Abstract
ATI-5261 is a 26-mer peptide that stimulates cellular cholesterol efflux with high potency. This peptide displays high aqueous solubility, despite having amphipathic α-helix structure and a broad non-polar surface. These features suggested to us that ATI-5261 may adopt a specific form in solution, having favorable structural characteristics and dynamics. To test this, we subjected ATI-5261 to a series of biophysical studies and correlated self-association with secondary structure and activity. Gel-filtration chromatography and native gel electrophoresis indicated ATI-5261 adopted a discrete self-associated form of low molecular weight at concentrations > 1 mg/ml. Formation of a discrete molecular species was verified by small angle X-ray scattering (SAXS), which further revealed the peptide formed a tetrameric assembly having an elongated shape and hollow central core. This assembly dissociated to individual peptide strands upon dilution to concentrations required for promoting high-affinity cholesterol efflux from cells. Moreover, the α-helical content of ATI-5261 was exceptionally high (74.1±6.8%) regardless of physical form and concentration. Collectively, these results indicate ATI-5261 displays oligomeric behavior generally similar to native apolipoproteins and dissociates to monomers of high α-helical content upon dilution. Optimizing self-association behavior and secondary structure may prove useful for improving the translatability and efficacy of apolipoprotein mimetic peptides.
Keywords: HDL mimetic peptide, small-angle X-ray scattering, oligomeric behavior, secondary structure, cholesterol efflux, ABCA1
Plasma apolipoproteins play diverse roles in biology and medicine, including the development of atherosclerosis (1–10). Members of this protein family can be found associated with lipoproteins in blood and play prominent roles in lipid metabolism as well as anti-inflammatory processes. Their broad biological importance is underscored by expanding roles in diabetes, neurobiology, blood coagulation, and host defense, suggesting apolipoproteins may be useful for diagnostic and therapeutic applications in a variety of disease settings.
Apolipoprotein(apo)A-I is the most abundant protein of HDL, and is thought to protect against atherosclerosis (11–14). ApolipoproteinE found in VLDL, chylomicrons and HDL and is also considered anti-atherogenic (15–17). The beneficial effects of apoA-I and E are generally related, in part, to activity in mediating reverse cholesterol transport (RCT) (18–23). This process involves removal of excess cholesterol from macrophage foam-cells in arterial plaque followed by the transport of cholesterol to the liver for excretion in feces. ApoE also mediates lipid transport in the brain and is considered a major risk factor for Alzheimer’s disease (10, 24–26). Despite therapeutic potential (27–32), there is uncertainty whether full-length apolipoproteins will be suitable for wide-spread clinical applications. Recombinant proteins are generally costly to manufacture and chemically complex, posing formidable challenges with regards to formulation and stability.
Small peptides represent an attractive alternative to proteins for routine use. However, designing peptides based on the plasma apolipoproteins is not without difficulties. Apolipoproteins generally consist of a series of amphipathic α-helices. Often several of these α-helices linked via proline are required to support potent biological activity (33–36). As a result, short sequences taken directly from native proteins typically possess poor structure and activity (33, 36, 37). Poor structure can predispose to peptide aggregation via non-specific hydrophobic interactions, thereby decreasing solubility (37, 38). These problems complicate production/purification, limit biological applications and confound interpretations of experimental results.
Recently we designed an apolipoprotein mimetic peptide (ATI-5261) that displays hydrophobic character as well as potent cholesterol efflux- and lipid binding- activities (39). These features together with its high aqueous solubility suggested to us that ATI-5261 may possess favorable structural characteristics in solution. Therefore, we tested whether ATI-5261 adopted a discrete molecular form at high concentrations, as suggested by its high aqueous solubility. We also sought to determine whether specific self-associated species were seen at low concentrations, which correlated with secondary structure and activity. We found that ATI-5261 formed a unique assembly of four non-covalently bound α-helical strands that dissociated to trimers, dimers and monomers in a concentration-dependent manner, similar to that reported for apoA-I (40, 41). Moreover, ATI-5261 retained high α-helical content upon extensive dilution, regardless of specific self-associated form. The data suggest ATI-5261 constitutes a new model for indentifying factors to improve structure and activity of HDL mimetic therapies.
EXPERIMENTAL PROCEDURES
Peptides
Peptides were synthesized (Biosynthesis Inc., Lewisville, TX) using all L-amino acids, capped with N-terminal acetyl and C-terminal amide groups, isolated by HPLC and used at a purity of >95%. The sequence EVRSKLEEWFAAFREFAEEFLARLKS corresponded to that of ATI-5261 and DWFKAFYDKVAEKFKEAF peptide 4F (38). Peptide 4F was used as a positive control (where applicable), because it represents a prototypical class A consensus peptide and its biophysical properties (42, 43) have been characterized. Unless otherwise stated, lyophilized peptides were dissolved in 10 mM phosphate buffered (pH=7.4) saline (150 mM NaCl), referred to as PBS. Peptide concentrations were determined by absorbance at 280 nm.
Self -association properties of peptides
Non-denaturing gradient gel electrophoresis was performed using premade Novex® 4–20% Tris-Glycine gels (Invitrogen). Gels were stained with SimplyBlue™ SafeStain (Invitrogen) and destained with distilled water to visualize peptides. Distribution of ATI-5261 physical forms was evaluated by fast protein liquid chromatography (FPLC), using either Superdex 75 10/300GL or Superdex ™ Peptide columns (GE Healthcare) pre-equilibrated with PBS. The system was operated at a flow rate of 0.5 ml/min at room temperature and 0.2 ml of peptide stock solutions injected onto columns. Elution of peptides was monitored by either 280 or 215 nm absorbance using a UVis920 GE detector. No differences were noted in ability of ATI-5261 to adopt a single self-associated species when FPLC experiments were performed at room temperature versus 4°C (data not shown). Columns were calibrated with the appropriate molecular weight standards indicated by the manufacturer and shown in figure legends.
Small angle X-ray scattering (SAXS) was used to verify the size and oligomeric status of ATI-5261 in solution. X-ray scattering data were collected at SIBYLS (44) (beamline 12.3.1 - sibyls.als.lbl.gov) at the Advanced Light Source, Lawrence Berkeley National Laboratory. Fifteen micro-liters of each of three concentrations of ATI-5261 and matching buffer (PBS) were used for collecting scattering data at 20°C. Shape was determined by the program GASBOR (45). Ten individual and highly consistent GASBOR runs were averaged to determine the final shape of the ATI-5261 assembly. Two independent preparations of ATI-5261 yielded identical results.
