, Webb E, Houlston R, Tops C, Vasen HFA, Devilee P, et al. Comprehensive genetic analysis of seven large families with mismatch repair proficient colorectal cancer. Genes Chromosomes Cancer. 2010;49:539–548. doi: 10.1002/gcc.20763. [DOI] [PubMed] [Google Scholar]
- 8.Kemp ZE, Carvajal-Carmona LG, Barclay E, Gorman M, Martin L, Wood W, Rowan A, Donohue C, Spain S, Jaeger E, et al. Evidence of linkage to chromosome 9q22.33 in colorectal cancer kindreds from the United Kingdom. Cancer Res. 2006;66:5003–5006. doi: 10.1158/0008-5472.CAN-05-4074. [DOI] [PubMed] [Google Scholar]
- 9.Skoglund J, Djureinovic T, Zhou XL, Vandrovcova J, Renkonen E, Iselius L, Bisgaard ML, Peltomaki P, Lindblom A. Linkage analysis in a large Swedish family supports the presence of a susceptibility locus for adenoma and colorectal cancer on chromosome 9q22.32-31.1. J Med Genet. 2006;43:e7. doi: 10.1136/jmg.2005.033928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiesner GL, Daley D, Lewis S, Ticknor C, Platzer P, Lutterbaugh J, MacMillen M, Baliner B, Willis J, Elston RC, et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. Proc Natl Acad Sci USA. 2003;100:12961–12965. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray-McGuire C, Guda K, Adrianto I, Lin CP, Natale L, Potter JD, Newcomb P, Poole EM, Ulrich CM, Lindor N, et al. Confirmation of linkage to and localization of familial colon cancer risk haplotype on chromosome 9q22. Cancer Res. 2010;70:5409–5418. doi: 10.1158/0008-5472.CAN-10-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neklason DW, Kerber RA, Nilson DB, Anton-Culver H, Schwartz AG, Griffin CA, Lowery JT, Schildkraut JM, Evans JP, Tomlinson GE, et al. Common familial colorectal cancer linked to chromosome 7q31: a genome-wide analysis. Cancer Res. 2008;68:8993–8997. doi: 10.1158/0008-5472.CAN-08-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jass JR, Cottier DS, Pokos V, Parry S, Winship IM. Mixed epithelial polyps in association with hereditary non-polyposis colorectal cancer providing an alternative pathway of cancer histogenesis. Pathology. 1997;29:28–33. doi: 10.1080/00313029700169494. [DOI] [PubMed] [Google Scholar]
- 14.Jeevaratnam P, Cottier DS, Browett PJ, Van De Water NS, Pokos V, Jass JR. Familial giant hyperplastic polyposis predisposing to colorectal cancer: a new hereditary bowel cancer syndrome. J Pathol. 1996;179:20–25. doi: 10.1002/(SICI)1096-9896(199605)179:1<20::AID-PATH538>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Rashid A, Houlihan PS, Booker S, Petersen GM, Giardiello FM, Hamilton SR. Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology. 2000;119:323–332. doi: 10.1053/gast.2000.9361. [DOI] [PubMed] [Google Scholar]
- 16.Young J, Barker MA, Simms LA, Walsh MD, Biden KG, Buchanan D, Buttenshaw R, Whitehall VL, Arnold S, Jackson L, et al. Evidence for BRAF mutation and variable levels of microsatellite instability in a syndrome of familial colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:254–263. doi: 10.1016/S1542-3565(04)00673-1. [DOI] [PubMed] [Google Scholar]
- 17.Frazier ML, Xi LX, Zong JH, Viscofsky N, Rashid A, Wu EF, Lynch PM, Amos CI, Issa JPJ. Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer. Cancer Res. 2003;63:4805–4808. [PubMed] [Google Scholar]
- 18.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6070. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 19.Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VLJ, Pike T, Simms LA, Young J, James M, Montgomery GW, et al. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology. 2006;131:1400–1407. doi: 10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 20.Pai RK, Hart J, Noffsinger AE. Sessile serrated adenomas strongly predispose to synchronous serrated polyps in non-syndromic patients. Histopathology. 2010;56:581–588. doi: 10.1111/j.1365-2559.2010.03520.x. [DOI] [PubMed] [Google Scholar]
- 21.Pai RK, Mackinnon AC, Joseph L, Noffsinger A, Hart J. Identification of histologically distinct conventional adenomas that arise predominately in patients with sessile serrated adenomas. Am J Surg Pathol. 2010;34:355–363. doi: 10.1097/PAS.0b013e3181c6b9dd. [DOI] [PubMed] [Google Scholar]
- 22.Buchanan DD, Sweet K, Drini M, Jenkins MA, Win AK, Gattas M, Walsh MD, Clendenning M, McKeone D, Walters R, et al. Phenotypic diversity in patients with multiple serrated polyps: a genetics clinic study. Int J Colorectal Dis. 2010;25:703–712. doi: 10.1007/s00384-010-0907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leggett BA, Devereaux B, Biden K, Searle J, Young J, Jass J. Hyperplastic polyposis—Association with colorectal cancer. Am J Surg Pathol. 2001;25:177–184. doi: 10.1097/00000478-200102000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Rubio CA, Stemme S, Jaramillo E, Lindblom A. Hyperplastic polyposis coli syndrome and colorectal carcinoma. Endoscopy. 2006;38:266–270. doi: 10.1055/s-2006-925026. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan DD, Roberts A, Walsh MD, Parry S, Young JP. Lessons from Lynch syndrome: a tumor biology-based approach to familial colorectal cancer. Future Oncol. 2010;6:539–549. doi: 10.2217/fon.