Abstract
The t(8;21) abnormality occurs in a minority of acute myeloid leukemia (AML) patients. The translocation results in an in-frame fusion of two genes, resulting in a fusion protein of one N-terminal domain from the AML1 gene and four C-terminal domains from the ETO gene. This protein has multiple effects on the regulation of the proliferation, the differentiation, and the viability of leukemic cells. The translocation can be detected as the only genetic abnormality or as part of more complex abnormalities. If t(8;21) is detected in a patient with bone marrow pathology, the diagnosis AML can be made based on this abnormality alone. t(8;21) is usually associated with a good prognosis. Whether the detection of the fusion gene can be used for evaluation of minimal residual disease and risk of leukemia relapse remains to be clarified. To conclude, detection of t(8;21) is essential for optimal handling of these patients as it has both diagnostic, prognostic, and therapeutic implications.
1. Introduction
Acute myeloid leukemia (AML) is a heterogeneous bone marrow malignancy, and patients with the cytogenetic t(8;21) abnormality represent a subset with specific clinical and biological characteristics [1]. The translocation fuses the AML1 gene (also called RUNX1) on chromosome 21 with the ETO gene (also referred to as the RUNX1T1 gene that encodes the CBFA2T1 protein) on chromosome 8. The criteria for the diagnosis differs from other AML patients; the leukemia cells showbiological characteristics that are uncommon in other AML subsets, and the prognosis after intensive chemotherapy is better for these patients than for the majority of AML patients [1]. t(8;21) was the first cytogenetic abnormality discovered in AML [2], and today it offers a unique example of how a cytogenetic abnormality is used to define a distinct subgroup of patients. The AML1 gene has p.d.d. been reported to be involved in 39 different rearrangements and most of them have been detected in myeloid malignancies. In the present paper we describe the biology and clinical characteristics of the most common t(8;21) abnormality.
2. The t(8;21) Abnormality in Human AML
2.1. Frequency and Predisposition
The t(8;21) abnormality is found in approximately 5%–10% of all AML cases and 10%–22% of AML cases with maturation corresponding to the previous FAB class M2 [3–8]. The incidence of AML with favourable cytogenetic abnormalities decreases with age [9, 10]; this is also true for t(8;21) that is most common in children/younger patients [11] and uncommon in patients above 60 years of age [10]. Approximately 10%–20% of children with AML have this translocation [12–15]. The detection of t(8;21) transcripts in Guthrie cards suggests that the initiating events can occur in utero [16]. This is also supported by studies of umbilical cord leukocytes that have described an association between prenatal pesticide exposure (i.e., detection of pesticides in meconium) and the detection of t(8;21) in the leukocytes and the level of the fusion transcript then correlated with the pesticide level [17].
Tissue-specific genomic organization probably contributes to the formation of this disease-specific translocation. Studies of the nuclear architecture have concluded that chromosomes 8 and 21 tend to colocalize in myeloid cells [18]. Furthermore, the chromatin organization at intron 5 of the RUNX1/AML1 gene, where the sequenced breakpoints have been mapped, are characterized by reduced histone H1 levels and increased levels of hyperacetylated H3 for AML1 expressing cells. Induction of DNA damage can then induce formation of t(8;21) in the HL-60 AML cell line but not in nonhematopoietic HeLa cells [19]. Thus, both the nuclear microarchitecture and epigenetic mechanisms seem to be important for the risk of t(8;21) formation in myeloid cells. The AML1-ETO fusion protein alone is not sufficient for leukemia development (see Section 9), but the protein downregulates the expression of DNA repair enzyme 8-oxoguanine DNA glycosylase (OGG1), which may then lead to additional genetic abnormalities required for development of AML [20].
2.2. AML1-ETO: Variant Rearrangements and Combination with Other Genetic Abnormalities
The t(8;21) generates two fusion genes, AML1-ETO and ETO-AML1, but only the AML1-ETO transcript transcribed from the derivative 8 chromosome is detectable by reverse transcriptase polymerase chain reaction (RT-PCR). Simple reciprocal translocation is by far the most common abnormality for creating AML1-ETO fusions, but the fusion can also occurs through variant rearrangements (Figures 1(a), 2(a)). The translocation can involve several chromosomes and occur together with inversion of the derivative 8 chromosome (e.g., inv(8)(p21;q22)t(8;21), inv(8)(q22q24)) [21, 22]. AML1-ETO fusion can also be a result of insertion, and both ins(21;8) and ins(8;21) have been described [23] (Figure 2(b)). These variant rearrangements can be cryptic and easily overlooked by conventional G-banding, and their frequencies are therefore unknown.
Figure 1.
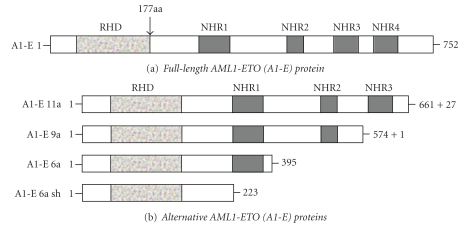
Domain organization of the full-length and alternative AML1-ETO fusion proteins. (a) The full-length AML1-ETO (A1-E) protein is shown, where most of the ETO (RUNX1T1) gene is fused into the N-terminal 177aa of AML1 (RUNX1) gene giving rise to a transcript coding for a protein of 752 amino acids (aa). The AML1 gene encodes the Runt homology domain (RHD) which is a DNA-binding protein, while ETO encodes four highly conserved functional domains called nervy homology domains (NHR1-4). (b) Different fusion transcripts arise due to alternative exon usage and splicing, which give rise to truncated proteins lacking NHR domains. Protein size (i.e., number of aa) is shown on the right with the number of additional aa that were not included in the original sequence. These alternative A1-E transcripts can be coexpressed alongside the full-length transcript and have different leukemogenic capabilities.
Figure 2.
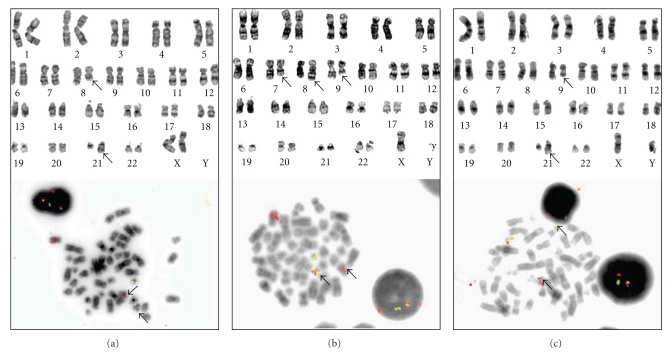
Cytogenetic analysis of AML blasts by G-banding and FISH. (a) The derivative chromosomes from a simple reciprocal translocation between 8q22 and 21q22 are detectable by G-banding (upper panel) and the translocation can be verified using FISH probes (lower panel) against ETO and AML1. With this particular probe (Vysis LSI ETV6(TEL)/RUNX1(AML1) ES Dual Color) fusion signals will appear both on derivative 8 and derivative 22. (b) In rare cases, AML1-ETO fusion occurs as a result of insertion. Small insertions can only be detected using FISH probes. In this case, the translocation t(7;8)(q11;q22) between chromosome 7 and 8 and the 9q deletion del(9)(q12q22) were detected. As deletion 9q is rare in AML and can coexist with AML1-ETO fusion, FISH analysis was, therefore, performed. Only one fusion signal on derivative 22 was detected, indicating an ins(21;8)(q22;q22q22) insertion. (c) AML1 can also be involved in translocations with other partners mimicking complex t(8;21). In this case, there is a translocation between 9q22 and 21q22. FISH analysis using in-house split-signal probes against AML1 verified the involvement of these genes, whereas the gene on derivative 9 is unknown.
The t(8;21) abnormality is often detected together with additional cytogenetic or molecular genetic abnormalities; a majority of the patients seem to have additional abnormalities (Table 1) [3]. These abnormalities are often numerical, but other translocations or deletions can also be detected.
Table 1.
Genetic abnormalities commonly detected in combination with t(8;21).
| Abnormality | Frequency in t(8;21) AML | Documented prognostic impact | Reference |
|---|---|---|---|
| Chromosomal abnormalities | |||
| -X in female patients | 30%–40% | None | [7, 24, 25] |
| -Y in male patients | 50%–60% | Possible improved | [7, 24, 25] |
| Del(9q) | 15%–35%; most studies state 15%–20% | None | [7, 24–26] |
| Trisomy 8 | 8% | [27] | |
| Complex abnormalities | 9%–23% | Adverse prognosis | [27–29] |
| Molecular abnormalities | |||
| KIT mutations | 25%–50% | Possible adverse prognosis | [30–41] |
| JAK2V617F | 6%–8% | [36, 42] | |
| Flt3 -ITD | 5% | Adverse prognosis | [24, 33, 43] |
| Flt3 D853 | 3%–7% | [24, 43] |
Chromosome 9 Alternations —
Deletion of chromosome 9q, the del(9q) abnormality, is recurrent but uncommon in AML, and it was detected only in 81 out of more than 5000 patients that entered 3 MRC studies [26]. The deletion was then detected in combination with t(8;21) for 29 out of these 81 patients, and the karyotyping indicated that a common area of deletion in region 9q21-22 was present in more than 90% of the cases. Targeted analysis against AML1-ETO was not performed in this study. del(9q) has also been found cooccurring with fusion due to insertion [44]. Based on the results from several clinical studies, the del(9q) abnormality seems to be present in 15%–35% of AML patients with t(8;21) [24, 26]. There was no indication from the MRC data for an adverse prognostic impact of this additional abnormality; patients with t(8;21) together with del(9q) had a 5-year overall survival of 75% in this study [26].
Sweetser et al. investigated the molecular genetics of 43 patients with del(9q) [45]. They described a commonly deleted region of 2.4 Mb containing 11 genes, 7 of these being downregulated in del(9q) AML compared with normal CD34+ hematopoietic cells or AML cells with normal karyotype. Two of these genes, TLE1 and TLE4, are most likely tumor suppressor since, loss of expression in AML1-ETO expressing cells leads to increased proliferation and cell survival.
Numerical Abnormalities —
Loss of sex chromosomes is especially common in t(8;21) AML patients (Table 1). Loss of one X chromosome does not seem to have any prognostic impact, whereas a recent study described a weak good prognostic impact for -Y [7].
Other Cytogenetic Abnormalities —
Several abnormalities have been described in patients with t(8;21) AML such as trisomy 4 and 8, but these combinations are uncommon, and their possible prognostic impact remains to be investigated. Other abnormalities include tetraploid or near-tetraploid clones [46].
Mutations of KIT —
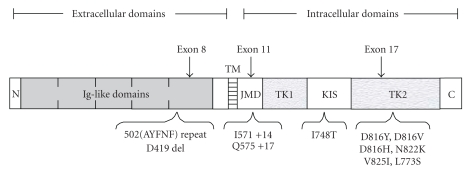
Wang et al. characterized c-kit mutations in a group of 54 AML patients with t(8;21), and they observed mutations for 26 of these patients (Figure 3) [30].
21 patients had abnormalities in the tyrosine kinase domain, four patients had mutations in the juxtamembrane or extracellular domains, and the last patient had a mutation in the kinase insert sequence between the adenosine triphosphate binding and phosphotransferase regions of the tyrosine kinase domain.
Seven different point mutations, three internal tandem duplications (ITD), and one amino acid deletion were detected.
The most common KIT abnormality was the N822K mutation (10/26) followed by three variants of D816 mutations (9/26); this is similar to another study [31].
Figure 3.
General architecture of the c-kit receptor and the mutations described in combination with the t(8;21) abnormality in the study by Wang et al. [30]. The c-kit type III receptor tyrosine kinase consists of an extracellular ligand-binding portion comprising five immunoglobulin-(Ig-) like repeats, a single transmembrane (TM) domain, a juxtamembrane domain (JMD), and a cytoplasmic portion containing and a split tyrosine kinase domain (TK1 and TK2) with a kinase insert sequence (KIS). Locations of c-kit abnormalities found in t(8;21) AML are indicated by the arrows. c-kit mutations are found more frequently within the extracellular fifth immunoglobulin-like domain (exon 8) and the second tyrosine kinase domain which contains the activation loop (exon 17).
Thus, many different KIT mutations have been detected in combination with t(8;21), but mutations in the tyrosine kinase domain predominate [30–32]. The incidence of KIT mutations in t(8;21) AML varies between 6%–31% in most clinical studies [30–41], and one exceptional study in children reported an incidence of 43% [47].
The biological impact of different KIT mutations was investigated in a recent experimental study [48]. These authors compared the effects of coexpressing either (i) the more common KITD 814V mutation within the tyrosine kinase domain or (ii) the less common deletion within Exon 8 (E8D419) with AML1-ETO in an experimental animal model. Their observations suggested that the KIT mutations were important for the disease phenotype and the KIT deletion was associated with a less aggressive disease.
The possible prognostic impact of KIT mutations has been investigated in several relatively small studies including 33–54 patients [30, 32, 34, 36] that all have described an adverse impact on relapse rate and/or long-term survival. Relapse rates as high as 70%–80% have been observed [31, 33, 34]. An adverse impact on overall survival was also described in the study reported by Boissel et al. that included 56 patients [33]. However, several recent studies have described no prognostic impact of the KIT mutations in t(8;21) AML both for adult patients [35, 38, 39] and in children AML [40, 47].
Flt3-Abnormalities —
Flt3-ITD occurs at a relatively low frequency in t(8;21) AML and seems to have an adverse prognostic impact also in these patients (Table 1) [33]. Animal models suggest that both AML1-ETO and Flt3-ITD alone are insufficient to cause leukemogenesis, but they may cooperate in inducing AML [49]. Flt3 mutations seem to be associated with an adverse prognosis in patients with t(8;21); a recent preliminary report described a 3-year overall survival of only 26% [35].
JAK2V617F Mutations —
Only three studies have investigated these mutations in t(8;21) AML, and they occurred in less than 10% of the patients [35, 36, 42].
Ras Mutations —
Mutations in NRas and KRas appear to be more frequent in pediatric than in adult patients, but they do not seem to have any prognostic impact in either group [11, 33, 35, 38]. Even though Ras mutations, thus, do not seem to have any impact on chemosensitivity, experimental studies suggest that these mutations are important in leukemogenesis and promote progression towards transformation in cells expressing the t(8;21) fusion protein [50].
PDGF Receptor Mutations —
Results from animal models suggest that PDGF-receptor (PDGFR) mutations may cooperate with AML1-ETO in leukemogenesis (see below), and PDGFRA seems to be frequently expressed in t(8;21) AML cells at least for pediatric patients [51]. However, PDGFRA mutations seem to be very uncommon in these patients although a N870S mutation has been described in the homologous domain of the activating KIT and Flt3 mutations.
Other Molecular-Genetic Abnormalities —
Mutations in AML1 and in the hematopoietic transcription factor PU-1 have been described in exceptional patients with t(8;21) AML [25]. The t(8;21) abnormality does not seem to coexist with CEBPA mutations [52], or any of the recently described mutations in IDH 1/2 [53, 54] or DNMT3A [54].