DMS cross-linking of ATI-5261
Cross-linking of ATI-5261 was achieved with dimethylsuberimidate (DMS, Pierce Biotechnolgy, Rockford, IL), dissolved in 0.2M triethanolamine, pH= 8.0. Peptide samples were reacted with 20-fold molar excess of DMS for 3 hrs at 25°C. The reaction was terminated by addition of 50 mM Tris-HCl. Reaction products were mixed with 3× SDS loading buffer (Cell Signaling Technology, Inc.), then boiled for 1 min. The supernatants were loaded onto a 10–20% Tris-Tricine SDS-PAGE gel and electrophoresed. Spectra™ Multicolor Low Range Protein standards (1.7 – 40 kDa, Fermentas) were used to identify the size of cross-linked species, visualized with SimplyBlue™ SafeStain. A substantial number of studies were performed to ensure optimal conditions were used to produce maximal cross-linking of peptide samples. These experiments included variation of incubation time and concentration of DMS as well as use of alternative cross-linking reagents. The latter included use of glutaraldehyde and acrolein, which produced essentially identical results as those obtained with DMS (data not shown).
CD Spectroscopy
Circular dichroism (CD) spectroscopy was carried out on a Jasco 810 spectropolarimeter at 25°C, using lipid-free peptide in 10 mM sodium phosphate buffer (pH=7.4) and averaging 4 scans (20 nm/min/scan) per peptide per run (i.e. each concentration) as described (46).
Fluorescence Spectroscopy
Fluorescence emission spectra of ATI-5261 (1 mg/ml) in the presence/absence of 50% trifluoroethanol (TFE) in sodium phosphate buffer, pH= 7.4 were recorded at 25°C. The Trp fluorescence emission was monitored following excitation at 280 nm and was recorded between 300 and 500 nm on a Perkin–Elmer spectrofluorimeter. A blank of buffer alone was subtracted from all spectra.
Cell-culture and cholesterol efflux assay
J774 mouse macrophages were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). Cells were plated onto 24-well culture plates and labeled with [3H]cholesterol (1 μCi/mL) in RPMI-1640 with 1% FBS for 48 h. A cAMP analogue (cpt-cAMP) was added (final concentration of 0.3 mM) to upregulate ABCA1 expression. Cells were extensively rinsed with serum-free RPMI-1640 medium followed by an extended rinse (2 h) with RPMI-1640 containing 0.2% bovine serum albumin (BSA). Lipid-free peptides were added to cells in serum-free RPMI-1640 medium to initiate cholesterol efflux. The amount of [3H]cholesterol appearing in the medium was expressed as a percentage of the radioactivity initially present in cells at time zero (47). The background release of [3H]cholesterol to serum-free medium alone was subtracted from the values obtained with lipid-free peptides.
Lipid clearance assay
A turbid solution of dimyristoylphosphatidylcholine (DMPC) was used to assess relative capacity of peptides to solubilize phospholipid (47). The DMPC was used at a final concentration of 0.16 mg/ml PBS (pH=7.4). The mass ratio of DMPC to peptides was 2:1 or 4:1. The absorbance (400 nm) of samples was monitored continuously over a period of 20 min at 25°C.
Statistics
Where appropriate, data were expressed as means ± SD of at least 3 independent experiments and statistical analyses performed using Student’s t-test.
RESULTS
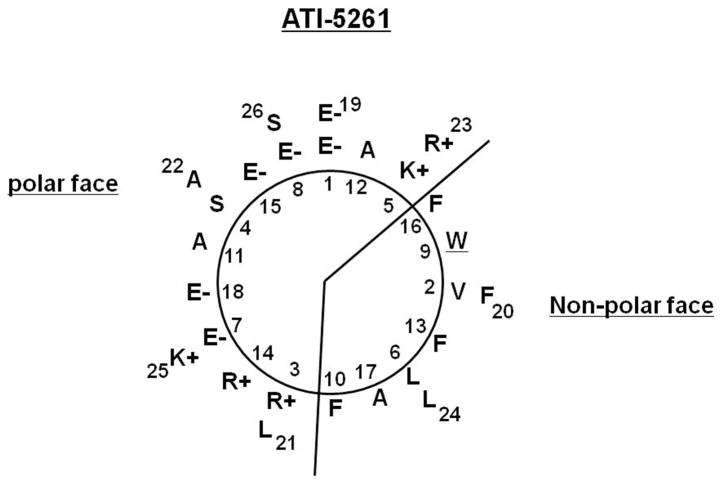
ATI-5261 is composed of a linear sequence of 26 amino acids that form a class A amphipathic α-helix with opposing polar and non-polar surfaces (Figure 1). The amphipathic nature of the peptide suggests it may self-associate to shield hydrophobic residues from water. Therefore, we performed a series of biophysical studies to evaluate the oligomeric behavior of the peptide in solution. ATI-5261 migrated as a low molecular weight (LMW) band on non-denaturing gels (Figure 2A). This contrasted the behavior of peptide 4F, which aggregated as high molecular weight (HMW) species under identical conditions, i.e. stock solutions = 1 mg/ml PBS, pH=7.4.
Figure 1. Helical-wheel projection of ATI-5261.
Linear sequence of amino acids is denoted by numbers, beginning with residue 1 in the intended foreground. Cationic residues are found at the polar/non-polar interface and negatively charged amino acids down the center of the polar surface, characteristic of a class A α-helix. A single tryptophan (W) residue is located on the non-polar surface.
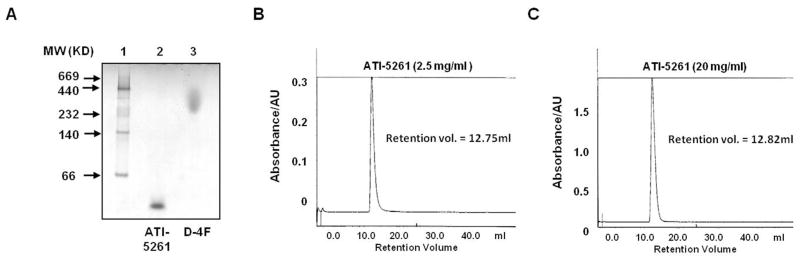
Figure 2. ATI-5261 adopts a low molecular weight form in aqueous buffer.
Stock solutions of ATI-5261 and peptide 4F were prepared in PBS (pH=7.4). Panel A- Native-PAGE (4–20% Tris-glycine gel) of peptides taken from 1 mg/ml stock solutions: lane 1, mobility of molecular weight (MW) standards including thyroglobulin (669 KD), ferritin (440 KD), catalase (232 KD), lactate dehydrogenase (140 KD) and albumin (66 KD); lane 2, ATI-5261 and lane 3, 4F. Peptide loads were 3 μg per well. Panels B and C- Chromatographic behavior of ATI-5261 in PBS on Superdex 75 columns at a flow rate of 0.5 ml/min. Peptide elution was monitored by absorbance at 280 nm. Representative profiles are shown for stock solution of 2.5 (B) and 20 mg/ml (C).