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, Hall D, Hopper JL, Jass J, Le Marchand L, et al. Colon cancer family registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 27.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 28.Walsh MD, Buchanan DD, Cummings MC, Pearson SA, Arnold ST, Clendenning M, Walters R, McKeone DM, Spurdle AB, Hopper JL, et al. Lynch syndrome-associated breast cancers: clinicopathologic characteristics of a case series from the colon cancer family registry. Clin Cancer Res. 2010;16:2414–2424. doi: 10.1158/1078-0432.CCR-09-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, Young J, Jenkins MA, Hopper JL, Baron JA, et al. Molecular Characterization of MSI-H Colorectal Cancer by MLH1 Promoter Methylation, Immunohistochemistry, and Mismatch Repair Germline Mutation Screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruglyak L, Daly MJ, ReeveDaly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 32.Ruschendorf F, Nurnberg P. ALOHOMORA: a tool for linkage analysis using 10 K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 33.Pendergrass CJ, Edelstein DL, Hylind LM, Phillips BT, Iacobuzio-Donahue C, Romans K, Griffin CA, Cruz-Correa M, Tersmette AC, Offerhaus GJA, et al. Occurrence of colorectal adenomas in younger adults: an epidemiologic necropsy study. Clin Gastroenterol Hepatol. 2008;6:1011–1015. doi: 10.1016/j.cgh.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiele H, Nürnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2004;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 35.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 36.Lindner TH, Hoffman K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 37.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 38.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MWR, Pooley KA, Scollen S, Baynes C, Ponder BAJ, Chanock S, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 39.MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MWR, Pharoah PDP, Ponder BAJ, Meath M, Bhattacharyya NP, Cox A. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96:1866–1869. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- 40.Sun T, Gao Y, Tan W, Ma SF, Shi YK, Yao JR, Guo YL, Yang M, Zhang XM, Zhang QR, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 41.Daley D, Lewis S, Platzer P, MacMillen M, Willis J, Elston RC, Markowitz SD, Wiesner GL. Identification of susceptibility genes for cancer in a genome-wide scan: results from the colon neoplasia sibling study. Am J Hum Genet. 2008;82:723–736. doi: 10.1016/j.ajhg.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Djureinovic T, Skoglund J, Vandrovcova J, Zhou XL, Kalushkova A, Iselius L, Lindblom A. A genome wide linkage analysis in Swedish families with hereditary non-familial adenomatous polyposis/non-hereditary non-polyposis colorectal cancer. Gut. 2006;55:362–366. doi: 10.1136/gut.2005.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young J, Jass JR. The case for a genetic predisposition to serrated neoplasia in the colorectum: hypothesis and review of the literature. Cancer Epidemiol Biomarkers Prev. 2006;15:1778–1784. doi: 10.1158/1055-9965.EPI-06-0164. [DOI] [PubMed] [Google Scholar]
- 44.Thorburn A. Death receptor-induced cell killing. Cell Signal. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Lee JW, Kim MR, Soung YH, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. Mutational analysis of the CASP6 gene in colorectal and gastric carcinomas. Apmis. 2006;114:646–650. doi: 10.1111/j.1600-0463.2006.apm_417.x. [DOI] [PubMed] [Google Scholar]
- 46.Pittman AM, Broderick P, Sullivan K, Fielding S, Webb E, Penegar S, Tomlinson I, Houlston RS. CASP8 variants D302H and-652 6 N ins/del do not influence the risk of colorectal cancer in the United Kingdom population. Br J Cancer. 2008;98:1434–1436. doi: 10.1038/sj.bjc.6604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haiman CA, Garcia RR, Kolonel LN, Henderson BE, Wu AH, Le Marchand L. A promoter polymorphism in the CASP8 gene is not associated with cancer risk. Nat Genet. 2008;40:259–260. doi: 10.1038/ng0308-259. [DOI] [PubMed] [Google Scholar]
- 48.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- 49.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 50.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 51.Ueno K, Hiura M, Suehiro Y, Hazama S, Hirata H, Oka M, Imai K, Dahiya R, Hinoda Y. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia. 2008;10:697–705. doi: 10.1593/neo.08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beppu H, Mwizerwa ON, Beppu Y, Dattwyler MP, Lauwers GY, Bloch KD, Goldstein AM. Stromal inactivation of BMPRII leads to colorectal epithelial overgrowth and polyp formation. Oncogene. 2008;27:1063–1070. doi: 10.1038/sj.onc.1210720. [DOI] [PubMed] [Google Scholar]
- 53.Kodach LL, Wiercinska E, De Miranda N, Bleuming SA, Musler AR, Peppelenbosch MP, Dekker E, van den Brink GR, Van Noesel CJM, Morreau H, et al. The bone morphogenetic protein pathway is inactivated in the majority of sporadic colorectal cancers. Gastroenterology. 2008;134:1332–1341. doi: 10.1053/j.gastro.2008.02.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.