2.3. t(8;21) in Secondary AML
Approximately 10%–20% of all cases of t(8;21) AML are secondary [25, 27] and can be detected after treatment for hematologic malignancies (e.g., T-ALL, Hodgkin's disease, and non-Hodgkin's lymphoma) as well as solid tumors (e.g., breast, lung, prostate, esophageal, and thyroid cancer) [55–57]. Secondary AML seems most common after chemotherapy, but it has also been described after radiotherapy alone [55]. The median latency until diagnosis was 37 months in one study, varying from 11 to 126 months [55].
The secondary form is associated with significantly higher age (median 59 versus 41 years) and higher peripheral blood blast counts than the de novo variant [55]. Secondary forms are also of the M2-subtype according to the FAB classification, Auer rods are detected only for certain patients, and the immunophenotype is similar to de novo forms usually being CD33+CD34+CD117+CD19+CD56+ [55]. A recent study described loss of the Y-chromosome in only 12% of all secondary cases compared with 36% of the de novo t(8;21) subset, whereas the frequencies of other abnormalities were similar for secondary and de novo forms [25]. The overall survival of patients with secondary t(8;21) seems to be significantly inferior to patients with de novo t(8;21) AML [55]. However, even though secondary t(8;21) AML has a reduced frequency of -Y, this decreased frequency of a favorable marker cannot explain the adverse prognosis of secondary t(8;21) AML, because the prognostic impact of -Y in the de novo group is relatively weak [7].
3. Molecular Genetics of t(8;21)
3.1. The AML1-ETO Fusion Gene
As described above (Section 2.2, Figure 2), simple reciprocal translocation is the most common abnormality. The fusion gene can also be formed through complex genetic abnormalities that can be detected by cytogenetic analysis, but these abnormalities may also be cryptic and easily overlooked by conventional G-banding.
The in-frame fusion of AML1 (RUNX1) to ETO (RUNX1T1) generally occurs with break-point in AML1 in intron 5-6 and in ETO in intron 1b-2 [24, 58]. The molecular effects on the fusion protein are given by its structure as illustrated in Figure 1(a) and Table 2. AML1 is a transcription factor that is crucial for hematopoietic differentiation, and binds to enhancers and promoters through its aminoterminal Runt domain; this domain is also present in the AML1-ETO fusion-protein and the protein can, therefore, bind to AML1 target gene promoters. However, even though the fusion protein has binding similarities with AML1, it causes an altered transcriptional activation and has another subnuclear localization than the normal AML1 transcription factor [59]. The ETO/RUNX1T1-encoded protein referred to as CBFA2T is also a nuclear protein and functions as a transcriptional repressor through its binding to both histone deacetylases and transcription factors [60, 61]. The full-length fusion protein contains all except the 31 N-terminal amino acids. The NHR-2 domain is important for homodimerization and interactions with components of the repressor complexes and seems to be responsible for the reduced intranuclear mobility of the fusion protein [62–64]. Thus, the final result of this combination will often be binding to AML1 target gene promoters resulting in suppression [63]. In addition, the fusion protein is directed to nuclear microenvironments distinct from those where the AML1 molecule resides and for this reason may not bind to all AML1 targets [18, 19]. the fusion protein also colocalizes with core-binding factor β (CBFβ) within the nucleus; this colocalization results in a reduced intranuclear mobility of CBFβ that probably disturbs myeloid differentiation [64].
Table 2.
Molecular structure of the t(8;21) fusion protein, the origin of various domains, and the localization of important molecular interactions.
| Origin | Domain (alternative nomenclature) | Molecular interactions |
|---|---|---|
| N-terminal | ||
| AML1 | Runt | DNA binding |
| Binding to CBFβ with formation of heterodimers | ||
| Binding of other transcriptional regulators | ||
| RUNX1T1-derived domains | NHR1 (eTAFH) | Interacts with the nuclear hormone receptor corepressor |
| Interaction with the activation domain of E-proteins (E2A and HEB) | ||
| NHR2 (HHR) | Mediates oligomerization with itself or other ETO molecules | |
| Interacts with the corepressors Sin3, Gfi1, and histone deacetylases 1 and 3 | ||
| NHR3 (Nervy) | Interacts with the regulatory subunit of type II cAMP-dependent protein kinase | |
| This domain together with the NHR4 domain is absent in the leukemogenic AE9a splice variant that naturally occurs in primary human AML cells with t(8;21) | ||
| NHR4 (MYND) | N-CoR and the silencing mediator of retinoid and thyroid hormone receptor (SMRT); these are associated with HDACs | |
| SON, an RNA/DNA-binding protein | ||
| This domain is absent in the leukemogenic AE9a splice variant | ||
| C-Terminal | ||
3.2. AML1-ETO Has Several Splice Variants
Several splice variants of the AML1-ETO gene are present in t(8;21) patient cells, and these are all variants in the ETO part of the molecule [65]. It is generally agreed that the full-length variant alone does not have a leukemogenic effect; this conclusion is based on studies using cell lines as well as in animal models (see Section 6). However, analyses of primary AML cells with t(8;21) have demonstrated that numerous in-frame and out-of-frame transcript variants exist, and these variants seem to result from alternative splicing, internal deletions, or breakpoint region insertions [65]. The full-length molecule encodes a protein of 752 amino acids, whereas the variants are generally shorter (Figure 1(b)).
The AML1-ETO9a variant includes the alternative ETO9a exon and results in a truncated molecule of 575 amino acids that lacks the NHR3 and NHR4 domains [66, 67]. In contrast to the full-length variant, this molecule alone is leukemogenic in a mouse model [66, 67], but this does not seem to be true for an alternative model [68]. A possible explanation for this difference could be that the levels of transcript/fusion protein seem to be lower in the nonleukemic model. The AML1-derived domains together with the ETO-derived NHR-2 domain seem to be critical for this leukemogenic activity. Coexpression of the full-length molecule together with the AML1-ETO9a variant results in earlier onset of leukemia and blockade of myeloid differentiation at an earlier stage. Thus, the various splice variants seem to cooperate in leukemogenesis.
Another variant contains an alternative exon at the C-terminal end instead of NH4R/Mynd; this exon encodes 27 amino acids in-frame. The variant is expressed in primary human t(8;21) AML cells and the encoded protein seems to reduce repressor activity and tends to form multimeres [69].
ETO-exon 6a can also be incorporated, potentially giving rise to two different truncated transcripts predicted to encode proteins of 223 and 395 amino acids, respectively [70]. The shorter variant lacks all four NHR domains whereas the longer variant (referred to as AML1-ETO6a) retains the NHR-1 domain. The AML1-ETO6a variant alone is not leukemogenic in animal models but seems to modulate the activity of the full-length fusion protein.
The two variants A1bETO and A1cETO were identified in primary human AML cells and have additional sequences upstream to the first AML1 exon. Several in-frame variants of these forms have been identified, and their final effects on the expression of AML1 responsive genes seem to vary from repressive to activating [65].
To summarize, several molecular variants of the AML1-ETO fusion protein exist, and the final effect of this chromosomal translocation thus depends on the balance between these various isoforms.
3.3. The Fusion Protein Has Several Binding Partners
The AML1/ETO fusion molecule has several partner molecules [60, 71–78]; nine important partners are listed in the upper part of Table 3. Seven of these partner molecules are involved in the regulation of gene transcription; the molecular mechanisms behind these effects vary between the partners, but involve (i) altered histone deacetylation, for example, regulation of histone acetylation through recruitment of histone deacetylases or the E-protein interaction with the histone acetyltransferase; and (ii) altered DNA methylation through interaction with DNA methyltransferase. Several of these partner molecules are expected to increase viability, and proliferation and/or decrease differentiation, but their relative contribution to leukemogenesis seems to vary and for example the E-protein interaction seems to be less important [79]. The last two partner molecules mentioned in Table 3 include SON that shows a cytoplasmic localization and is also involved in regulation of proliferation and apoptosis, and protein kinase A (PKA) that based on the initial study does not seem to have any major role in leukemogenesis [78]. Finally, the biological functions and thereby the leukemic activity of the fusion protein can probably be modulated through two mechanisms as described in the lower part of Table 3, either through proteolytic cleavage by calpains or through alternative splicing.
Table 3.
Important proteins that directly interact with or modulate the t(8;21) fusion protein.
| Molecule | Function of the molecule | Structure: mechanism of interaction | Function/biological effect |
|---|---|---|---|
| Molecular interaction with the fusion molecule as a partner molecules | |||
| GFI1 [71] | Transcription repressor | GF35N and GF36S variant alleles have repressor activity. The fusion protein colocalizes and interacts with the more common GFI136S, and its repressor activity is thereby inhibited; the fusion protein does not colocalize or inhibit the GFI135N repressor activity | GFI1 is a regulator of myeloid differentiation, and the interaction and effect of the fusion protein vary between patients and depend on genetic differences |
| CBPβ [72] | Transcription repressor | Forms heterodimers with the t(8;21) fusion protein through binding to the Runt domain from AML1/RUNX1 | Animal models suggest that CBPβ is important for contribution to the fusion proteins inhibition of neutrophil differentiation, is essential for its growth-enhancing effect and cooperation with receptor-initiated signaling |
| E-proteins [60] | Transcription factor | DNA-bound E-proteins interact with the histone acetyl transferase p300/CREB binding protein, leading to histone acetylation and initiation of transcription. This effect is silenced by the fusion protein through (i) preventing E-protein/p300 activation and (ii) local recruitment of HDACs | E-proteins are important regulators of growth, differentiation and apoptosis and these functions are probably inhibited through a stable binding to the NHR4 domain of the fusion protein |
| SMRT/N-CoR complex [73] | Transcriptional regulators | Binding of the complex together with their associated HDACs to AML1 target genes through the fusion protein causes aberrant repression of transcription | Contributes to the differentiation block and attenuates the effect of the fusion protein on cell proliferation |
| UBF1 [74] | Transcription factor | UBF1 binds ribosomal DNA and regulates RNA polymerase 1 activity (see below); the fusion protein associates with UBF1 | Modulation of RNA polymerase 1-mediated ribosomal RNA transcription during interphase |
| SON [75] | Growth regulation | Binding to the NHR4 domain and is possibly involved in the antiproliferative signaling mediated by this domain | SON shows an abnormal cytoplasmic localization in t(8;21) cells; the functions are largely unknown but it seems to be involved in regulation of proliferation and apoptosis |
| Histone deacetylases [73] | Acetylation of histones | Direct recruitment of HDACs with silencing of AML1-target hematopoietic genes | There is physical binding between the fusion protein and HDAC1, the final functional effect being regarded as a leukemia-enhancing effect |
| DNA methyltransferase 1 [76] | DNA methylation | Silencing of gene expression through methylation, probably functionally linked to HDACs. | Contributes to the silencing of gene expression, and is involved in the reduced IL3 expression |
| Protein kinase A (PKA) [78] | Protein phosphorylation | Binding to the NHR-3 domain of the fusion protein | Even though PKA is important for regulation of cell proliferation, the interaction with AML1-ETO does not seem to have any major impact on proliferation or in vivo leukemogenesis |
| Modulation of the fusion molecule | |||
| Calpains [77] | Proteolytic cleavage | Calpain is required for the induction of blood disorders by the fusion protein in Drosophilia | Calpains cleave a restricted set of protein substrates; one hypothesis is that the enzyme cleaves the fusion protein, and thereby generates a more potent inducer of leukemia similar to the leukemic splice variant. Alternatively calpains may affect leukemic cell migration |
| Alternative AML1/ETO splicing [62] | One of the splice variants lacks the two carboxyterminal ETO domains | In contrast to the full-length variant this alternatively spliced molecule alone can induce leukemic transformation in experimental models without additional genetic abnormalities | |
Even though many biological effects of the fusion molecule seem to depend on altered expression of AML1-regulated genes [62–64], other mechanisms involving the ETO portion of the fusion protein may also be important. For example, ETO2(MTG16) has corepressor activity and binds to the transcriptional repressor N-CoR; this binding is inhibited by the fusion protein probably through occupation of the ETO2 binding site by the Mynd/NHR4 domain [80]. This effect seems to be important for the myeloid differentiation block. Another example is the oligomerization domain of ETO that is important for the AML1/ETO-mediated regulation of cell-cycle progression and apoptosis [81].
3.4. The Fusion Protein Affects a Wide Range of Biological Functions
The fusion molecule affects a wide range of cellular molecules, and the data summarized in Table 4 illustrates the complexity of the AML1/ETO effects [20, 62, 74, 76, 82–103]. Firstly, gene expression as well as ribosomal function are affected. Secondly, the fusion protein can reduce DNA repair [20]; this is combined with decreased expression of the p53 tumor suppressor, and the final effect is probably an increased risk of new leukemogenic events [104]. A recent study in Drosophila described that the fusion molecule induced increased levels of reactive oxygen species (ROS); these high ROS levels are important for the development of the AML1-ETO associated phenotype and may also contribute to leukemogenesis through an increased risk of developing additional genetic abnormalities [105]. Thirdly, the responses to hematopoietic growth factors are altered; this is due to altered cytokine release, receptor expression, and probably also downstream intracellular signaling. These events, together with altered cell-cycle regulation, will alter the proliferative capacity of the cells. Finally, the regulation of apoptosis is altered, and the cells show activation of stress responses. Most of these alterations are in favor of increased proliferation and survival and decreased differentiation (Tables 3 and 4), but the fusion protein also has opposite effects, and this may explain why the fusion protein alone cannot induce leukemic transformation (see above).
Table 4.