A single sharp peak of ATI-5261 was consistently obtained by gel-filtration chromatography, using a Pharmacia FPLC system fitted with a Superdex 75 column (Figure 2B). Elution profiles and retention volumes (12.8 ml) were identical using ATI-5261 from stock solutions of either 2.5 or 20 mg peptide/ml (Figure 2B and 2C). Similar results were also obtained using stock solutions of 140 mg peptide/ml (highest concentration tested), indicating ATI-5261 adopted a relatively consistent and stable form over a wide-range of concentrations with little or no apparent aggregation tendency. Similarly, gel-filtration profiles were not altered by prolonged storage (2 years) in PBS at 4° C under sterile conditions or following incubation at 37° C for 24 h (data not shown). In contrast, peptide 4F could not be resolved on the Superdex 75 column and typically eluted with extensive rinsing and/or with 0.5 N NaOH, consistent with its tendency to form HMW aggregates as shown on native PAGE (Figure 2A).
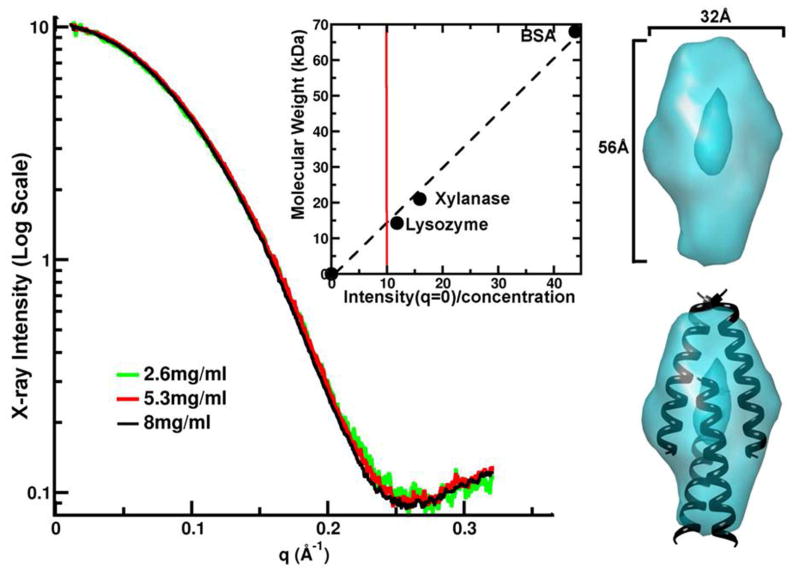
The ability of ATI-5261 to maintain a constant physical form over a broad-range of concentrations (>1 mg/ml) suggested the peptide may form a discrete structure in solution. This was evaluated by small-angle X-ray scattering (Figure 3). Little or no aggregation was detected from the X-ray intensity profiles using peptide at 2 – 8 mg/ml PBS, consistent with results obtained by gel-filtration chromatography (Figure 2). Thus the X-ray scattering curves for ATI-5261 were highly reproducible over the concentration range examined, yielding an estimated MW of 13 KDa. This corresponded to nearly 4-times the MW (3230 Da) of monomeric peptide. Scattering curves also revealed the ATI-5261 tetramer displayed an elongated shape (height/width ratio of 1.75) and hollow central region. These results suggest inter-helical interactions were greater toward the ends of the assembly. Peptide 4F failed to meet the acceptance criteria for SAXS analysis at the advanced light source LBNL, because the solubility was too low in PBS (maximum solubility was approximately 1 mg/ml) and because we could not detect a single homogeneous peak by FPLC (http://bl1231.als.lbl.gov/saxs_protocols/saxs_sample_prep.php).
Figure 3. Small-angle X-ray scattering (SAXS) reveals ATI-5261 forms a tetrameric assembly.
Peptide stock solutions were prepared in PBS (pH=7.4). X-ray scattering curves for ATI-5261 at three concentrations are shown. Once scaled for concentration, the X-ray intensity at q = 0 (inset, red line) was determined to be 13.1kDa or 4.1 times the monomeric weight of the peptide when calibrated against molecular weight standards lysozyme, xylanase and BSA. The elongated shape determined from the SAXS curves (teal) has a hollow central core. Picture at bottom right shows hypothetical fit of 4 α-helical strands within volume estimates, illustrating how overlapping segments may account for the elongated shape of the assembly.
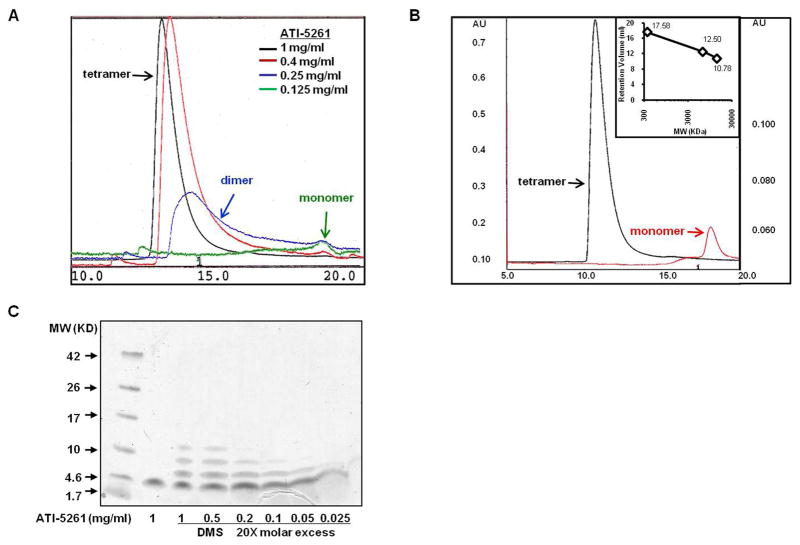
More detailed FPLC experiments were conducted to determine if the ATI-5261 assembly remained intact upon extensive dilution or whether the complex dissociated to individual peptide strands, as concentrations approached the active biological range (39). A characteristic peak (retention volume=12.8 ml) corresponding to the ATI-5261 tetramer was observed using stock solutions of 1 mg peptide/ml (Figure 4A). Dilution of peptide to 0.25 mg/ml produced a broad-peak on the Superdex 75 column, corresponding to a heterogeneous mixture of various low molecular weight species; whereas more extensive dilutions (<0.25 mg/ml) mostly monomeric peptide (Figure 4A). The latter was verified using a high resolution Superdex ™ Peptide column (Figure 4B). Moreover, the MW (12.8 kDa) of the peptide assembly was found to be identical to that revealed by SAXS, i.e. using 1 mg peptide/ml (Figure 4B). Close examination of the FPLC profiles (base-width) and consideration of injection volume (0.2 ml) suggest ATI-5261 is subjected to ~10-fold dilution on the columns, indicating peptide monomers were predominant at concentrations <0.025 mg/ml. This was verified by SDS-PAGE/chemical cross-linking assay, which revealed dissociation of the tetrameric assembly to monomers at 0.5 to 0.025 mg/ml concentrations (Figure 4C). Collectively, these results indicate the ATI-5261 assembly dissociates to peptide monomers at low concentrations, where the cholesterol efflux response is sensitive to peptide concentration in the extra-cellular medium (39).
Figure 4. The tetrameric assembly of ATI-5261 dissociates to lower molecular weight forms upon dilution.
Stock solution of ATI-5261 prepared in PBS (pH=7.4) were analyzed by gel-filtration chromatography. Panel A. Separation of various physical forms of ATI-5261 on a Superdex 75 column; 0.2 ml of each stock solution (indicated) was injected onto the column; flow-rate was 0.5 ml/min; elution buffer was PBS. Representative chromatographic profiles of ATI-5261 stock solutions: 1 mg/ml (black), 0.4 mg/ml (red), 0.25 mg/ml (blue) and 0.125 mg/ml (green) are shown. Elution was monitored using absorbance at 280 nm. Results are arbitrary absorbance units (AU) scaled to clearly illustrate peak positions; actual AU= 1.0, 0.25, 0.08, and 0.02 for stock solutions of 1, 0.4, 0.25, 0.125 mg ATI-5261/ml, respectively. Panel B- FPLC using Superdex™ Peptide 10/300 GL columns to separate ATI-5261 forms. Representative profiles of ATI-5261 at 1 mg/ml (black) and 0.1 mg/ml (red) are shown. Calibration curve (inset) shows retention volume of 17.58 ml for (Gly)6 (0.36 KDa), 12.5 ml for aprotinin (6.5 KDa) and 10.78 ml for ribonuclease A (13.6 KDa). Elution was monitored by absorbance at 215 nm. Panel C- ATI-5261 at various concentrations cross-linked with DMS and separated by SDS-PAGE (10–20% Tricine gel). Cross-linking patterns were visualized by staining with SimplyBlue™ SafeStain.
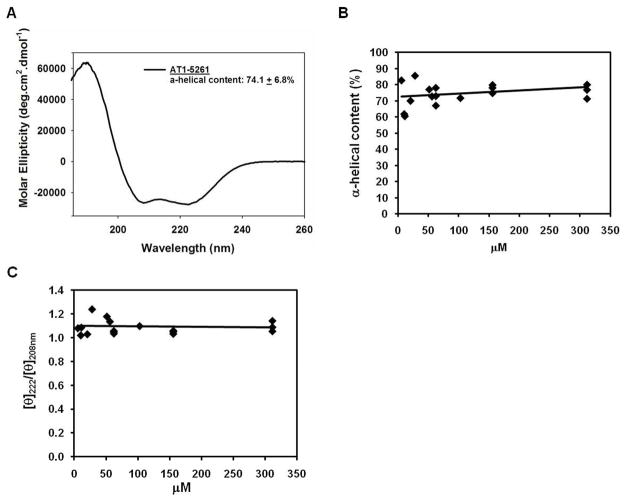
Figure 5A shows a representative CD scan of ATI-5261 in 10 mM phosphate buffer (pH=7.4). The lipid-free peptide possessed high α-helical content (74.1±6.8%), as judged by molar ellipticity at 222 nm. Moreover, α-helical content remained relatively constant over a wide-range of peptide concentrations (6 - 310 μM, i.e. 18 – 1000 μg/ml), indicating ATI-5261 exhibited a strong propensity to form an α-helix (Figure 5B). CD scans were further characterized by a prominent peak at 208 nm of equivalent ellipticity to the 222 nm peak (Figure 5A). This pattern (222/208 ellipticity ratio) also remained constant with increasing peptide concentrations, indicating secondary structure was well maintained (Figure 5C). The latter was particularly noted for extensively diluted samples (6 μM, i.e. 18 μg/ml of ATI-5261), wherein α-helical content remained relatively high (82.6%). The C-terminal (CT) domain of apoE, from which ATI-5261 was derived, also possessed relatively high α-helical content (65 and 68%, two separate experiments at 0.1 mg/ml concentration) that similarly remained constant upon dilution, as described (48, 49).
Figure 5. ATI-5261 maintains high α-helical content and structural integrity over a wide-range of concentrations.
Peptide stock solutions of 2 mg/ml were prepared in 10 mM phosphate buffer (pH=7.4), and diluted with this same buffer as indicated. Panel A. Representative circular dichroism (CD) spectrum of lipid-free ATI-5261 at a concentration of 0.2 mg/ml (62 μM) is shown. Mean α-helicity was calculated from the molar ellipticity at 222 nm (n=4). Panel B. α-helical content of ATI-5261 at various concentrations, ranging from 6 μM to 310 μM (18 – 1000 μg/ml). Panel C. Molar ellipticity ratio ([θ]222/[θ]208nm) plotted against concentration of ATI-5261.
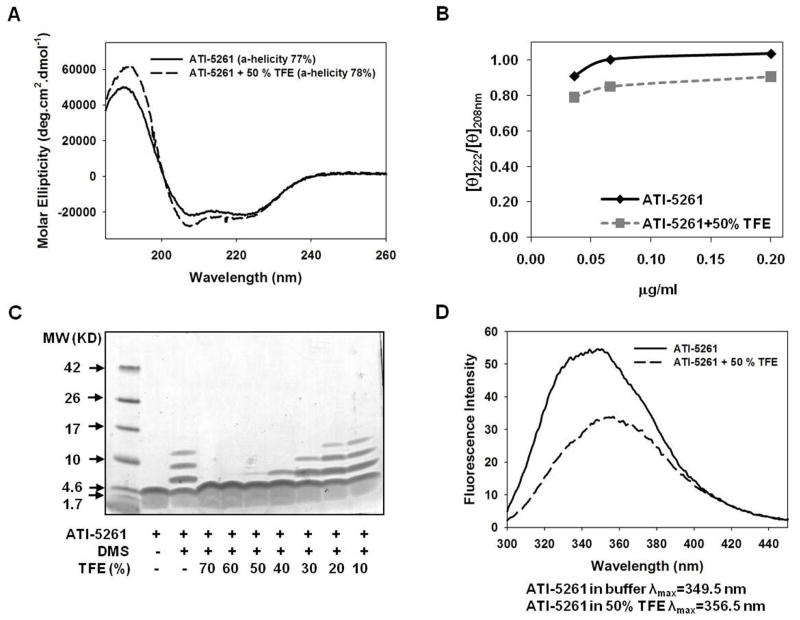
Treatment with the membrane mimetic trifloroethanol (TFE) had little impact of peptide secondary structure, as α-helical content remained unchanged by the presence of 50% TFE compared to controls with no TFE (Figure 6A and 6B). However, TFE markedly increased the 208 nm molar ellipticity of ATI-5261 and, hence, the 222/208 ellipticity ratio was <1. This behavior is consistent with formation of single-stranded α-helix and generally mimics the behavior of apoE CT domain (48). Disruption of strand-strand interactions was verified by SDS-PAGE/chemical cross-linking assay, which demonstrated 50% TFE was sufficient to produce mostly monomeric peptide (Figure 6C). The latter was accompanied by a marked decrease in tryptophan fluorescence intensity, accompanied by a 2 or 3 nm shift in the wavelength of maximal fluorescence emission towards higher wavelengths with 50% TFE, reflecting exposure of the non-polar surface of the peptide to the aqueous environment (Figure 6D). In contrast, the α-helical content of peptide 4F was generally low at 0.2 mg/ml, but was greatly enhanced upon addition of 50% TFE (28 vs. 49%, absence and presence of 50% TFE respectively), consistent with the known behavior of the peptide (50).