Important molecular mechanisms involved in AML1/ETO-induced leukemic transformation.
| Molecule | Function | Effect of AML1/ETO on the molecule | Final effect on t(8;21) cells |
|---|---|---|---|
| Altered gene transcription | |||
| PU.1 [82] | Transcription factor | Decreased expression by AML1/ETO | Inhibition of differentiation |
| C/EBPα [83, 84] | Transcription factor | Downregulation of C/EBPα; the normal function of this factor is regulation of differentiation and inhibition of proliferation | Increased proliferation and inhibition of differentiation |
| C/EBP β [85] | Transcription factor | The normal function is transcriptional upregulation of C/EBPα | Altered regulation of differentiation and proliferation through reduced expression of C/EBPα |
| POU4F1 [86] | Transcription factor | The POU4F1 levels are significantly correlated with the fusion protein levels. One study described differential regulation of 140 genes by this factor, and half of them are also AML1/ETO targets | POU4F1 probably contribute to the gene expression signature associated with t(8;21) AML |
| PAX5 [87, 88] | Transcription factor | Increased expression at the mRNA and protein level | Aberrant expression of B lymphocyte markers, including CD19, CD79a |
| RNA-rependent mechanisms and ribosomal functions | |||
| μRNA [62, 89] | Regulatory RNA molecules | The fusion protein selects a set of μRNAs (miR); it occupies the miR-24-23-27 locus and upregulates their expression | Modulation of proliferation and differentiation through the effects on miR24 |
| miR24 downregulates the mitogen-activated protein kinase phosphatase 7 and enhances the downstream signaling through phosphorylation of c-jun and p38 kinases | |||
| Silencing of miR223 through epigenetic mechanisms | Altered regulation of myelopoiesis through effects on mir223 | ||
| RNA polymerase I [74] | Transcriptional regulators | The fusion protein seems to localize to the nucleolar organizing regions during mitosis, associates with metaphase chromosomes and occupies ribosomal DNA repeats during interphase together with UBF1 (see Table 3) | In contrast to AML1 the fusion protein seems to be a positive regulator of rDNA transcription. Transcription regulated by RNA polymerase 1 seems to increase the proliferation of transformed cells (discussed in [74]) |
| DNA damage and repair | |||
| OGG1[20] | DNA repair | OGG1 is an important part of the DNA base excision repair pathway, its expression is downregulated by the fusion protein | High OGG1 levels are associated with an adverse prognosis; the downregulation may increase chemosensitivity |
| DNA damage [90] | Carcinogen-DNA adducts | Increased formation of aromatic hydrocarbon-DNA adducts and upregulation of the cytochrome P450 1A1 enzyme that metabolizes polycyclic aromatic hydrocarbons | This effect may contribute to an increased susceptibility to additional genetic damage |
| Increased intracellular ROS [105] | Altered signalling. DNA damage? | AML1-ETO causes increased intracellular levels of reactive oxygen species (ROS) in Drosophila | ROS are important for induction of the AML1-ETO associated phenotype and may also increase the risk of additional genetic abnormalities |
| Increased mutation frequency [91] | Predisposition to leukemia progression | Predisposed for additional genetic effects that are required for leukemogenesis. | |
| Cytokine-mediated growth regulation | |||
| IL3 [76] | Hematopoietic growth factor | Decreased gene expression | Decreased growth factor-dependent proliferation |
| M-CSF receptor [92] | Growth factor | M-CSF is a growth-enhancing hematopoietic growth factor | Increased cytokine-dependent AML cell proliferation |
| G-CSF receptor [93] | Growth factor | G-CSF is a growth-enhancing hematopoietic growth factor | Increased cytokine-dependent AML cell proliferation |
| BCL2 [94] | Antiapoptotic signaling | Upregulation by the AML1-ETO fusion protein | Increased antiapoptotic signaling |
| C/EBPε [92, 93] | Transcriptional regulator | Induction of G-CSF receptor expression; upregulation of the myeloid-specific promoter for the M-CSF receptor | Increased growth factor receptor expression and thereby increased cytokine-dependent proliferation by t(8;21) cells |
| NF1 [95] | Tumor suppressor | Decreased expression of the Neurofibromatosis1 (NF1) tumor suppressor | Decreased protein levels are associated with increased response of primary AML cells to GM-CSF |
| Tyrosine receptor kinase A [96] | A part of the nerve growth factor receptor (NGF) | Upregulation of this growth factor both at the mRNA and protein level | NGF is released by bone marrow stromal cells; in addition, AML1-ETO expressing cells show increased proliferation in response to growth factors |
| Cell-cycle regulation | |||
| p21 [97] | Negative cell-cycle regulator | Increased mRNA and protein levels of p21 | p21 is a cell-cycle inhibitor, this effect may contribute to the absence of leukemogenesis in the presence of t(8;21) alone |
| p27kip [98] | Negative cell-cycle regulator | Increased expression caused by either a direct effect of the fusion protein or by Cx43 | Cell-cycle inhibition |
| SSX21P [99] | Cell-cycle regulation? | Low expression of this molecule is associated with low expression of CDC20; possibly causing attenuation of the spindle checkpoint | Altered cell-cycle regulation, increased risk of aneuploidy? |
| Disrupted spindle checkpoint [100] | Aneuploidy | Disruption of the spindle checkpoint during cell-cycle progression | Increased risk of aneuploidy. |
| Regulation of apoptosis and stress responses | |||
| Annexin A1 [101] | Proapoptotic, antiproliferative | Downregulated at the gene expression level by the fusion protein | The molecule has proapoptotic and antiproliferative effects; these functions are thus inhibited |
| Connexin 43 (Cx43) [98, 102] | Gap junction component | Increased expression of Connexin 43 in cells with t(8;21), possibly both a direct and an indirect effect mediated via c-Jun | Cx43 often inhibits cell proliferation both through gap junction dependent and independent mechanisms; this effect may contribute to the lack of leukemogenesis by the full-length fusion protein |
| p53 [91] | Tumor suppressor | Activation of the p53 pathway | Possibly increased chemosensitivity and thereby contribution to the good prognosis of these patients |
| TXNIP [103] | Part of stress responses | Involved in reactive oxygen stress responses, AML cells with t(8;21) have increased protein levels of this molecule. The mechanism is not known | High levels inhibit the proliferation of myeloid progenitor cells; this may contribute to the good prognosis of these patients |
Two studies have shown that the fusion protein upregulates Connexin 43 (Cx43) (Table 4). Cx43 forms gap junctions and is thereby involved in communication between cells; it has a role both in normal and leukemic hematopoiesis and may also function as an intracellular signaling molecule independent of its role in gap junction formation [106, 107]. Thus, the effect on Cx43 may represent an additional leukemogenic mechanism in t(8;21) AML.
The fusion protein seems to preferentially bind those AML1 target genes with duplicated binding sites in the regulatory elements, and this selectivity may be an important mechanism for dysregulated gene expression and leukemogenesis [108]. Another selective regulatory mechanism is possibly the epigenetic structure at the fusion protein's target sites; a recent study described that the most downregulated genes were characterized by aberrant repressive histone tail changes both at the AML1 consensus as well as the transcription start site [109].
Intracellular signaling is altered in t(8;21) AML. WT1-mediated signaling may contribute to leukemogenesis, and these patients seem to have increased WT1 levels [110]. Proteomic studies suggest that the t(8;21) abnormality also interacts with the p53 network [111]. The survival and growth of t(8;21) AML cells depend on autocrine loops between vascular endothelial growth factor (VEGF) and its receptors (VEGFR) that activate various downstream pathways like PI-3 kinase, Akt, or MEK cascades [112, 113]. Both VEGFR1 and VEGFR2 monoclonal antibodies suppressed the growth of primary AML cells with t(8;21), an effect mainly mediated through reduced phosphorylation of Akt and MEK [114]. In addition, inhibition of VEGFR2 potentiated the growth inhibitory effect of idarubicin for the t(8;21) Kasumi-1 cell line though the mechanisms of this effect are not fully elucidated yet [114]. Another study suggests that VEGF receptor type-2-mediated signaling stimulates proliferation of t(8;21) AML cells, an effect mediated through increased phosphorylation of Akt, and inhibition of this signaling seems to potentiate the effect of cytarabine [115].
The global gene expression profiles of U937 AML cells when expressing different AML-associated fusion proteins were compared in a recent study [116]. The analysis revealed a role of AML1-ETO in downstream pathways known to regulate DNA repair and stem-cell maintenance, including activation of the Notch signaling pathway through Jagged-1 ligand [116, 117]. On the other hand, the effect of AML1-ETO depletion has been investigated by electroporating the t(8;21) carrying Kasumi-1 cell line with specific siRNA [118]. This depletion severely diminished the cell clonogenicity, inhibited G1-S transition, reduced apoptosis and induced senescence. The presence of exogenous G-CSF or GM-CSF could not rescue these cells from senescence but partly counteracted the antiproliferative effect.
The effects of AML1-ETO on leukemic stem-cells have not been investigated in detail. However, the effect of AML1 alone on normal hematopoietic stem-cell homeostasis was investigated in a recent study [119], and an enrichment of quiescent stem-cells was observed in AML1-deficient bone marrow. These results suggest a negative regulatory effect of AML1 on normal stem-cells. Thus, the AML1-ETO fusion molecule may then have an opposite effect of AML1 on leukemic stem-cells and cause an enrichment of quiescent AML stem-cells similar to the AML1 deficiency through its suppression of AML1 target genes (see Section 3.1).
4. Alternative Methods for Detection of t(8;21) in Human AML
Cytogenetic analysis by G-banding is highly recommended at the time of diagnosis of all AML patients [120]. Although this time-consuming method requires the leukemic cells to be captured in mitosis, it is still the best screening method for detection of leukemia-specific chromosomal aberrations. t(8;21) is readily detectable by G-banding (Figure 2), but it can also occur due to variant rearrangements (complex translocations, inversion, and insertion) that may be overlooked if additional fusion-specific analyses like fluorescence in situ hybridization (FISH) or reverse transcriptase polymerase chain reaction (RT-PCR) are not used (Figure 2) [121, 122]. Detection of the fusion protein is less common; the protein can then be detected by Western blotting, but a recently described method using immunobeads for flow cytometric detection seems promising and may represent a future alternative [123].
FISH analysis for AML1-ETO fusion should be performed using locus-specific probes. The advantage with this method is that there is no need for cells in metaphase, and it can, therefore, be performed also in bone marrow biopsies or bone marrow/blood smears. Commercially available dual color probes against AML1 and ETO resulting in fusion-signals are most commonly used (Figure 2). Particular care has to be taken when interpreting the signaling patterns from interphase analyses, and dual-color double-fusion probes should be used in order to reduce the false-positive rate. Although rare, one has to be aware that variant AML1-ETO rearrangements can give only one fusion signal (Figure 2). Probes covering the whole AML1 gene will also detect other AML1 rearrangements, and instead of fusion signals, the AML1 rearrangements will then be seen as two weaker signals that can be misinterpreted as trisomy 21 in interphase analyses. The chromosomal band 21q22 where the AML1 gene is situated, has been reported involved in 55 different rearrangements, and several of these can be overlooked by G-banding, and the majority of these abnormalities have been found in AML/MDS [124]. If only interphases are available, for analyses, the methodological alternative is to verify AML1 rearrangements by using AML1 dual color split-signal probes (Figure 2(c)). Thus, due to the possibility of other less common AML1 rearrangements, the detection of t(8;21) may require a careful interpretation of additional molecular analyses before a final conclusion can be reached.
RT-PCR is a rapid and sensitive method for detection of the fusion transcript. Although simple to perform, the method is not without challenges. RNA is readily degraded, and tests for RNA quality have to be performed [125]. RT-PCRs are capable to detect one leukemic cell in 105-106 normal cells [126], and this demands precautions to avoid cross-contamination leading to false positive results. Furthermore, the presence of leukemia-specific fusion transcripts has been detected in tissue from healthy donors when using nested PCR. Single round PCR should, therefore, be sufficient at diagnosis [127]. False negative results can occur if the assay is not designed to detect all different known fusion and transcript variants. As described above (Section 3.2), several AML1-ETO variants have been described due to alternative promoters and splicing, and different variants can occur in the same patient. However, most variants include exon 3–5 in AML1 and exon 2–4 in ETO [17]. These regions can thus be used for design of the primers; the problem of false-negative results can thereby be reduced, and standardized protocols for RT-PCR have now been published [128–131]. RT-PCR should thus be regarded as a rapid and sensitive methodology, and it can even be used for simultaneous screening for several prognostically important fusion transcripts [132].
5. The Biological and Clinical Characteristics of Human t(8;21) AML
5.1. The Diagnostic Criteria for AML in Patients with t(8;21)
The general diagnostic criterium for AML is detection of at least 20% blasts in the bone marrow. However, t(8;21) AML shows morphological signs of neutrophil maturation. Rare cases can, therefore, be seen with bone marrow blast counts below 20%; according to the WHO classification, such cases should also be classified and treated as AML and not as myelodysplastic syndrome [3].
5.2. Morphological and Immunophenotypic Characteristics of t(8;21) AML
The morphology of the leukemic cells has been described by Arber et al. [3], the most common features being relatively large blasts with basophilic cytoplasm, often numerous azurophilic granules and a perinuclear clearing. Auer rods are common and may be detected in blasts or immature neutrophils. The cells show maturing to promyelocytes and myelocytes, and mature neutrophils, possibly with morphological signs of dysplasia, are also present in the marrow. Among the dysplastic signs are abnormal nuclear segmentation (e.g., pseudo-Pelger-Huet anomaly) and cytoplasmic staining abnormalities like pink staining in mature neutrophils. Eosinophilic precursors are often increased; basophils and/or mast cells are also increased sometimes. The erythroid cells and megakaryocytes are morphologically normal. The abnormal differentiation of the leukemic cells may even cause a morphological picture similar to chronic myeloproliferative neoplasias. Lee et al. [133] described a patient with variant t(8;21) as a complex t(8;10;21) (q22;q24;q22) abnormality; this patient presented with morphological findings in blood and bone marrow mimicking atypical chronic myeloid leukemia.
The blasts express myeloperoxidase and are typically CD13+CD34+HLA-DR+ [134]. There are also immunophenotypic signs of granulocyte maturation with subpopulations expressing CD15 or CD65, eventually as a part of asynchronous maturation with concomitant expression of CD34 [134, 135]. Aberrant expression of the lymphoid markers CD19, PAX5, and eventually cytoplasmic CD79a is common [87]. CD56 can also be expressed especially for patients with KIT mutations [134, 135], whereas CD19 expression is uncommon for patients with this mutation [136].
5.3. Extramedullary Manifestations of AML
Myeloid sarcoma is a rare condition that can involve almost any site of the body, but especially lymph nodes and skin (for references, see [137]). Other common sites are head and neck soft and subcutaneous tissue and the orbits, whereas intrathoracic manifestation is uncommon [138, 139]. Such tumors can be the first manifestation and precede the primary bone marrow manifestation by several months, or it can represent the first manifestation of a relapse [137–141]. Myeloid sarcoma has been reported in 15% of patients with t(8;21) AML [27]. However, t(8;21) is a relatively rare cytogenetic abnormality in myeloid sarcomas; a recent cytogenetic study of 74 patients with such sarcomas reported that the t(8;21) abnormality was detected only in 2%-3% of these cases [137]. The sarcomas in patients with t(8;21) can be located to uncommon regions, including (i) intracerebral tumors [142] or intraspinal sarcoma with spinal cord compression [143] (ii) abdominal affection either as ovarial infiltration with ascites [144] or compression of nerves or nerve plexuses (e.g., presacral tumors) with neurological symptoms [145]; (iii) skeletal or heart muscle affection [146]; or (iv) pulmonary involvement with initial symptoms resembling nodular or interstitial pneumonia [133, 139]. Even though these lesions may give serious local symptoms at the time of diagnosis, at least in children detection of granulocytic sarcoma in t(8;21) AML does not seem to have a major impact on the long-term survival [147].
5.4. Myelomastocytosis and Myelomastocytic Leukemia
Systemic mastocytosis and AML, including the t(8;21) variant of AML, can show similarities. Firstly, activating KIT mutations are common both in t(8;21) AML and in systemic mastocytosis [1]. Secondly, systemic mastocytosis can occur in combination with other hematological malignancies, usually myeloid malignancies (AML, myelodysplastic syndrome or chronic myeloproliferative neoplasms) and in the WHO classification, this is termed systemic mastocytosis with associated hematological nonmast cell disease [1]. Thirdly, even though the combination of mastocytosis and t(8;21) AML seems rare [148], the t(8;21) AML variant can show phenotypic similarities with mastocytosis and has increased serum tryptase levels (see below). The KIT mutations are detected both in AML and mast cells in those rare cases when mastocytosis is associated with AML; this observation demonstrate that both cell types then are derived from the same clone [149–151].