Figure 6. Effect of membrane mimetic trifluoroethanol (TFE) on self-association and secondary structure of ATI-5261.
Stock solution of ATI-5261 were prepared in 10 mM phosphate (pH=7.4) and treated with and without TFE for 30 minutes at 25°C before analysis. Panel A. CD spectra of lipid-free ATI-5261 (0.2 mg/ml) in the presence or absence of 50% TFE, showing mean α-helicity calculated from molar ellipticity at 222 nm (n=3). Panel B. [θ]222/[θ]208 ratio of ATI-5261 in the presence and absence of 50% TFE, plotted against concentration of peptide. Panel C. DMS cross-linking/SDS-PAGE of ATI-5261 following treatment (30 min.) with increasing amounts of TFE. A representative stained gel is shown, indicating predominantly monomeric peptide at concentrations > 40% TFE. Panel D. Fluorescence emission spectra of ATI-5261 in the presence or absence of 50% TFE. Results are representative of three separate experiments with different batches of peptide.
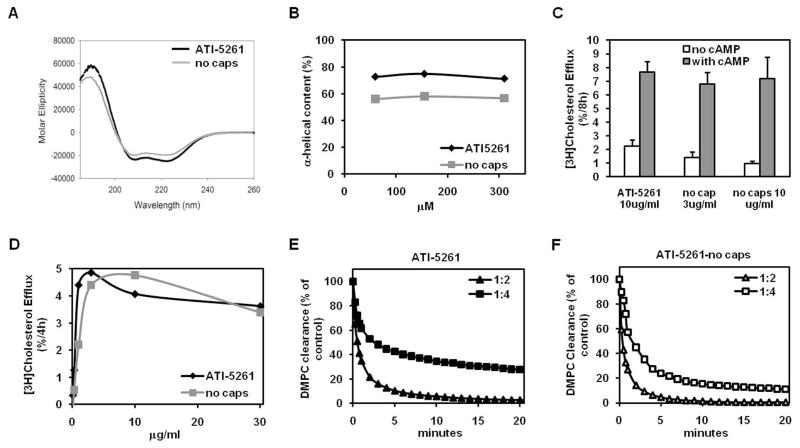
To determine whether the high α-helical content of ATI-5261 was related to end-group blockage, the peptide was synthesized with and without N-terminal acetyl and C-terminal amide groups. ATI-5261 lacking end-group modifications retained high (56.9±0.8%) α-helical content over the experimental range of concentrations, indicating secondary structure was minimally dependent on the end-group modifications (Figure 7A and 7B). Moreover, ATI-5261 lacking end-group modifications retained selective and potent cholesterol efflux activity (Figure 7C and 7D) as well as the ability to bind phospholipid (Figure 7E vs. 7F). These results suggest secondary structure and activity of ATI-5261 are largely dependent on intrinsic properties of the peptide and, thus, minimally impacted by end group modification.
Figure 7. Impact of end-group modifications on secondary structure, cholesterol efflux- and lipid-binding- activities of ATI-5261.
ATI-5261 was synthesized with and without N-terminal acetyl and C-terminal amide groups and dissolved in 10 mM phosphate buffer (pH=7.4). Panel A. Representative CD scan and mean α-helicity of ATI-5261 (62 μM) lacking end-group modifications. Panel B. Relationship between α-helicity and concentration of ATI-5261 with and without end-group modifications. Panel C. Ability of ATI-5261 to promote cholesterol efflux from J774 macrophages treated with and without cAMP. Cells labeled with [3H]cholesterol were incubated (4 h) with lipid-free ATI-5261 and ATI-5261 lacking end-group modifications (3 μg/ml each) to monitor cholesterol efflux. Values are means ± SD, n=5. Panel D. Dependence of cholesterol efflux on the concentration of lipid-free ATI-5261 with/without end group modifications, determined using J774 cells treated with cAMP. Results are representative of 2 experiments. Panel E. Lipid-binding activity of ATI-5261, prepared with N-terminal acetyl and C-terminal amide groups. Lipid-binding activity was assessed as clearance of turbid solutions of DMPC, using peptide:lipid mass ratios of 1:2 and 1:4 at 25°C and following absorbance at 400 nm. Panel F. Lipid-binding activity of ATI-5261, lacking end-group modifications; conditions identical to panel E. Results are representative of two separate experiments.
DISCUSSION
Recently we created a single α-helix peptide designated ATI-5261 that stimulates cellular cholesterol efflux with apolipoprotein molar potency (39). This peptide is unique because previous studies have suggested several α-helical segments linked via proline are required for mediating cholesterol efflux efficiently via ABCA1 (33–35). Potent activity coupled with high aqueous solubility suggested to us that ATI-5261 may possess favorable structural features. Thus we evaluated biophysical properties of ATI-5261 to better understand peptide structure and activity. We found that ATI-5261 a) formed a discrete assembly of four non-covalently bound α-helices with little or no tendency to aggregate as high molecular weight species, b) dissociated to trimers, dimmers and monomers upon extensive dilution, and c) retained high α-helical content regardless of concentration, specific self-associated form, presence of N- and C-terminal capping and stabilizing agent TFE. These features distinguish ATI-5261 from other apolipoproteins mimetics in the literature, indicating the peptide constitutes a new platform for studies of α-helix structure and for optimizing the biophysical properties of HDL mimetic therapies.