Due to the low number of published cases, it is not possible to give general guidelines about the treatment of the myelomastocytic leukemia variants. The bone marrow mast cells seem to become more prominent after intensive AML therapy, but this may simply be due to mast cell chemoresistance as evidenced by their persistence after chemotherapy. Some patients have been treated with allogeneic stem-cell transplantation, and the very limited experience with this therapeutic strategy suggests that the type of conditioning therapy may be important for eradication of both AML and mast cells [149, 150].
A recent study investigated serum tryptase levels in AML [152]. Increased levels were detected in nearly half of AML patients and were associated with t(8;21) and KIT mutations. The levels decreased after chemotherapy [150, 152], suggesting that serum tryptase can be used as a marker of treatment response in these patients. Thus, altered tryptase levels seem relatively common in t(8;21) AML, but morphological myelomastocytosis is rare. One possible explanation for the increased levels could be tryptase release by the AML cells as an ectopic phenotypic characteristic. Alternatively, the levels may reflect an activation/stimulation of tryptase release by normal mast cells. The association with KIT mutations suggests that AML cell release is most likely.
5.5. Differences between Core-Binding Factor AML with t(8;21) and inv(16)/t(16;16)
Even though t(8;21) and inv(16)/t(16;16) are both regarded as core-binding factor AMLs with good prognosis, the two forms show several differences in their pretreatment features. This has been reviewed in detail by Mrózek and Bloomfield [43]. Firstly, t(8;21) AML is more frequent in African than white Americans, it has lower white blood cell counts and lower percentages of blasts in the bone marrow. Secondly, extramedullary disease is less frequent in t(8;21), especially lymphadenopathy, splenomegaly, gingival hypertrophy, and skin/mucosa involvement. Thirdly, the frequency and patterns of secondary cytogenetic abnormalities differ. A secondary chromosomal abnormality is detected in 70% of patients with t(8;21) but only in one-third of inv(16) patients. As described above (Section 2.2), the most frequent secondary abnormalities in t(8;21) is loss of sex chromosomes and deletion of 9q, whereas in inv(16), the most common are +22, +8, del7q, and +21. For mutations detected by molecular genetic methods, both KIT and Ras mutations occur, but they are more frequent in inv(16), than in t(8;21) leukemia [33]. These pretreatment differences suggest that the molecular mechanisms in the leukemogenesis differ between these two groups.
Mrózek and Bloomfield [43] also discussed possible prognostic differences between t(8;21) and inv(16)/t(16;16) AML. Observations from several studies suggest that relapse of t(8;21) is less responsive to salvage treatment, and these patients, therefore, have a lower overall survival. This difference may be caused by the additional genetic abnormalities; the KIT mutations associated with t(8;21) AML seem to have adverse prognostic effects whereas +22 in inv(16) has been associated with lower relapse risk in some studies. Finally, there is possibly also an influence of genetic factors/race on the response to chemotherapy in t(8;21) patients.
6. Animal Models of t(8;21) AML
Several animal models of t(8;21) AML have been developed, including transgenic and knock-in models, conditional knock-in, as well as chimeric models [153]. The models are important for our current understanding of the role of the AML1-ETO fusion protein in leukemogenesis (for detailed discussion and references see [153]). AML1 knock-out causes embryonic death due to bleeding complications, and knock-in models of the AML1-ETO fusion gene resulted in a similar embryonic phenotype, suggesting that repression of AML1-regulated genes are important in the fusion model. Furthermore, one of the transgenic models directed the fusion gene into the stem-cell compartment and its expression was then significantly lower in lymphoid cells than in myeloid progenitors. This observation underlines the association between AML1-ETO expression and myelopoiesis. Furthermore, transgenic mice that express the AML1-ETO fusion molecule specifically in myeloid cells appeared to be healthy and developed AML only if they in addition were exposed to a mutagen [154]. These observations are consistent with the hypothesis that the AML1-ETO fusion protein alone is not sufficient for development of AML; additional abnormalities have to be present [154]. Similar observations have been made in other conditional knock-in models and models based on retroviral transduction to the stem-cell compartment; the fusion protein alone often results only in increased proliferative capacity and altered regulation of differentiation. In one of these abnormalities an increase in immature eosinophils similar to the human t(8;21) AML was observed (discussed in detail in [153]).
Other chimeric models based on retroviral transduction have also been published (reviewed in [153]). Transfection of the AML1-domain alone from the fusion protein had no effect on hematopoiesis, showing that the ETO-derived part of the molecule is required for development of the hematopoietic abnormalities. Furthermore, the AML1-ETO molecule has also been transfected to mice with other abnormalities, including (i) deficiency of the myelosuppressive Interferon regulatory factor (IRF), (ii) mutations in receptor tyrosine kinases such as TEL/PDGFβR and Flt3; and (iii) downregulation of the p21 cell-cycle inhibitor. All these combinations resulted in development of AML. Thus, the additional abnormalities cooperated with AML1-ETO in leukemogenesis.
7. The Global Gene Expression Profile, microRNA (MIR) Expression, and Epigenomic Profile in Human t(8;21) AML
Global gene expression profiling in AML has revealed that major prognostic subgroups based on genetic markers are recapitulated in large-scale gene expression patterns [4, 155, 156]. These global profiles have identified specific signatures for patients with the t(8;21) abnormality [4, 155, 156], and its prediction can be made with almost 100% specificity and sensitivity [157]. There is an overlap between the expression profiles for the t(8;21) and inv(16) abnormalities [119], but approximately one third of the transcripts are specific for the t(8;21)-associated profile [119]. RUNX1T1/ETO itself has been identified as the most discriminative gene for the t(8;21) cluster [4, 155], but several other genes are also frequently up- or downregulated. Of special interest is probably the transcription factor POU4F1 that is important for embryonic brain development but without any known role in normal or leukemic hematopoiesis. This gene is frequently upregulated in t(8;21) AML [156]; this upregulation is probably not directly caused by the AML1/ETO fusion protein [86] but the unique transcription profile of t(8;21) AML is probably largely attributed to POU4F1 [86]. Finally, AML1-ETO downregulates genes involved in multiple DNA repair pathways, a possible explanation for the increased in vitro DNA damage and p53 activation in these cells [91, 104]. Based on the results summarized in Table 4 we conclude that the t(8;21) fusion protein alters the expression levels of a wide range of molecules, and thereby affects the regulation of several intracellular processes; this conclusion is also supported by the t(8;21)-associated microarray profile (e.g., altered transcriptional regulation and DNA repair).
Lück and coworkers identified two distinct gene expression signatures among t(8;21) and inv(16) AML based on the presence of KIT mutations [158]. The KIT mutated cases were then characterized by deregulation of genes belonging to the NFκB signaling pathway [159]. On the other hand, Bullinger and coworkers classified t(8;21) and inv(16) AMLs in favorable and unfavorable prognostic subsets based on supervised analysis of gene expression, the two distinct groups being characterized by altered expression of genes involved in the MAP-kinase and the mTOR pathways, respectively [160]. All these pathways are suggested to play a role in leukemogenesis and are considered as potential therapeutic targets [161, 162].
A major part of the published clinical studies do not find any significant prognostic impact of KIT mutations (Section 2.2). The observations from the studies of global gene expression profiles described above and the different effects on intracellular signaling pathways for KIT mutations (the NFκB pathway is affected) compared with chemosensitivity (MAP kinase)/chemoresistance (mTOR) are consistent with the hypothesis that KIT mutations do not have any major prognostic impact. This may be true at least for the chemotherapy regimen used in these German/Austrian studies [158, 160]. A possible explanation for the different prognostic impact between various studies may be that the prognostic impact depends on differences between the chemotherapy regimen.
The global gene expression analyses have identified additional genes than those summarized in Table 4, whose altered expression may be clinically relevant [4, 155, 156]. Firstly, PRAME (preferentially expressed antigen of melanoma) is upregulated, and this AML-associated antigen is now used in vaccination trials and may also be a candidate marker for detection of minimal residual disease (MRD) [163, 164]. Secondly, the structural membrane protein CAV1 is also upregulated, and this molecule is possibly important for chemosensitivity [165]. Among the down-regulated genes are the cystein protease CTSW cathepsin W, the cancer-associated actin-bundling protein LCP1 (lymphocyte cytosolic protein 1), the actin-regulatory protein CAPG (capping protein, gelsolin-like) and the semaphorin receptor PLXNB2 (plexin B2). These genes seem to be involved in cancer cell migration/invasion, cancer-associated angiogenesis or tumor progression [166–169].
It has been demonstrated that patient-derived AML cells express a specific signature of microRNA, a class of small noncoding RNAs involved in regulation of protein coding mRNA [170, 171]. Most microRNAs seem to be downregulated in t(8;21) AML except the miR126/126∗ that is upregulated [170]. miR126/126∗ can inhibit apoptosis, increase cell viability, and enhance colony formation possibly by interacting with AML1-ETO itself [171]. Finally, studies in the t(8;21) positive Kasumi-1 cell line suggest that the microRNA signature seems to be different in the CD34+CD38− AML cell subset which is believed to harbor a major part of leukemic stem-cells [172].
DNA methylation and histone modifications are important epigenetic mechanisms of gene regulation [173]. These mechanisms seem to play essential roles both independently and cooperatively in malign transformation and progression [173]. This is also true for AML, where different cytogenetic subgroups, including t(8;21), seem to be characterized by distinct epigenetic modifications [174–176]. However, the clustering seems to be less pronounced based on methylation data compared to gene expression data [174]. The t(8;21) DNA methylation cluster harbors patients not having the AML1-ETO fusion gene, even though they seem to have a similar prognosis and also share other features with t(8;21) patients [175]. Furthermore, human hematopoietic stem-cells transduced with the AML1/ETO fusion gene failed to reproduce the epigenetic signature [176], supporting the theory that other mutations are needed to create the fully malign phenotype [154]. In contrast to data from methylation studies, modification of histone H3 Lysine 9 methylation was recently demonstrated to show only minor differences between different cytogenetic groups [177].
8. The Prognostic Impact of t(8;21) in Human AML
8.1. Patients Receiving Conventional Intensive Chemotherapy
t(8;21) is usually associated with a relatively low risk of relapse. A recent MRC report analyzed the survival data for 5876 AML patients (median age 44 years) including 421 patients with t(8;21) [7]. The long-term disease-free survival for this subset was 61%, and a similar high survival has also been observed in other studies [178, 179]. No difference in overall survival was then observed when comparing patients with t(8;21) alone versus patients with additional cytogenetic abnormalities the only possible exception being loss of the Y chromosome that was associated with an improved survival of borderline significance in the MRC study. A French study investigated elderly patients (median age 67 years) with CBF-AML, including 60 patients with t(8;21) [27]. These patients received induction treatment with an anthracycline combined with cytarabine; 80% of the patients achieved complete remission after one induction cycle and 88% after two cycles. However, despite this high remission rate, the median 5-year overall survival for these patients was only 31%. A high white blood cell count (WBC) at diagnosis, poor performance status, and del(9q) were all associated with an adverse prognosis, whereas administration of intensive consolidation treatment was associated with better survival. High WBC is in general considered as an adverse prognostic factor in t(8;21); this is mainly based on clinical studies indicating an unfavorable outcome for patients with high WBC alone [27] or high WBC plus a high percentage of bone marrow blasts [180]. AML t(8;21) with high WBC (usually defined as >20 × 109/L) is often considered to have a less favorable prognosis similar to the intermediate prognostic group in clinical studies [181] (e.g., the HOVON 102 AML/SAKK 30/09 study; EudraCT number 2009-011613-24). Finally, it should also be mentioned that the adverse prognostic impact of KIT mutations that is observed in certain studies may be explained by an association between KIT mutations and high peripheral blood blast counts [37].
A recent study described an adverse prognostic impact of high bone marrow cellularity in patients with t(8;21) [182]. The authors found that the bone marrow cellularity was the single most important prognostic parameter in these patients, and they classified their patients in three groups with (i) neither leukocytosis nor increased bone marrow cellularity (ii) only leukocytosis (cutoff 9.1 × 109/L),and (iii) patients with increased marrow cellularity with or without leukocytosis. The survival after chemotherapy was lowest (32% overall 5-years survival) for the last group. These results strongly suggest that bone marrow cellularity should be further evaluated as a possible prognostic parameter in these patients.
t(8;21) is also regarded as a good prognostic marker in pediatric AML. The complete remission rate close to 100%, eventfree survival exceeding 69% and overall survival exceeding 80% have recently been reported for children included in the MRC-AML10, MRC-AML12 and AML-BFM 98 studies and with no effect of additional chromosomal abnormalities on prognosis [183, 184]. Other pediatric protocols have reported eventfree survival below 50% [15, 185, 186], and the reasons for this discrepancy remain to be identified. However, children with relapsed t(8;21) AML seem to have a chance to be cured by salvage therapy [183, 184].
8.2. Experiences with Allogeneic Stem-Cell Transplantation in t(8;21) AML
Two large studies each including more than 300 patients have investigated the outcome after allogeneic stem-cell transplantation for AML patients with inv(16) and t(8;21) [187, 188]. Kuwatsuka et al. described no difference between patients in first remission receiving allogeneic and autologous stem-cell transplantation, and a long-term disease-free survival of 15%–20% was seen even for 85 patients undergoing allotransplantation not in complete remission [187]. As for patients receiving conventional therapy, additional cytogenetic abnormalities did not have any prognostic impact for these patients either. Similarly, Gorin et al. observed a comparable survival for auto- and allotransplanted patients of approximately 60%, and additional cytogenetic abnormalities had no significant prognostic impact, but high white blood cell counts had an adverse impact [188].
The smaller study by Schlenk et al. showed no advantage of allotransplantation compared with conventional intensive chemotherapy [189], thus supporting the two studies described above. In contrast, Shin et al. observed that the survival for allotransplanted patients had improved during the last years and was significantly better than for patients receiving high-dose cytarabine in a retrospective multicenter study based on distribution of questionnaires to each participating center [190]. In our opinion, most of the currently available data suggest that allogeneic stem-cell transplantation usually should not be recommended for t(8;21) patients in first complete remission. However, the presence of additional KIT mutations, high WBC counts and secondary leukemia have been associated with an adverse prognosis (see Sections 2.2, 2.3, and 8.1), suggesting that allotransplantation should be considered even in first complete remission for the younger subsets of these patients at least when an optimal donor is available.
The possible prognostic impact of KIT mutations in combination with t(8;21) was discussed in detail in Section 2.2. This question has been addressed in several clinical studies, and both adverse prognosis and no prognostic impact have been observed [30–36, 38–40, 47]. It was recently recommended that core-binding factor AML with KIT mutations should be classified as intermediate with regard to prognosis [191]. However, it is in our opinion, difficult to recommend allogeneic transplantation for these patients with its risk of early transplant-related mortality as long as the results are conflicting and several studies show no prognostic impact of the KIT mutations after conventional chemotherapy.
8.3. Disease-Stabilizing Therapy in t(8;21) AML
The combination of all-trans retinoic acid (ATRA) together with valproic acid or another histone deacetylase inhibitor is now tried for disease-stabilization in human AML [192, 193]. Most of the patients included in these studies have not been suitable for intensive chemotherapy; many of them had relapsed disease, and only a small minority had t(8;21). Experiments in the t(8;21) positive Kasumi-1 cell line have demonstrated that the AML1/ETO fusion protein recruits an HDAC-containing repressor complex to the promoters of AML1 target genes [73]. Valproic acid and probably also other HDAC inhibitors cause a dissociation of the fusion molecule from the HDACs. Histone acetylation thereby leads to transcriptional reactivation and increased proapoptotic signaling [73, 101].
Only a few clinical studies have investigated the effect of ATRA in t(8;21) AML, and the results are conflicting. Treatment with ATRA alone for 40 days induced complete hematological remission in one patient [194], whereas others have reported that coexpression of the t(15;17), and the t(8;21) encoded fusion proteins can be associated with ATRA resistance [195].
9. Detection of Minimal Residual Disease in t(8;21) AML
9.1. Methodological Strategies for Detection of MRD
Although detection of the AML1-ETO fusion transcripts represent a favorable prognostic marker, up to 30% of patients will experience a relapse of the disease [7, 178, 179]. MRD means remaining leukemia cells in a patient judged to be in complete hematological remission according to conventional morphological criteria. AML1-ETO-transcripts can persist in patients with t(8;21) even after stem-cell transplantation with GVHD and in long-time CR [196–200], but an increase in the fusion transcript expression seems to be predictive of relapse [201].
RT-PCR is a sensitive approach which identifies leukemic cells in 105-106 normal cells. By using quantitative RT-PCR it is easy to follow the patient through the course of the disease [202]. Leroy et al. assessed the prognostic value of real-time quantitative PCR in 21 AML1-ETO patients treated by the same protocol and who all achieved CR. Blood and bone marrow were collected at diagnosis, at CR, after intensive consolidation therapy and every 3–6 months thereafter; the median followup time being 15 months. The relapse rate was higher in patients with high pretreatment fusion transcript expression, and the absence of recurrent disease correlated with posttherapeutic absolute transcript levels below 10−3 compared to the Kasumi-1 cell line, or more than 3 log decrease of transcript levels compared to the levels at the time of diagnosis [202].
In a study by Ommen et al. various chromosomal aberrations appeared to have different relapse kinetics and therefore optimal sampling intervals might differ; the best MRD sampling interval for AML1-ETO seems to be every fourth month [203]. However, clinical utility of monitoring MRD with such high sensitivity is still under investigation and standardization of sampling procedures, handling and shipment of samples as well as the PCR analyses are required. Standardization of the assays is also necessary to allow comparison of results in different studies and for setting a threshold for AML1-ETO-transcript expression that defines molecular relapse [120, 204].
Multiparameter flow cytometry is less sensitive, but can be applied in the evaluation of MRD in most AML-cases and provides additional information about remaining cells (reviewed in [204]). The method is rapid and detects the presence of 10−4 leukemic cells, but it is not as specific as PCR due to the possibility of phenotypic shifts in relapsed disease. Tandem analysis with RT-PCR and flow cytometry can improve MRD detection [205]. Interphase FISH may also have a potential as an adjunct analysis to cytomorphology, cytogenetics, or multiparameter flow cytometry in the identification of MRD, since strong agreement between these methods has been described in large cohorts [206–208].
9.2. Clinical Consequences of MRD Detection
Benefit from preemptive treatment has been shown in PML-RARA acute promyelocytic leukemias [209, 210], but for other acute myeloid leukemia variants, few investigators have taken clinical action on detection of molecular relapse. For clinical utility, it has been shown that kinetics of the AML1-ETO decline correlate to relapse rate and outcome [202, 205, 211]. Monitoring the different alternative splice variants of the fusion transcript has also shown that persistence of the exon9a variant is indicative for later relapse [131].
We previously described that the translocation can be detected on Guthrie cards and may thus occur in utero (Section 2.1), the fusion protein alone is not sufficient for leukemogenesis (Section 6) and (iii) the translocation can persist for years after allotransplantation without any signs of leukemia relapse (Section 9.1). Taken together, these observations further support the hypothesis that detection of the fusion transcript is not sufficient for evaluation of the relapse risk. One should rather use serial determinations; the detection of increasing AML-ETO transcript levels or eventually the kinetics of the increase may be more reliable to evaluate the risk of an imminent relapse.
10. Concluding Remarks
The t(8;21) variant of human AML is a heterogeneous subset characterized by a common disease-specific molecular translocation. This variant is often referred to as core-binding factor AML together with the inv(16)/t(16;16) variants, but these two cytogenetically identified AML subsets show several biological and clinical differences. In contrast to other AML patients the diagnosis of t(8;21) AML can be made even when less than 20% leukemic blasts are present in the bone marrow. The disease is characterized by (i) ectopic expression of B-cell associated molecules; (ii) additional genetic abnormalities are common; (iii) the leukemic cells show specific global gene expression and microRNA profiles; and (iv) usually there is a low risk of leukemia relapse after high-dose cytarabine therapy. Despite the common fundamental cytogenetic characteristics, it should always be remembered that this specific cytogenetic abnormality identifies a heterogeneous subset of patients.
Conflict of Interests
The authors report no potential conflict of interest.
Acknowledgment
The authors received financial support for their scientific work from the Norwegian Cancer Society and the Helse-Vest Foundation.
References
- 1.WHO. World Health Organization Classification of Tumors of Haematopoietic and Lymphoid Tissues. 4th edition. Lyon, France: International Agency for Cancer; 2008. [Google Scholar]
- 2.Rowley JD. Identification of a translocation with quinacrine fluorescence in a patient with acute leukemia. Annales de Genetique. 1973;16(2):109–112. [PubMed] [Google Scholar]
- 3.Arber DA, Stein AS, Carter NH, Ikle D, Forman SJ, Slovak ML. Prognostic impact of acute myeloid leukemia classification: importance of detection of recurring cytogenetic abnormalities and multilineage dysplasia on survival. American Journal of Clinical Pathology. 2003;119(5):672–680. doi: 10.1309/EM7K-CQR4-GLMH-RCX4. [DOI] [PubMed] [Google Scholar]
- 4.Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. The New England Journal of Medicine. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y, Wang Y, Wang H, et al. Cytogenetic profile of de novo acute myeloid leukemia: a study based on 1432 patients in a single institution of China. Leukemia. 2009;23(10):1801–1806. doi: 10.1038/leu.2009.107. [DOI] [PubMed] [Google Scholar]
- 6.Sanderson RN, Johnson PRE, Moorman AV, et al. Population-based demographic study of karyotypes in 1709 patients with adult acute myeloid leukemia. Leukemia. 2006;20(3):444–450. doi: 10.1038/sj.leu.2404055. [DOI] [PubMed] [Google Scholar]
- 7.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 8.Klaus M, Haferlach T, Schnittger S, Kern W, Hiddemann W, Schoch C. Cytogenetic profile in de novo acute myeloid leukemia with FAB subtypes M0, M1, and M2: a study based on 652 cases analyzed with morphology, cytogenetics, and fluorescence in situ hybridization. Cancer Genetics and Cytogenetics. 2004;155(1):47–56. doi: 10.1016/j.cancergencyto.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Bacher U, Kern W, Schnittger S, Hiddemann W, Haferlach T, Schoch C. Population-based age-specific incidences of cytogenetic subgroups of acute myeloid leukemia. Haematologica. 2005;90(11):1502–1510. [PubMed] [Google Scholar]
- 10.Bruserud Ø, Hovland R, Wergeland L, Huang TS, Gjertsen BT. Flt3-mediated signaling in human acute myelogenous leukemia (AML) blasts: a functional characterization of the effects of Flt3-ligand in AML cell populations with and without genetic Flt3 abnormalities. Haematologica. 2003;88(4):416–428. [PubMed] [Google Scholar]
- 11.Mrózek K, Marcucci G, Paschka P, Bloomfield CD. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Current Opinion in Oncology. 2008;20(6):711–718. doi: 10.1097/CCO.0b013e32831369df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubnitz JE, Raimondi SC, Halbert AR, et al. Characteristics and outcome of t(8;21)-positive childhood acute myeloid leukemia: a single institution’s experience. Leukemia. 2002;16(10):2072–2077. doi: 10.1038/sj.leu.2402633. [DOI] [PubMed] [Google Scholar]
- 13.Raimondi SC, Chang MN, Ravindranath Y, et al. Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a Cooperative Pediatric Oncology Group study-POG 8821. Blood. 1999;94(11):3707–3716. [PubMed] [Google Scholar]
- 14.Pession A, Rondelli R, Basso G, et al. Treatment and long-term results in children with acute myeloid leukaemia treated according to the AIEOP AML protocols. Leukemia. 2005;19(12):2043–2053. doi: 10.1038/sj.leu.2403869. [DOI] [PubMed] [Google Scholar]
- 15.Perel Y, Auvrignon A, Leblanc T, et al. Treatment of childhood acute myeloblastic leukemia: dose intensification improves outcome and maintenance therapy is of no benefit—multicenter studies of the French LAME (Leucémie Aiguë Myéloblastique Enfant) Cooperative Group. Leukemia. 2005;19(12):2082–2089. doi: 10.1038/sj.leu.2403867. [DOI] [PubMed] [Google Scholar]
- 16.Wiemels JL, Xiao Z, Buffler PA, et al. In utero origin of t(8;21) AML1-ETO translocations in childhood acute myeloid leukemia. Blood. 2002;99(10):3801–3805. doi: 10.1182/blood.v99.10.3801. [DOI] [PubMed] [Google Scholar]
- 17.LaFiura KM, Bielawski DM, Posecion NC, et al. Association between prenatal pesticide exposures and the generation of leukemia-associated t(8;21) Pediatric Blood and Cancer. 2007;49(5):624–628. doi: 10.1002/pbc.21283. [DOI] [PubMed] [Google Scholar]
- 18.Manvelyan M, Kempf P, Weise A, et al. Preferred co-localization of chromosome 8 and 21 in myeloid bone marrow cells detected by three dimensional molecular cytogenetics. International Journal of Molecular Medicine. 2009;24(3):335–341. doi: 10.3892/ijmm_00000237. [DOI] [PubMed] [Google Scholar]
- 19.Stuardo M, Martinez M, Hidalgo K, et al. Altered chromatin modifications in AML1/RUNX1 breakpoint regions involved in (8;21) translocation. Journal of Cellular Physiology. 2009;218(2):343–349. doi: 10.1002/jcp.21599. [DOI] [PubMed] [Google Scholar]
- 20.Liddiard K, Hills R, Burnett AK, Darley RL, Tonks A. OGG1 is a novel prognostic indicator in acute myeloid leukaemia. Oncogene. 2010;29(13):2005–2012. doi: 10.1038/onc.2009.462. [DOI] [PubMed] [Google Scholar]
- 21.Vieira L, Oliveira V, Ambrósio AP, et al. Translocation (8;17;15;21)(q22;q23;q15;q22) in acute myeloid leukemia (M2): a four-way variant of t(8;21) Cancer Genetics and Cytogenetics. 2001;128(2):104–107. doi: 10.1016/s0165-4608(01)00404-6. [DOI] [PubMed] [Google Scholar]
- 22.Anelli L, Albano F, Zagaria A, et al. Pericentric chromosome 8 inversion associated with the 5′ RUNX1/3′CBFA2T1 gene in acute myeloid leukemia cases. Annals of Hematology. 2005;84(4):245–249. doi: 10.1007/s00277-004-0960-x. [DOI] [PubMed] [Google Scholar]
- 23.Specchia G, Albano F, Anelli L, et al. Insertions generating the 5′RUNX1/3′C8FA2T1 gene in acute myeloid leukemia cases show variable breakpoints. Genes Chromosomes and Cancer. 2004;41(1):86–91. doi: 10.1002/gcc.20061. [DOI] [PubMed] [Google Scholar]
- 24.Peterson LF, Boyapati A, Ahn EY, et al. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110(3):799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchenbauer F, Schnittger S, Look T, et al. Identification of additional cytogenetic and molecular genetic abnormalities in acute myeloid leukaemia with t(8;21)/AML1-ETO. British Journal of Haematology. 2006;134(6):616–619. doi: 10.1111/j.1365-2141.2006.06229.x. [DOI] [PubMed] [Google Scholar]
- 26.Peniket AJ, Wainscoat J, Side L, et al. Del(9q) AML: clinical and cytological characteristics and prognostic implications. British Journal of Haematology. 2005;129(2):210–220. doi: 10.1111/j.1365-2141.2005.05445.x. [DOI] [PubMed] [Google Scholar]
- 27.Prébet T, Boissel N, Reutenauer S, et al. Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: a collaborative study of the French CBF-AML intergroup. Journal of Clinical Oncology. 2009;27(28):4747–4753. doi: 10.1200/JCO.2008.21.0674. [DOI] [PubMed] [Google Scholar]
- 28.Appelbaum FR, Kopecky KJ, Tallman MS, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. British Journal of Haematology. 2006;135(2):165–173. doi: 10.1111/j.1365-2141.2006.06276.x. [DOI] [PubMed] [Google Scholar]
- 29.Narimatsu H, Iino M, Ichihashi T, et al. Clinical significance of minimal residual disease in patients with t(8;21) acute myeloid leukemia in Japan. International Journal of Hematology. 2008;88(2):154–158. doi: 10.1007/s12185-008-0108-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang YY, Zhou GB, Yin T, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(4):1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paschka P, Marcucci G, Ruppert AS, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B study. Journal of Clinical Oncology. 2006;24(24):3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 32.Shimada A, Taki T, Tabuchi K, et al. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): a study of the Japanese Childhood AML Cooperative Study Group. Blood. 2006;107(5):1806–1809. doi: 10.1182/blood-2005-08-3408. [DOI] [PubMed] [Google Scholar]
- 33.Boissel N, Leroy H, Brethon B, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML) Leukemia. 2006;20(6):965–970. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 34.Cairoli R, Beghini A, Grillo G, et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood. 2006;107(9):3463–3468. doi: 10.1182/blood-2005-09-3640. [DOI] [PubMed] [Google Scholar]
- 35.Paschka P, Du J, Schlenk RF, et al. Mutations in the Fms-related tyrosine kinase 3 (FLT3) gene independently predict poor outcome in acute myeloid leukemia (AML) with t(8;21): a study of the outcome in acute myeloid leukemia (AML) with t(8;21): a study of the German-Austrian AML Study Group (AMLSG) ASH Annual Meeting Abstracts. 2009;114(22) abstract no 825. [Google Scholar]
- 36.Schnittger S, Bacher U, Kern W, Haferlach T, Haferlach C. JAK2V617F as progression marker in CMPD and as cooperative mutation in AML with trisomy 8 and t(8;21): a comparative study on 1103 CMPD and 269 AML cases. Leukemia. 2007;21(8):1843–1845. doi: 10.1038/sj.leu.2404707. [DOI] [PubMed] [Google Scholar]