Non-specific aggregation of peptides generally produces high molecular weight forms with limited solubility (38). This can create difficulties when using peptides and confound interpretations of experimental results, thereby hindering scientific- and clinical- applications. In contrast, ATI-5261 readily formed a single low molecular weight (LMW) species over a broad-range of concentrations ≥1 mg/ml, as determined by FPLC using a Superdex 75 column. Similar results were consistently seen using a high resolution Superdex™ Peptide column, which revealed ATI-5261 exhibited an apparent molecular weight (~12.8 kDa) roughly 4-times the size of the monomeric peptide (MW= 3230). Formation of single LMW species was verified by SAXS. These analyses employed peptide at concentrations (2 – 8 mg/ml) that overlapped FPLC experiments using ATI-5261 ≥ 1 mg/ml. This behavior to adopt a tetrameric form likely accounts for the high aqueous solubility of ATI-5261 in the absence of lipid, providing favorable solution properties in physiological buffers with little or no apparent aggregation tendency.
To our knowledge, ATI-5261 is the first HDL/apolipoprotein mimetic amenable to high-resolution structural analysis in the absence of lipid. Such studies may prove useful for identifying determinants that control peptide aggregation and solubility as well as new features not yet considered to optimize structure/activity of HDL mimetics. This is made possible because of the uniform nature of the ATI-5261 self-associated form, which provides a highly homogenous sample for detailed analysis. The present studies employing SAXS offers an example of this, where 15–20 Å resolution of ATI-5261 assembly was achieved. This technique revealed ATI-5261 formed a tetrameric assembly of elongated shape under physiological conditions of salt and pH. Formation of a tetrameric assembly was generally consistent with various cross-linked forms generated upon exposure to DMS. These cross-linked forms ranged in size from monomers to tetramers and likely reflect overlapping peptide strands characteristic of the assembly predicted by SAXS. These assertions are based on the height/width ratio of 1.75 for the assembly as well as the length (56Å) of its long-axis, which is considerably greater than that predicted (32Å) for single stranded peptide of 7 helical turns (i.e. 26-mer peptide of high α-helical content). Exposure of ATI-5261 to 50% TFE produced monomeric peptide with concomitant quenching of tryptophan fluorescence. This reflects exposure of the non-polar surface to water upon dissociation of the assembly and suggests ATI-5261 self-associates via interactions between non-polar surfaces of adjacent helical strands, thereby shielding hydrophobic amino acids from the bulk water-phase. Such a mechanism would be consistent with the peptide’s high solubility, maximizing exposure of polar residues to the aqueous environment.
ATI-5261 stimulates cellular cholesterol efflux with a Km=0.86 μg/ml (0.27 μM), displaying saturation in efflux at 3 μg/ml. Presently, we found that the ATI-5261 tetramer dissociated to lower molecular weight species upon dilution, in keeping with studies of native apolipoproteins (40, 41, 51). This was verified by FPLC using Superdex 75 and Peptide columns as well as SDS-PAGE/chemical cross-linking assays with DMS, visualizing peptide with a stain (Figure 4). These techniques revealed that monomeric ATI-5261 predominated at concentrations ≤ 25 μg/ml, i.e. far above the concentration required for mediating cholesterol efflux efficiently. Therefore, we believe activity of the peptide may be attributed to its lower molecular weight forms, reflecting dissociation of “inactive” tetramers to expose non-polar surfaces for binding lipid.
The α-helical content of apoA-I mimetic peptides is generally thought to increase with increasing peptide concentration, indicating helix-helix interactions can induce α-helical formation/stability (50). Results presented in Figure 5 indicate, however, that ATI-5261 retained high α-helical content over a broad-range of concentrations (6– 310 μM). This includes several independent experiments using ATI-5261 at concentrations as little as 18 μg/ml (lowest concentration currently tested), where the monomeric form of the peptide is produced and saturation of cholesterol efflux is expected. This may explain the potent cholesterol efflux activity of ATI-5261, as secondary structure is thought to be a key determinant for mediating ABCA1 cholesterol efflux (35, 52). Moreover absence of end group capping or presence of membrane mimetic TFE had little impact on secondary structure, indicating ATI-5261 exhibited a strong propensity to adopt an α-helix conformation. The former also indicates that an α-helical content of at least 50–60% is sufficient to stimulate cellular cholesterol efflux efficiently. The lack of strict dependence of ATI-5261 α-helical content on concentration further suggests that intra-molecular forces may influence secondary structure and activity. Thus it is likely the primary amino acid sequence of ATI-5261 greatly favors formation of secondary structure and, perhaps, self-association and solubility properties.
The molecular features of ATI-5261 that govern self-association are currently not known. However, this could involve numerous factors such as peptide size, charge, and/or hydrophobic residues, which direct peptide-peptide interactions. This is supported by data derived from SAXS and tryptophan flouresence, whereby the ATI-5261 oligomer appears to adopt a unique shape with distinct features toward the ends and central regions. Also noteworthy is that ATI-5261 was derived from a segment of apoE that possesses reasonable good secondary structure and self-association properties (39, 53), suggesting that putative determinants governing formation of the tetrameric assembly may be inherent to sequences from which the peptide was derived. Although speculative, increased secondary structure may also play a role minimizing potential for non-specific or random interactions (i.e. aggregation tendency) between peptide molecules.
In summary, ATI-5261 represents a novel HDL/apolipoprotein mimetic with greatly improved drugability features, such as high aqueous solubility and limited aggregation tendency. Additional noteworthy features include ability to adopt a specific tertiary form in solution composed of a limited number of α-helical peptide strands as well as high α-helical content upon dilution, i.e. in the absence of lipid. These features may enable more detailed structural analysis, providing models to make further design improvements and/or platforms for creating highly efficient cholesterol efflux peptides. Thus, ATI-5261 appears to constitute an attractive new model to define structure-activity relationships and anti-atherosclerosis mechanisms. Such a model takes into account oligomerization/dissociation behavior and secondary structure of peptide monomers. A deeper understanding of apolipoprotein self-association/dynamics may prove useful for optimizing overall structure and translatability of HDL/apolipoprotein mimetic therapies.
Acknowledgments
The work was supported by funds from the Tobacco-Related Disease Research Program (TRDRP) of the state of California grant 17RT-0082 (JKB) and 17RT-0165 (VN) and NIH grant R21-HL085791 (JKB).
The authors would like to thank Dr. Peter Walian for providing expert advice optimizing FPLC instrumentation. The work was conducted, in part, at Lawrence Berkeley National Laboratory through the United States Department of Energy, Office of Science, Office of Biological and Environmental Research under contract DE-AC02-05CH11231. Dr. Gregory Hura’s contribution was supported by this contract.