- 37.Sritana N, Auewarakul CU. KIT and FLT3 receptor tyrosine kinase mutations in acute myeloid leukemia with favorable cytogenetics: two novel mutations and selective occurrence in leukemia subtypes and age groups. Experimental and Molecular Pathology. 2008;85(3):227–231. doi: 10.1016/j.yexmp.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Goemans BF, Zwaan CHM, Miller M, et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19(9):1536–1542. doi: 10.1038/sj.leu.2403870. [DOI] [PubMed] [Google Scholar]
- 39.Care RS, Valk PJM, Goodeve AC, et al. Incidence and prognosis of c-KIT and FLT3 mutations in core binding factor (CBF) acute myeloid leukaemias. British Journal of Haematology. 2003;121(5):775–777. doi: 10.1046/j.1365-2141.2003.04362.x. [DOI] [PubMed] [Google Scholar]
- 40.Pollard JA, Alonzo TA, Gerbing RB, et al. Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood. 2010;115(12):2372–2379. doi: 10.1182/blood-2009-09-241075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnittger S, Kohl TM, Haferlach T, et al. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107(5):1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 42.Döhner K, Du J, Corbacioglu A, Scholl C, Schlenk RF, Döhner H. JAK2V617F mutations as cooperative genetic lesions in t(8;21)-positive acute myeloid leukemia. Haematologica. 2006;91(11):1569–1570. [PubMed] [Google Scholar]
- 43.Mrózek K, Bloomfield CD. Clinical significance of the most common chromosome translocations in adult acute myeloid leukemia. Journal of the National Cancer Institute—Monographs. 2008;(39):52–57. doi: 10.1093/jncimonographs/lgn003. [DOI] [PubMed] [Google Scholar]
- 44.Urioste M, Martínez-Ramírez A, Cigudosa JC, et al. Identification of ins(8;21) with AML1/ETO fusion in acute myelogenous leukemia M2 by molecular cytogenetics. Cancer Genetics and Cytogenetics. 2002;133(1):83–86. doi: 10.1016/s0165-4608(01)00555-6. [DOI] [PubMed] [Google Scholar]
- 45.Sweetser DA, Peniket AJ, Haaland C, et al. Delineation of the minimal commonly deleted segment and identification of candidate tumor-suppressor genes in del(9q) acute myeloid leukemia. Genes Chromosomes and Cancer. 2005;44(3):279–291. doi: 10.1002/gcc.20236. [DOI] [PubMed] [Google Scholar]
- 46.Xiao Z, Liu S, Liu X, Yu M, Hao Y. Tetraploidy or near-tetraploidy clones with double 8;21 translocation: a non-random additional anomaly of acute myeloid leukemia with t(8;21)(q22;q22) Haematologica. 2005;90(3):413–414. [PubMed] [Google Scholar]
- 47.Shih LY, Liang DC, Huang CF, et al. Cooperating mutations of receptor tyrosine kinases and Ras genes in childhood core-binding factor acute myeloid leukemia and a comparative analysis on paired diagnosis and relapse samples. Leukemia. 2008;22(2):303–307. doi: 10.1038/sj.leu.2404995. [DOI] [PubMed] [Google Scholar]
- 48.Coppersmith HJ, Kimand H-G, Klug C. Differential contributions of c-kit activating mutations to promotion of AML1-ETO associated neoplasia. ASH Annual Meeting Abstracts. 2010;116(21) abstract no. 4195. [Google Scholar]
- 49.Schessl C, Rawat VPS, Cusan M, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. Journal of Clinical Investigation. 2005;115(8):2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou F-S, Wunderlich M, Griesinger A, Mulloy JC. N-RasG12D induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood. 2011;117(7):2237–2240. doi: 10.1182/blood-2010-01-264119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiwatari M, Taki T, Tsuchida M, et al. Novel missense mutations in the tyrosine kinase domain of the platelet-derived growth factor receptor α (PDGFRA) gene in childhood acute myeloid leukemia with t(8;21) (q22;q22) or inv(16) (p13q22) [10] Leukemia. 2005;19(3):476–477. doi: 10.1038/sj.leu.2403638. [DOI] [PubMed] [Google Scholar]
- 52.Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19(3):329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 53.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. Journal of Clinical Oncology. 2010;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 54.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. New England Journal of Medicine. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gustafson SA, Lin P, Chen SUS, et al. Therapy-related acute myeloid leukemia with t(8;21) (q22;q22) shares many features with de novo acute myeloid leukemia with t(8;21)(q22;q22) but does not have a favorable outcome. American Journal of Clinical Pathology. 2009;131(5):647–655. doi: 10.1309/AJCP5ETHDXO6NCGZ. [DOI] [PubMed] [Google Scholar]
- 56.Chang H, Chuang CL, Hsien KC. Therapy-related acute myeloid leukemia after concurrent chemoradiotherapy for esophageal cancer: report of two cases. Tumori. 2009;95(3):371–373. doi: 10.1177/030089160909500317. [DOI] [PubMed] [Google Scholar]
- 57.Owatari S, Otsuka M, Uozumi K, Takeshita T, Hanada S. Two cases of secondary acute myeloid leukemia accompanying adult T-cell leukemia/lymphoma. International Journal of Hematology. 2007;85(1):32–35. doi: 10.1532/IJH97.06145. [DOI] [PubMed] [Google Scholar]
- 58.Erickson P, Gao J, Chang KS, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80(7):1825–1831. [PubMed] [Google Scholar]
- 59.Dowdy CR, Xie R, Frederick D, et al. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Human Molecular Genetics. 2009;19(6):1048–1057. doi: 10.1093/hmg/ddp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plevin MJ, Zhang J, Guo C, Roeder RG, Ikura M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10242–10247. doi: 10.1073/pnas.0603463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hug BA, Lazar MA. ETO interacting proteins. Oncogene. 2004;23(24):4270–4274. doi: 10.1038/sj.onc.1207674. [DOI] [PubMed] [Google Scholar]
- 62.Zaidi SK, Dowdy CR, Van Wijnen AJ, et al. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Research. 2009;69(21):8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development. 2005;132(10):2263–2272. doi: 10.1242/dev.01824. [DOI] [PubMed] [Google Scholar]
- 64.Qiu J, Wong J, Tweardy DJ, Dong S. Decreased intranuclear mobility of acute myeloid leukemia 1-containing fusion proteins is accompanied by reduced mobility and compartmentalization of core binding factor β. Oncogene. 2006;25(28):3982–3993. doi: 10.1038/sj.onc.1209431. [DOI] [PubMed] [Google Scholar]
- 65.Lafiura KM, Edwards H, Taub JW, et al. Identification and characterization of novel AML1-ETO fusion transcripts in pediatric t(8;21) acute myeloid leukemia: a report from the Children’s Oncology Group. Oncogene. 2008;27(36):4933–4942. doi: 10.1038/onc.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan M, Kanbe E, Peterson LF, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nature Medicine. 2006;12(8):945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 67.Yan M, Ahn EY, Hiebert SW, Zhang DE. RUNX1/AML1 DNA-binding domain and ETO/MTG8 NHR2-dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood. 2009;113(4):883–886. doi: 10.1182/blood-2008-04-153742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsoulaki O, Lacaudand G, Kouskoff V. Characterisation of AML1-ETO9A leukaemiogenesis using an inducible murine model. Haematologica. 2010;95(s2) abstract no 0025. [Google Scholar]
- 69.Kozu T, Fukuyama T, Yamami T, Akagi K, Kaneko Y. MYND-less splice variants of AML1-MTG8 (RUNX1-CBFA2T1) are expressed in leukemia with t(8;21) Genes Chromosomes and Cancer. 2005;43(1):45–53. doi: 10.1002/gcc.20165. [DOI] [PubMed] [Google Scholar]
- 70.Mannari D, Gascoyne D, Dunne J, Chaplin T, Young B. A novel exon in AML1-ETO negatively influences the clonogenic potential of the t(8;21) in acute myeloid leukemia. Leukemia. 2010;24(4):891–894. doi: 10.1038/leu.2009.288. [DOI] [PubMed] [Google Scholar]
- 71.Khandanpour C, Thiede C, Valk PJM, et al. Avariant allele of growth factor independence 1 (GFI1) is associated with acute myeloid leukemia. Blood. 2010;115(12):2462–2472. doi: 10.1182/blood-2009-08-239822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roudaia L, Cheney MD, Manuylova E, et al. CBFβ is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113(13):3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S, Klisovic RB, Vukosavljevic T, et al. Targeting AML1/ETO-histone deacetylase repressor complex: a novel mechanism for valproic acid-mediated gene expression and cellular differentiation in AML1/ETO-positive acute myeloid leukemia cells. Journal of Pharmacology and Experimental Therapeutics. 2007;321(3):953–960. doi: 10.1124/jpet.106.118406. [DOI] [PubMed] [Google Scholar]
- 74.Bakshi R, Zaidi SK, Pande S, et al. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. Journal of Cell Science. 2008;121(23):3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahn EY, Yan M, Malakhova OA, et al. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(44):17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu S, Shen T, Huynh L, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Research. 2005;65(4):1277–1284. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 77.Osman D, Gobert V, Ponthan F, Heidenreich O, Haenlin M, Waltzer L. A Drosophila model identifies calpains as modulators of the human leukemogenic fusion protein AML1-ETO. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(29):12043–12048. doi: 10.1073/pnas.0902449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Corpora T, Roudaia L, Oo M, et al. Structure of the AML1-ETO NHR3-PKA(RIIalpha) complex and its contribution to AML1-ETO activity. Journal of Molecular Biology. 2010;402(3):560–577. doi: 10.1016/j.jmb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park S, Chen W, Cierpicki T, et al. Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity. Blood. 2009;113(15):3558–3567. doi: 10.1182/blood-2008-06-161307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ibañez V, Sharma A, Buonamici S, et al. AML1-ETO decreases ETO-2 (MTG16) interactions with nuclear receptor corepressor, an effect that impairs granulocyte differentiation. Cancer Research. 2004;64(13):4547–4554. doi: 10.1158/0008-5472.CAN-03-3689. [DOI] [PubMed] [Google Scholar]
- 81.Wichmann C, Chen L, Heinrich M, et al. Targeting the oligomerization domain of ETO interferes with RUNX1/ETO oncogenic activity in t(8;21)-positive leukemic cells. Cancer Research. 2007;67(5):2280–2289. doi: 10.1158/0008-5472.CAN-06-3360. [DOI] [PubMed] [Google Scholar]
- 82.Vangala RK, Heiss-Neumann MS, Rangatia JS, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101(1):270–277. doi: 10.1182/blood-2002-04-1288. [DOI] [PubMed] [Google Scholar]
- 83.Fuchs O, Provaznikova D, Kocova M, et al. CEBPA polymorphisms and mutations in patients with acute myeloid leukemia, myelodysplastic syndrome, multiple myeloma and non-Hodgkin’s lymphoma. Blood Cells, Molecules, and Diseases. 2008;40(3):401–405. doi: 10.1016/j.bcmd.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 84.Westendorf JJ, Yamamoto CM, Lenny N, Downing JR, Selsted ME, Hiebert SW. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription, and blocks granulocytic differentiation. Molecular and Cellular Biology. 1998;18(1):322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rochford JJ, Semple RK, Laudes M, et al. ETO/MTG8 is an inhibitor of C/EBPβ activity and a regulator of early adipogenesis. Molecular and Cellular Biology. 2004;24(22):9863–9872. doi: 10.1128/MCB.24.22.9863-9872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fortier JM, Payton JE, Cahan P, Ley TJ, Walter MJ, Graubert TA. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia. 2010;24(5):950–957. doi: 10.1038/leu.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiacci E, Pileri S, Orleth A, et al. PAX5 expression in acute leukemias: higher B-lineage specificity than CD79a and selective association with t(8;21)-acute myelogenous leukemia. Cancer Research. 2004;64(20):7399–7404. doi: 10.1158/0008-5472.CAN-04-1865. [DOI] [PubMed] [Google Scholar]
- 88.Valbuena JR, Medeiros LJ, Rassidakis GZ, et al. Expression of B cell-specific activator protein/PAX5 in acute myeloid leukemia with t(8;21)(q22;q22) American Journal of Clinical Pathology. 2006;126(2):235–240. doi: 10.1309/LG0Q-0VXY-BETJ-4VHE. [DOI] [PubMed] [Google Scholar]
- 89.Fazi F, Racanicchi S, Zardo G, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12(5):457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 90.Xu M, Li D, Lu Y, Chen GQ. Leukemogenic AML1-ETO fusion protein increases carcinogen-DNA adduct formation with upregulated expression of cytochrome P450-1A1 gene. Experimental Hematology. 2007;35(8):1249–1255. doi: 10.1016/j.exphem.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 91.Krejci O, Wunderlich M, Geiger H, et al. P53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008;111(4):2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhoades KL, Hetherington CJ, Rowley JD, et al. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shimizu K, Kitabayashi I, Kamada N, et al. AML1-MTG8 leukemic protein induces the expression of granulocyte colony-stimulating factor (G-CSF) receptor through the up-regulation of CCAAT/enhancer binding protein epsilon. Blood. 2000;96(1):288–296. [PubMed] [Google Scholar]
- 94.Klampfer L, Zhang J, Zelenetz AO, Uchida H, Nimer SD. The AML1/ETO fusion protein activates transcription of BCL-2. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(24):14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang G, Khalaf W, Van De Locht L, et al. Transcriptional repression of the neurofibromatosis-1 tumor suppressor by the t(8;21) fusion protein. Molecular and Cellular Biology. 2005;25(14):5869–5879. doi: 10.1128/MCB.25.14.5869-5879.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mulloy JC, Jankovic V, Wunderlich M, et al. AML1-ETO fusion protein up-regulates TRKA mRNA expression in human CD34+ cells, allowing nerve growth factor-induced expansion. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(11):4016–4021. doi: 10.1073/pnas.0404701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peterson LF, Yan M, Zhang DE. The p21 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007;109(10):4392–4398. doi: 10.1182/blood-2006-03-012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li XI, Xu YAB, Wang Q, et al. Leukemogenic AML1-ETO fusion protein upregulates expression of connexin 43: the role in AML 1-ETO-induced growth arrest in leukemic cells. Journal of Cellular Physiology. 2006;208(3):594–601. doi: 10.1002/jcp.20695. [DOI] [PubMed] [Google Scholar]