ABBREVIATIONS
- SAXS
Small angle X-ray scattering
- Apo
Apolipoprotein
- RCT
Reverse cholesterol transport
- FPLC
Fast protein liquid chromatography
- DMS
Dimethylsuberimidate
- CD
Circular dichroism
- TFE
Trifluoroethanol
- FBS
Fetal bovine serum
- BSA
Bovine serum albumin
- DMPC
Dimyristoylphosphatidylcholine
- LMW
Low molecular weight
- HMW
High molecular weight
References
- 1.Segrest JP, Li L, Anantharamaiah GM, Harvey SC, Liadaki KN, Zannis V. Structure and function of apolipoprotein A-I and high-density lipoprotein. Curr Opin Lipidol. 2000;11:105–115. doi: 10.1097/00041433-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 3.Rifkind BM. High-density lipoprotein cholesterol and coronary artery disease: survey of the evidence. Am J Cardiol. 1990;66:3A–6A. doi: 10.1016/0002-9149(90)90561-e. [DOI] [PubMed] [Google Scholar]
- 4.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc Natl Acad Sci U S A. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladu MJ, Reardon C, Van Eldik L, Fagan AM, Bu G, Holtzman D, Getz GS. Lipoproteins in the central nervous system. Ann N Y Acad Sci. 2000;903:167–175. doi: 10.1111/j.1749-6632.2000.tb06365.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilhelm AJ, Zabalawi M, Grayson JM, Weant AE, Major AS, Owen J, Bharadwaj M, Walzem R, Chan L, Oka K, Thomas MJ, Sorci-Thomas MG. Apolipoprotein A-I and its role in lymphocyte cholesterol homeostasis and autoimmunity. Arterioscler Thromb Vasc Biol. 2009;29:843–849. doi: 10.1161/ATVBAHA.108.183442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kattan OM, Kasravi FB, Elford EL, Schell MT, Harris HW. Apolipoprotein E-mediated immune regulation in sepsis. J Immunol. 2008;181:1399–1408. doi: 10.4049/jimmunol.181.2.1399. [DOI] [PubMed] [Google Scholar]
- 9.Smith JD. Apolipoprotein E4: an allele associated with many diseases. Ann Med. 2000;32:118–127. doi: 10.3109/07853890009011761. [DOI] [PubMed] [Google Scholar]
- 10.Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:4725–4727. doi: 10.1073/pnas.92.11.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin EM, Krauss RM, Spangler EA, Verstuyft JG, Clift SM. Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature. 1991;353:265–267. doi: 10.1038/353265a0. [DOI] [PubMed] [Google Scholar]
- 12.Tangirala RK, Tsukamoto K, Chun SH, Usher D, Pure E, Rader DJ. Regression of atherosclerosis induced by liver-directed gene transfer of apolipoprotein A-I in mice. Circulation. 1999;100:1816–1822. doi: 10.1161/01.cir.100.17.1816. [DOI] [PubMed] [Google Scholar]
- 13.Linsel-Nitschke P, Tall AR. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 14.Navab M, Anantharamaiah GM, Reddy ST, Van Lenten BJ, Fogelman AM. HDL as a biomarker, potential therapeutic target, and therapy. Diabetes. 2009;58:2711–2717. doi: 10.2337/db09-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellosta S, Mahley RW, Sanan DA, Murata J, Newland DL, Taylor JM, Pitas RE. Macrophage-specific expression of human apolipoprotein E reduces atherosclerosis in hypercholesterolemic apolipoprotein E-null mice. J Clin Invest. 1995;96:2170–2179. doi: 10.1172/JCI118271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenow K, Pearce NJ, Ramji DP. The key role of apolipoprotein E in atherosclerosis. J Mol Med. 2005;83:329–342. doi: 10.1007/s00109-004-0631-3. [DOI] [PubMed] [Google Scholar]
- 17.Davignon J, Cohn JS, Mabile L, Bernier L. Apolipoprotein E and atherosclerosis: insight from animal and human studies. Clin Chim Acta. 1999;286:115–143. doi: 10.1016/s0009-8981(99)00097-2. [DOI] [PubMed] [Google Scholar]
- 18.Rothblat GH, Bamberger M, Phillips MC. Reverse cholesterol transport. Methods Enzymol. 1986;129:628–644. doi: 10.1016/0076-6879(86)29095-3. [DOI] [PubMed] [Google Scholar]
- 19.Fielding CJ, Fielding PE. Molecular physiology of reverse cholesterol transport. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 20.Tall AR. An overview of reverse cholesterol transport. Eur Heart J. 1998;19(Suppl A):A31–35. [PubMed] [Google Scholar]
- 21.Zhang Y, Zanotti I, Reilly MP, Glick JM, Rothblat GH, Rader DJ. Overexpression of apolipoprotein A-I promotes reverse transport of cholesterol from macrophages to feces in vivo. Circulation. 2003;108:661–663. doi: 10.1161/01.CIR.0000086981.09834.E0. [DOI] [PubMed] [Google Scholar]
- 22.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 23.Rothblat GH, Phillips MC. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr Opin Lipidol. 2010;21:229–238. doi: 10.1097/mol.0b013e328338472d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bookheimer S, Burggren A. APOE-4 genotype and neurophysiological vulnerability to Alzheimer’s and cognitive aging. Annu Rev Clin Psychol. 2009;5:343–362. doi: 10.1146/annurev.clinpsy.032408.153625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardif JC, Gregoire J, L’Allier PL, Ibrahim R, Lesperance J, Heinonen TM, Kouz S, Berry C, Basser R, Lavoie MA, Guertin MC, Rodes-Cabau J. Effects of reconstituted high-density lipoprotein infusions on coronary atherosclerosis: a randomized controlled trial. JAMA. 2007;297:1675–1682. doi: 10.1001/jama.297.15.jpc70004. [DOI] [PubMed] [Google Scholar]
- 28.Tardif JC, Heinonen T, Noble S. High-density lipoprotein/apolipoprotein A-I infusion therapy. Curr Atheroscler Rep. 2009;11:58–63. doi: 10.1007/s11883-009-0009-7. [DOI] [PubMed] [Google Scholar]
- 29.Shah PK, Nilsson J, Kaul S, Fishbein MC, Ageland H, Hamsten A, Johansson J, Karpe F, Cercek B. Effects of recombinant apolipoprotein A-I(Milano) on aortic atherosclerosis in apolipoprotein E-deficient mice. Circulation. 1998;97:780–785. doi: 10.1161/01.cir.97.8.780. [DOI] [PubMed] [Google Scholar]