- 99.Guinn BA, Mohamedali A, Mills KI, et al. Leukemia associated antigens: their dual role as biomarkers and immunotherapeutic targets for acute myeloid leukemia. Biomark Insights. 2007;2:69–79. [PMC free article] [PubMed] [Google Scholar]
- 100.Boyapati A, Yan M, Peterson LF, Biggs JR, Le Beau MM, Zhang DE. Aleukemia fusion protein attenuates the spindle checkpoint and promotes aneuploidy. Blood. 2007;109(9):3963–3971. doi: 10.1182/blood-2006-09-045583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tabe Y, Jin L, Contractor R, et al. Novel role of HDAC inhibitors in AML1/ETO AML cells: activation of apoptosis and phagocytosis through induction of annexin A1. Cell Death and Differentiation. 2007;14(8):1443–1456. doi: 10.1038/sj.cdd.4402139. [DOI] [PubMed] [Google Scholar]
- 102.Gao FH, Wang Q, Wu YL, Li XI, Zhao KEW, Chen GQ. c-Jun N-terminal kinase mediates AML1-ETO protein-induced connexin-43 expression. Biochemical and Biophysical Research Communications. 2007;356(2):505–511. doi: 10.1016/j.bbrc.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 103.Erkeland SJ, Palande KK, Valkhof M, Gits J, Oorschot ADV, Touw IP. The gene encoding thioredoxin-interacting protein (TXNIP) is a frequent virus integration site in virus-induced mouse leukemia and is overexpressed in a subset of AML patients. Leukemia Research. 2009;33(10):1367–1371. doi: 10.1016/j.leukres.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 104.Shikami M, Miwa H, Nish K, II, et al. Low p53 expression of acute myelocytic leukemia cells with t(8;21) chromosome abnormality: association with low p14 expression. Leukemia Research. 2006;30(4):379–383. doi: 10.1016/j.leukres.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 105.Sinenko SA, Hung T, Moroz T, et al. Genetic manipulation of AML1-ETO-induced expansion of hematopoietic precursors in a Drosophila model. Blood. 2010;116(22):4612–4620. doi: 10.1182/blood-2010-03-276998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Foss B, Hervig T, Bruserud Ø. Connexins are active participants of hematopoietic stem cell regulation. Stem Cells and Development. 2009;18(6):807–812. doi: 10.1089/scd.2009.0086. [DOI] [PubMed] [Google Scholar]
- 107.Foss B, Tronstad KJ, Bruserud Ø. Connexin-based signaling in acute myelogenous leukemia (AML) Biochimica et Biophysica Acta. 2010;1798(1):1–8. doi: 10.1016/j.bbamem.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 108.Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. T(8;21)(q22;q22) fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112(4):1392–1401. doi: 10.1182/blood-2007-11-124735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rossetti S, Hoogeveen AT, Liang P, Stanciu C, van der Spek P, Sacchi N. A distinct epigenetic signature at targets of a leukemia protein. BMC Genomics. 2007;8, article 38 doi: 10.1186/1471-2164-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rodrigues PC, Oliveira SN, Viana MB, et al. Prognostic significance of WT1 gene expression in pediatric acute myeloid leukemia. Pediatric Blood and Cancer. 2007;49(2):133–138. doi: 10.1002/pbc.20953. [DOI] [PubMed] [Google Scholar]
- 111.Balkhi MY, Trivedi AK, Geletu M, et al. Proteomics of acute myeloid leukaemia: cytogenetic risk groups differ specifically in their proteome, interactome and post-translational protein modifications. Oncogene. 2006;25(53):7041–7058. doi: 10.1038/sj.onc.1209689. [DOI] [PubMed] [Google Scholar]
- 112.Hiramatsu A, Miwa H, Shikami M, et al. Disease-specific expression of VEGF and its receptors in AML cells: possible autocrine pathway of VEGF/type1 receptor of VEGF in t(15;17) AML and VEGF/type2 receptor of VEGF in t(8;21) AML. Leukemia and Lymphoma. 2006;47(1):89–95. doi: 10.1080/10428190500270386. [DOI] [PubMed] [Google Scholar]
- 113.Dias S, Hattori K, Zhu Z, et al. Autocrine stimulation of VEGFR-2 activates human leukemic cell growth and migration. Journal of Clinical Investigation. 2000;106(4):511–521. doi: 10.1172/JCI8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Imai N, Miwa H, Shikami M, et al. Growth inhibition of AML cells with specific chromosome abnormalities by monoclonal antibodies to receptors for vascular endothelial growth factor. Leukemia Research. 2009;33(12):1650–1657. doi: 10.1016/j.leukres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 115.Imai N, Shikami M, Miwa H, et al. t(8;21) acute myeloid leukaemia cells are dependent on vascular endothelial growth factor (VEGF)/VEGF receptor type2 pathway and phosphorylation of Akt. British Journal of Haematology. 2006;135(5):673–682. doi: 10.1111/j.1365-2141.2006.06372.x. [DOI] [PubMed] [Google Scholar]
- 116.Alcalay M, Meani N, Gelmetti V, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. Journal of Clinical Investigation. 2003;112(11):1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ersvaer E, Hatfield K, Reikvam H, Bruserud O. Future perspectives: therapeutic targeting of NOTCH signalling may become a strategy in patients receiving stem cell transplantation for hematologic malignancies. Bone Marrow Research. 2011;2011 doi: 10.1155/2011/570796. Article ID 570796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martinez N, Drescher B, Riehle H, et al. The oncogenic fusion protein RUNX1-CBFA2T1 supports proliferation and inhibits senescence in t(8;21)-positive leukaemic cells. BMC Cancer. 2004;4, article 44 doi: 10.1186/1471-2407-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ichikawa H, Tanabe K, Mizushima H, et al. Common gene expression signatures in t(8;21)- and inv(16)-acute myeloid leukaemia. British Journal of Haematology. 2006;135(3):336–347. doi: 10.1111/j.1365-2141.2006.06310.x. [DOI] [PubMed] [Google Scholar]
- 120.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 121.Rowe D, Cotterill SJ, Ross FM, et al. Cytogenetically cryptic AML1—ETO and CBFβ—MYH11 gene rearrangements: incidence in 412 cases of acute myeloid leukaemia. British Journal of Haematology. 2000;111(4):1051–1056. doi: 10.1046/j.1365-2141.2000.02474.x. [DOI] [PubMed] [Google Scholar]
- 122.Sarriera JE, Albitar M, Estrov Z, et al. Comparison of outcome in acute myelogenous leukemia patients with translocation (8;21) found by standard cytogenetic analysis and patients with AML1/ETO fusion transcript found only by PCR testing. Leukemia. 2001;15(1):57–61. doi: 10.1038/sj.leu.2401951. [DOI] [PubMed] [Google Scholar]
- 123.Weerkamp F, Dekking E, Ng YY, et al. Flow cytometric immunobead assay for the detection of BCR-ABL fusion proteins in leukemia patients. Leukemia. 2009;23(6):1106–1117. doi: 10.1038/leu.2009.93. [DOI] [PubMed] [Google Scholar]
- 124.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris M-J, Férec C, De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncology. 2011;7(1):77–91. doi: 10.2217/fon.10.158. [DOI] [PubMed] [Google Scholar]
- 125.Gjertsen BT, Øyan AM, Marzolf B, et al. Analysis of acute myelogenous leukemia: preparation of samples for genomic and proteomic analyses. Journal of Hematotherapy and Stem Cell Research. 2002;11(3):469–481. doi: 10.1089/15258160260090933. [DOI] [PubMed] [Google Scholar]
- 126.Bruserud Ø, Tjønnfjord G, Gjertsen BT, Foss B, Ernst P. New strategies in the treatment of acute myelogenous leukemia: mobilization and transplantation of autologous peripheral blood stem cells in adult patients. Stem Cells. 2000;18(5):343–351. doi: 10.1634/stemcells.19-1-1. [DOI] [PubMed] [Google Scholar]
- 127.Rowe D, Bown NP, Albitar M, Estey E. Diagnostic detection of AML1/ETO gene fusion by polymerase chain reaction. Leukemia. 2002;16(8):1576–1577. doi: 10.1038/sj.leu.2402667. [DOI] [PubMed] [Google Scholar]
- 128.Van Dongen JJM, Macintyre EA, Gabert JA, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13(12):1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 129.Gabert J, Beillard E, van der Velden VHJ, et al. Standardization and quality control studies of 'real time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer Program. Leukemia. 2003;17(12):2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 130.Tobal K, Liu Yin JA. Diagnosis and monitoring of AML1-MTG8 (ETO)-positive acute myeloid leukemia by qualitative and real-time quantitative RT-PCR. Methods in molecular medicine. 2006;125:149–161. doi: 10.1385/1-59745-017-0:149. [DOI] [PubMed] [Google Scholar]
- 131.Ommen HB, Østergaard M, Yan M, Brændstrup K, Zhang DE, Hokland P. Persistent altered fusion transcript splicing identifies RUNX1-RUNX1T1+ AML patients likely to relapse. European Journal of Haematology. 2010;84(2):128–132. doi: 10.1111/j.1600-0609.2009.01371.x. [DOI] [PubMed] [Google Scholar]
- 132.Pallisgaard N, Hokland P, Riishøj DC, Pedersen B, Jørgensen P. Multiplex reverse transcription-polymerase chain reaction for simultaneous screening of 29 translocations and chromosomal aberrations in acute leukemia. Blood. 1998;92(2):574–588. [PubMed] [Google Scholar]
- 133.Lee DA, Harris CP, Gresik VM, Rao P, Lau CC. Granulocytic sarcoma presenting as pneumonia in a child with t(8;21) acute myelogenous leukemia: diagnosis by flourescent in situ hybridization. Journal of Pediatric Hematology/Oncology. 2004;26(7):431–434. doi: 10.1097/00043426-200407000-00006. [DOI] [PubMed] [Google Scholar]
- 134.Arber DA, Glackin C, Lowe G, Medeiros LJ, Slovak ML. Presence of t(8;21)(q22;q22) in myeloperoxidase-positive, myeloid surface antigen-negative acute myeloid leukemia. American Journal of Clinical Pathology. 1997;107(1):68–73. doi: 10.1093/ajcp/107.1.68. [DOI] [PubMed] [Google Scholar]
- 135.Zheng J, Wang X, Hu YU, et al. A correlation study of immunophenotypic, cytogenetic, and clinical features of 180 AML patients in China. Cytometry Part B—Clinical Cytometry. 2008;74(1):25–29. doi: 10.1002/cyto.b.20368. [DOI] [PubMed] [Google Scholar]
- 136.De J, Zanjani R, Hibbard M, Davis BH. Immunophenotypic profile predictive off KIT activating mutations in AML1-ETO leukemia. American Journal of Clinical Pathology. 2007;128(4):550–557. doi: 10.1309/JVALJNL4ELQMD536. [DOI] [PubMed] [Google Scholar]
- 137.Pileri SA, Ascani S, Cox MC, et al. Myeloid sarcoma: clinico-pathologic, phenotypic and cytogenetic analysis of 92 adult patients. Leukemia. 2007;21(2):340–350. doi: 10.1038/sj.leu.2404491. [DOI] [PubMed] [Google Scholar]
- 138.Neiman RS, Barcos M, Berard C. Granulocytic sarcoma: a clinicopathologic study of 61 biopsied cases. Cancer. 1981;48(6):1426–1437. doi: 10.1002/1097-0142(19810915)48:6<1426::aid-cncr2820480626>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 139.Kottaridis PD, Ketley N, Peggs K, et al. An unusual case of intrapulmonary granulocytic sarcoma presenting as interstitial pneumonitis following allogeneic bone marrow transplantation for acute myeloid leukaemia and responding to donor lymphocyte infusion. Bone Marrow Transplantation. 1999;24(7):807–809. doi: 10.1038/sj.bmt.1701974. [DOI] [PubMed] [Google Scholar]
- 140.Chevallier P, Mohty M, Lioure B, et al. Allogeneic hematopoietic stem-cell transplantation for myeloid sarcoma: a retrospective study from the SFGM-TC. Journal of Clinical Oncology. 2008;26(30):4940–4943. doi: 10.1200/JCO.2007.15.6315. [DOI] [PubMed] [Google Scholar]
- 141.Tsimberidou AM, Kantarjian HM, Wen S, et al. Myeloid sarcoma is associated with superior event-free survival and overall survival compared with acute myeloid leukemia. Cancer. 2008;113(6):1370–1378. doi: 10.1002/cncr.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Colović N, Colović M, Cemerikić V, et al. Granulocytic sarcoma of the brain in a patient with acute myeloid leukemia. Acta chirurgica Iugoslavica. 2004;51(3):129–131. [PubMed] [Google Scholar]
- 143.Al-Sobbi EM, Jeha TM, Al-Taher MI. Granulocytic sarcoma causing cord compression in a pregnant woman with acute myeloid leukemia and t(8;21) Saudi Medical Journal. 2008;29(11):1658–1661. [PubMed] [Google Scholar]
- 144.Tamaki H, Yoshihara S, Fujioka T, Kawakami M, Oka Y, Ogawa H. Molecular detection of AML1-MTG8-positive cells in peripheral blood from a patient with isolated extramedullary relapse of t(8;21) acute myeloid leukemia. Leukemia. 2009;23(2):424–426. doi: 10.1038/leu.2008.220. [DOI] [PubMed] [Google Scholar]
- 145.Wong KF, Siu LLP, Wong WS. Aleukaemic acute myeloid leukaemia with t(8;21)(q22;q22): images in haematology. British Journal of Haematology. 2009;146(4):p. 345. doi: 10.1111/j.1365-2141.2009.07580.x. [DOI] [PubMed] [Google Scholar]
- 146.Jost E, Lorenzen J, Haage P, et al. Heart and muscle involvement by extra-medullary myeloid leukemia: a case report and review of the literature. Leukemia and Lymphoma. 2005;46(12):1819–1824. doi: 10.1080/10428190500233830. [DOI] [PubMed] [Google Scholar]
- 147.Felice MS, Zubizarreta PA, Alfaro EM, et al. Good outcome of children with acute myeloid leukemia and t(8;21)(q22;q22), even when associated with granulocytic sarcoma: a report from a single institution in Argentina. Cancer. 2000;88(8):1939–1944. [PubMed] [Google Scholar]
- 148.Pullarkat ST, Pullarkat V, Kroft SH, et al. Systemic mastocytosis associated with t(8;21)(q22;q22) acute myeloid leukemia. Journal of Hematopathology. 2009;2(1):27–33. doi: 10.1007/s12308-009-0023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pullarkat V, Bedell V, Kim Y, et al. Neoplastic mast cells in systemic mastocytosis associated with t(8;21) acute myeloid leukemia are derived from the leukemic clone. Leukemia Research. 2007;31(2):261–265. doi: 10.1016/j.leukres.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 150.Sperr WR, Drach J, Hauswirth AW, et al. Myelomastocytic leukemia: evidence for the origin of mast cells from the leukemic clone and eradication by allogeneic stem cell transplantation. Clinical Cancer Research. 2005;11(19 I):6787–6792. doi: 10.1158/1078-0432.CCR-05-1064. [DOI] [PubMed] [Google Scholar]
- 151.Pullarkat ST, Sedarat F, Paquette R, Said J. Systemic mastocytosis with plasma cell dyscrasia: report of a case. Leukemia Research. 2008;32(7):1160–1163. doi: 10.1016/j.leukres.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 152.Cairoli R, Ripamonti CB, Beghini A, et al. Total serum tryptase: a predictive marker for KIT mutation in acute myeloid leukemia. Leukemia Research. 2009;33(9):1282–1284. doi: 10.1016/j.leukres.2009.03.043. [DOI] [PubMed] [Google Scholar]
- 153.McCormack E, Bruserud O, Gjertsen BT. Review: genetic models of acute myeloid leukaemia. Oncogene. 2008;27(27):3765–3779. doi: 10.1038/onc.2008.16. [DOI] [PubMed] [Google Scholar]
- 154.Yuan Y, Zhou L, Miyamoto T, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bullinger L, Döhner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. The New England Journal of Medicine. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 156.Miller BG, Stamatoyannopoulos JA. Integrative meta-analysis of differential gene expression in acute myeloid leukemia. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009466. Article ID e9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Verhaak RGW, Wouters BJ, Erpelinck CAJ, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lück SC, Russ AC, Du J, et al. KIT mutations confer a distinct gene expression signature in core binding factor leukaemia: research paper. British Journal of Haematology. 2010;148(6):925–937. doi: 10.1111/j.1365-2141.2009.08035.x. [DOI] [PubMed] [Google Scholar]
- 159.Reikvam H, Olsnes AM, Gjertsen BT, Ersvar E, Bruserud O. Nuclear factor-B signaling: a contributor in leukemogenesis and a target for pharmacological intervention in human acute myelogenous leukemia. Critical Reviews in Oncogenesis. 2009;15(1-2):1–41. doi: 10.1615/critrevoncog.v15.i1-2.10. [DOI] [PubMed] [Google Scholar]
- 160.Bullinger L, Rücker FG, Kurz S, et al. Gene-expression profiling identifies distinct subclasses of core binding factor acute myeloid leukemia. Blood. 2007;110(4):1291–1300. doi: 10.1182/blood-2006-10-049783. [DOI] [PubMed] [Google Scholar]
- 161.Reikvam H, Hatfield K, Ersvaer E, Ryningen A, Bruserud Ø. Pharmacological targeting of the PI3K-AKT/PKB-mTOR pathway alters local angioregulation in acute myelogenous leukemia. Haematologica. 2010;95(s2) abstract no 0634. [Google Scholar]
- 162.Doepfner KT, Boller D, Arcaro A. Targeting receptor tyrosine kinase signaling in acute myeloid leukemia. Critical Reviews in Oncology/Hematology. 2007;63(3):215–230. doi: 10.1016/j.critrevonc.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 163.Paydas S, Tanriverdi K, Yavuz S, Disel U, Baslamisli F, Burgut R. PRAME mRNA levels in cases with acute leukemia: clinical importance and future prospects. American Journal of Hematology. 2005;79(4):257–261. doi: 10.1002/ajh.20425. [DOI] [PubMed] [Google Scholar]
- 164.Liseth K, Ersvaer E, Hervig T, Bruserud Ø. Combination of intensive chemotherapy and anticancer vaccines in the treatment of human malignancies: the hematological experience. Journal of Biomedicine and Biotechnology. 2010;2010 doi: 10.1155/2010/692097. Article ID 692097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pang A, Au WY, Kwong YL. Caveolin-1 gene is coordinately regulated with the multidrug resistance 1 gene in normal and leukemic bone marrow. Leukemia Research. 2004;28(9):973–977. doi: 10.1016/j.leukres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 166.Stoeckle C, Gouttefangeas C, Hammer M, Weber E, Melms A, Tolosa E. Cathepsin W expressed exclusively in CD8+ T cells and NK cells, is secreted during target cell killing but is not essential for cytotoxicity in human CTLs. Experimental Hematology. 2009;37(2):266–275. doi: 10.1016/j.exphem.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 167.Park T, Zong Ping Chen , Leavitt J. Activation of the leukocyte plastin gene occurs in most human cancer cells. Cancer Research. 1994;54(7):1775–1781. [PubMed] [Google Scholar]
- 168.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cellular and Molecular Life Sciences. 2004;61(19-20):2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Neufeld G, Kessler O. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nature Reviews Cancer. 2008;8(8):632–645. doi: 10.1038/nrc2404. [DOI] [PubMed] [Google Scholar]
- 170.Jongen-Lavrencic M, Sun SUM, Dijkstra MK, Valk PJM, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 171.Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pedranzini L, Mottadelli F, Ronzoni S, et al. Differential cytogenomics and miRNA signature of the Acute Myeloid Leukaemia Kasumi-1 cell line CD34(+)38(-) compartment. Leukemia Research. 2010;34(10):1287–1295. doi: 10.1016/j.leukres.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 173.Esteller M. Molecular origins of cancer: epigenetics in cancer. The New England Journal of Medicine. 2008;358(11):1148–1096. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 174.Bullinger L, Ehrich M, Döhner K, et al. Quantitative DNA methylation predicts survival in adult acute myeloid leukemia. Blood. 2010;115(3):636–642. doi: 10.1182/blood-2009-03-211003. [DOI] [PubMed] [Google Scholar]
- 175.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alvarez S, Suela J, Valencia A, et al. DNA methylation profiles and their relationship with cytogenetic status in adult acute myeloid leukemia. PLoS ONE. 2010;5(8) doi: 10.1371/journal.pone.0012197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Müller-Tidow C, Klein H-U, Hascher A, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116(18):3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Narimatsu H, Yokozawa T, Iida H, et al. Clinical characteristics and outcomes in patients with t(8;21) acute myeloid leukemia in Japan. Leukemia. 2008;22(2):428–432. doi: 10.1038/sj.leu.2404905. [DOI] [PubMed] [Google Scholar]
- 179.Lin P, Chen L, Luthra R, Konoplev SN, Wang X, Medeiros LJ. Acute myeloid leukemia harboring t(8;21)(q22;q22): a heterogeneous disease with poor outcome in a subset of patients unrelated to secondary cytogenetic aberrations. Modern Pathology. 2008;21(8):1029–1036. doi: 10.1038/modpathol.2008.92. [DOI] [PubMed] [Google Scholar]
- 180.Nguyen S, Leblanc T, Fenaux P, et al. A white blood cell index as the main prognostic factor in t(8;21) acute myeloid leukemia (AML): a survey of 161 cases from the French AML intergroup. Blood. 2002;99(10):3517–3523. doi: 10.1182/blood.v99.10.3517. [DOI] [PubMed] [Google Scholar]
- 181.Cornelissen JJ, Van Putten WLJ, Verdonck LF, et al. Results of a HOVON/SAKK donor versus no-donor analysis of myeloablative HLA-identical sibling stem cell transplantation in first remission acute myeloid leukemia in young and middle-aged adults: benefits for whom? Blood. 2007;109(9):3658–3666. doi: 10.1182/blood-2006-06-025627. [DOI] [PubMed] [Google Scholar]
- 182.Shin H-J, Chung J, Kim HJ, et al. Bone marrow cellularity is a single most important independent prognostic factor in AML patients with t(8;21) ASH Annual Meeting Abstracts. 2010;116(21) abstract no 1707. [Google Scholar]
- 183.Harrison CJ, Hills RK, Moorman AV, et al. Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council Treatment Trials AML 10 and 12. Journal of Clinical Oncology. 2010;28(16):2674–2681. doi: 10.1200/JCO.2009.24.8997. [DOI] [PubMed] [Google Scholar]
- 184.Von Neuhoff C, Reinhardt D, Sander A, et al. Prognostic impact of specific chromosomal aberrations in a large group of pediatric patients with acute myeloid leukemia treated uniformly according to trial AML-BFM 98. Journal of Clinical Oncology. 2010;28(16):2682–2689. doi: 10.1200/JCO.2009.25.6321. [DOI] [PubMed] [Google Scholar]
- 185.Smith FO, Alonzo TA, Gerbing RB, Woods WG, Arceci RJ. Long-term results of children with acute myeloid leukemia: a report of three consecutive Phase III trials by the Children’s Cancer Group: CCG 251, CCG 213 and CCG 2891. Leukemia. 2005;19(12):2054–2062. doi: 10.1038/sj.leu.2403925. [DOI] [PubMed] [Google Scholar]
- 186.Entz-Werle N, Suciu S, van der Werff ten Bosch J, et al. Results of 58872 and 58921 trials in acute myeloblastic leukemia and relative value of chemotherapy vs allogeneic bone marrow transplantation in first complete remission: the EORTC Children Leukemia Group report. Leukemia. 2005;19(12):2072–2081. doi: 10.1038/sj.leu.2403932. [DOI] [PubMed] [Google Scholar]
- 187.Kuwatsuka Y, Miyamura K, Suzuki R, et al. Hematopoietic stem cell transplantation for core binding factor acute myeloid leukemia: T(8;21) and inv(16) represent different clinical outcomes. Blood. 2009;113(9):2096–2103. doi: 10.1182/blood-2008-03-145862. [DOI] [PubMed] [Google Scholar]
- 188.Gorin N-C, Labopin M, Frassoni F, et al. Identical outcome after autologous or allogeneic genoidentical hematopoietic stem-cell transplantation in first remission of acute myelocytic leukemia carrying inversion 16 or t(8;21): a retrospective study from the European Cooperative Group for Blood and Marrow Transplantation. Journal of Clinical Oncology. 2008;26(19):3183–3188. doi: 10.1200/JCO.2007.15.3106. [DOI] [PubMed] [Google Scholar]
- 189.Schlenk RF, Pasquini MC, Pérez WS, et al. HLA-identical sibling allogeneic transplants versus chemotherapy in acute myelogenous leukemia with t(8;21) in first complete remission: collaborative study between the German AML Intergroup and CIBMTR. Biology of Blood and Marrow Transplantation. 2008;14(2):187–196. doi: 10.1016/j.bbmt.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shin HJ, Kim HJ, Sohn SK, et al. Re-analysis of the outcomes of post-remission therapy for acute myeloid leukemia with core binding factor according to years of patient enrollment. Japanese journal of clinical oncology. 2010;40(6):556–566. doi: 10.1093/jjco/hyq007. [DOI] [PubMed] [Google Scholar]
- 191.Foran JM. New prognostic markers in acute myeloid leukemia: perspective from the clinic. Hematology American Society of Hematology Education Program. 2010;2010(1):47–55. doi: 10.1182/asheducation-2010.1.47. [DOI] [PubMed] [Google Scholar]
- 192.Bruserud Ø, Stapnes C, Ersvær E, Gjertsen BT, Ryningen A. Histone deacetylase inhibitors in cancer treatment: a review of the clinical toxicity and the modulation of gene expression in cancer cells. Current Pharmaceutical Biotechnology. 2007;8(6):388–400. doi: 10.2174/138920107783018417. [DOI] [PubMed] [Google Scholar]
- 193.Ryningen A, Stapnes C, Paulsen K, Lassalle P, Gjertsen BT, Bruserud Ø. In vivo biological effects of ATRA in the treatment of AML. Expert Opinion on Investigational Drugs. 2008;17(11):1623–1633. doi: 10.1517/13543784.17.11.1623. [DOI] [PubMed] [Google Scholar]
- 194.Chen Z, Wang Y, Wang W, Gong J, Xue Y. All-trans retinoic acid as a single agent induces complete remission in a patient with acute leukemia of M subtype. Chinese Medical Journal. 2002;115(1):58–61. [PubMed] [Google Scholar]
- 195.Abreu E Lima RS, Rozera Baruffi M, Gouveia De Lima AS, et al. The co-expression of PML/RARα and AML1/ETO fusion genes is associated with ATRA resistance. British Journal of Haematology. 2005;128(3):407–409. doi: 10.1111/j.1365-2141.2004.05343.x. [DOI] [PubMed] [Google Scholar]
- 196.Kusec R, Laczika K, Knöbl P, et al. AML1/ETO fusion mRNA can be detected in remission blood samples of all patients with t(8;21) acute myeloid leukemia after chemotherapy or autologous bone marrow transplantation. Leukemia. 1994;8(5):735–739. [PubMed] [Google Scholar]
- 197.Miyamoto T, Nagafuji K, Akashi K, et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87(11):4789–4796. [PubMed] [Google Scholar]
- 198.Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993;82(3):712–715. [PubMed] [Google Scholar]
- 199.Saunders MJ, Tobal K, Liu Yin JA. Detection of t(8;21) by reverse transcriptase polymerase chain reaction in patients in remission of acute myeloid leukaemia type M2 after chemotherapy or bone marrow transplantation. Leukemia Research. 1994;18(12):891–895. doi: 10.1016/0145-2126(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 200.Jurlander J, Caligiuri MA, Ruutu T, et al. Persistence of the AML1/ETO fusion transcript in patients treated with allogeneic bone marrow transplantation for t(8;21) leukemia. Blood. 1996;88(6):2183–2191. [PubMed] [Google Scholar]
- 201.Schnittger S, Weisser M, Schoch C, Hiddemann W, Haferlach T, Kern W. New score predicting for prognosis in PML-RARA+, AML1-ETO+, or CBFB-MYH11 acute myeloid leukemia based on quantification of fusion transcripts. Blood. 2003;102(8):2746–2755. doi: 10.1182/blood-2003-03-0880. [DOI] [PubMed] [Google Scholar]
- 202.Leroy H, de Botton S, Gradel-Duflos N, et al. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21) Leukemia. 2005;19(3):367–372. doi: 10.1038/sj.leu.2403627. [DOI] [PubMed] [Google Scholar]
- 203.Ommen HB, Schnittger S, Jovanovic JV, et al. Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood. 2010;115(2):198–205. doi: 10.1182/blood-2009-04-212530. [DOI] [PubMed] [Google Scholar]
- 204.Al-Mawali A, Gillis D, Lewis I. The role of multiparameter flow cytometry for detection of minimal residual disease in acute myeloid leukemia. American Journal of Clinical Pathology. 2009;131(1):16–26. doi: 10.1309/AJCP5TSD3DZXFLCX. [DOI] [PubMed] [Google Scholar]
- 205.Perea G, Lasa A, Aventín A, et al. Prognostic value of minimal residual disease (MRD) in acute myeloid leukemia (AML) with favorable cytogenetics [t(8;21) and inv(16)] Leukemia. 2006;20(1):87–94. doi: 10.1038/sj.leu.2404015. [DOI] [PubMed] [Google Scholar]
- 206.Cox MC, Panetta P, Venditti A, et al. Comparison between conventional banding analysis and FISH screening with an AML-specific set of probes in 260 patients. Hematology Journal. 2003;4(4):263–270. doi: 10.1038/sj.thj.6200262. [DOI] [PubMed] [Google Scholar]
- 207.Bacher U, Kern W, Schoch C, Schnittger S, Hiddemann W, Haferlach T. Evaluation of complete disease remission in acute myeloid leukemia: a prospective study based on cytomorphology, interphase fluorescence in situ hybridization, and immunophenotyping during follow-up in patients with acute myeloid leukemia. Cancer. 2006;106(4):839–847. doi: 10.1002/cncr.21665. [DOI] [PubMed] [Google Scholar]
- 208.Vance GH, Kim H, Hicks GA, et al. Utility of interphase FISH to stratify patients into cytogenetic risk categories at diagnosis of AML in an Eastern Cooperative Oncology Group (ECOG) clinical trial (E1900) Leukemia Research. 2007;31(5):605–609. doi: 10.1016/j.leukres.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 209.Grimwade D, Jovanovic JV, Hills RK, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. Journal of Clinical Oncology. 2009;27(22):3650–3658. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]
- 210.Esteve J, Escoda L, Martín G, et al. Outcome of patients with acute promyelocytic leukemia failing to front-line treatment with all-trans retinoic acid and anthracycline-based chemotherapy (PETHEMA protocols LPA96 and LPA99): benefit of an early intervention. Leukemia. 2007;21(3):446–452. doi: 10.1038/sj.leu.2404501. [DOI] [PubMed] [Google Scholar]
- 211.Morschhauser F, Cayuela JM, Martini S, et al. Evaluation of minimal residual disease using reverse-transcription polymerase chain reaction in t(8;21) acute myeloid leukemia: a multicenter study of 51 patients. Journal of Clinical Oncology. 2000;18(4):788–794. doi: 10.1200/JCO.2000.18.4.788. [DOI] [PubMed] [Google Scholar]