- 30.Alexander ET, Weibel GL, Joshi MR, Vedhachalam C, de la Llera-Moya M, Rothblat GH, Phillips MC, Rader DJ. Macrophage reverse cholesterol transport in mice expressing ApoA-I Milano. Arterioscler Thromb Vasc Biol. 2009;29:1496–1501. doi: 10.1161/ATVBAHA.109.191379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nissen SE, Tsunoda T, Tuzcu EM, Schoenhagen P, Cooper CJ, Yasin M, Eaton GM, Lauer MA, Sheldon WS, Grines CL, Halpern S, Crowe T, Blankenship JC, Kerensky R. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. JAMA. 2003;290:2292–2300. doi: 10.1001/jama.290.17.2292. [DOI] [PubMed] [Google Scholar]
- 32.Shah PK. Apolipoprotein A-I/HDL infusion therapy for plaque stabilization-regression: a novel therapeutic approach. Curr Pharm Des. 2007;13:1031–1038. doi: 10.2174/138161207780487520. [DOI] [PubMed] [Google Scholar]
- 33.Vedhachalam C, Narayanaswami V, Neto N, Forte TM, Phillips MC, Lund-Katz S, Bielicki JK. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 2007;46:2583–2593. doi: 10.1021/bi602407r. [DOI] [PubMed] [Google Scholar]
- 34.Wool GD, Vaisar T, Reardon CA, Getz GS. An apoA-I mimetic peptide containing a proline residue has greater in vivo HDL binding and anti-inflammatory ability than the 4F peptide. J Lipid Res. 2009;50:1889–1900. doi: 10.1194/jlr.M900151-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yancey PG, Bielicki JK, Johnson WJ, Lund-Katz S, Palgunachari MN, Anantharamaiah GM, Segrest JP, Phillips MC, Rothblat GH. Efflux of cellular cholesterol and phospholipid to lipid-free apolipoproteins and class A amphipathic peptides. Biochemistry. 1995;34:7955–7965. doi: 10.1021/bi00024a021. [DOI] [PubMed] [Google Scholar]
- 36.Gillotte KL, Zaiou M, Lund-Katz S, Anantharamaiah GM, Holvoet P, Dhoest A, Palgunachari MN, Segrest JP, Weisgraber KH, Rothblat GH, Phillips MC. Apolipoprotein-mediated plasma membrane microsolubilization. Role of lipid affinity and membrane penetration in the efflux of cellular cholesterol and phospholipid. J Biol Chem. 1999;274:2021–2028. doi: 10.1074/jbc.274.4.2021. [DOI] [PubMed] [Google Scholar]
- 37.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta G, Chaddha M, Hama S, Navab M, Fogelman AM, Garber DW, Mishra VK, Epand RM, Epand RF, Lund-Katz S, Phillips MC, Segrest JP, Anantharamaiah GM. Effects of increasing hydrophobicity on the physical-chemical and biological properties of a class A amphipathic helical peptide. J Lipid Res. 2001;42:1096–1104. [PubMed] [Google Scholar]
- 39.Bielicki JK, Zhang H, Cortez Y, Zheng Y, Narayanaswami V, Patel A, Johansson J, Azhar S. A new HDL mimetic peptide that stimulates cellular cholesterol efflux with high efficiency greatly reduces atherosclerosis in mice. J Lipid Res. 2010;51:1496–1503. doi: 10.1194/jlr.M003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitello LB, Scanu AM. Studies on human serum high density lipoproteins. Self-association of apolipoprotein A-I in aqueous solutions. J Biol Chem. 1976;251:1131–1136. [PubMed] [Google Scholar]
- 41.Ren X, Zhao L, Sivashanmugam A, Miao Y, Korando L, Yang Z, Reardon CA, Getz GS, Brouillette CG, Jerome WG, Wang J. Engineering mouse apolipoprotein A-I into a monomeric, active protein useful for structural determination. Biochemistry. 2005;44:14907–14919. doi: 10.1021/bi0508385. [DOI] [PubMed] [Google Scholar]
- 42.Van Lenten BJ, Wagner AC, Anantharamaiah GM, Navab M, Reddy ST, Buga GM, Fogelman AM. Apolipoprotein A-I mimetic peptides. Curr Atheroscler Rep. 2009;11:52–57. doi: 10.1007/s11883-009-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navab M, Anantharamaiah GM, Reddy ST, Hama S, Hough G, Grijalva VR, Yu N, Ansell BJ, Datta G, Garber DW, Fogelman AM. Apolipoprotein A-I mimetic peptides. Arterioscler Thromb Vasc Biol. 2005;25:1325–1331. doi: 10.1161/01.ATV.0000165694.39518.95. [DOI] [PubMed] [Google Scholar]
- 44.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel AB, Khumsupan P, Narayanaswami V. Pyrene fluorescence analysis offers new insights into the conformation of the lipoprotein-binding domain of human apolipoprotein E. Biochemistry. 2010;49:1766–1775. doi: 10.1021/bi901902e. [DOI] [PubMed] [Google Scholar]
- 47.Natarajan P, Forte TM, Chu B, Phillips MC, Oram JF, Bielicki JK. Identification of an apolipoprotein A-I structural element that mediates cellular cholesterol efflux and stabilizes ATP binding cassette transporter A1. J Biol Chem. 2004;279:24044–24052. doi: 10.1074/jbc.M400561200. [DOI] [PubMed] [Google Scholar]
- 48.Choy N, Raussens V, Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J Mol Biol. 2003;334:527–539. doi: 10.1016/j.jmb.2003.09.059. [DOI] [PubMed] [Google Scholar]
- 49.Chroni A, Pyrpassopoulos S, Thanassoulas A, Nounesis G, Zannis VI, Stratikos E. Biophysical analysis of progressive C-terminal truncations of human apolipoprotein E4: insights into secondary structure and unfolding properties. Biochemistry. 2008;47:9071–9080. doi: 10.1021/bi800469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datta G, Epand RF, Epand RM, Chaddha M, Kirksey MA, Garber DW, Lund-Katz S, Phillips MC, Hama S, Navab M, Fogelman AM, Palgunachari MN, Segrest JP, Anantharamaiah GM. Aromatic residue position on the nonpolar face of class a amphipathic helical peptides determines biological activity. J Biol Chem. 2004;279:26509–26517. doi: 10.1074/jbc.M314276200. [DOI] [PubMed] [Google Scholar]
- 51.Garai K, Frieden C. The Association-Dissociation Behavior of the ApoE Proteins: Kinetic and Equilibrium Studies. Biochemistry. 2010;49:9533–9541. doi: 10.1021/bi101407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brubaker G, Peng DQ, Somerlot B, Abdollahian DJ, Smith JD. Apolipoprotein A-I lysine modification: effects on helical content, lipid binding and cholesterol acceptor activity. Biochim Biophys Acta. 2006;1761:64–72. doi: 10.1016/j.bbalip.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J Biol Chem. 1998;263:6249–6258. [PubMed] [Google Scholar]