Abstract
Natural killer cells are a group of lymphocytes which function as tightly controlled surveillance operatives which identify transformed cells through a discrete balance of activating and inhibitory receptors ultimately leading to the destruction of incongruent cells. The understanding of this finely tuned balancing act has been aided by the high-resolution structure determination of activating and inhibitory receptors both alone and in complex with their ligands. This paper collates these structural studies detailing the aspects which directly relate to the natural killer cell function and serves to inform both the specialized structural biologist reader and a more general immunology audience.
1. Introduction
The functional outcome of Natural Killer (NK) cells is determined by integrating both activating and inhibitory signals resulting in a highly controlled response which mediates cytotoxicity against transformed cells and, in addition, the release of cytokines critical to the immune response. At the center of this balance resides a group of receptors that allow the relay of intracellular signaling via intrinsic or associated cytoplasmic molecular motifs for various kinases or phosphatases [1–4]. The list of identified NK cell activating and inhibitory receptors currently exceeds twenty. In humans, the activating NK receptors include CD16, the short-tail members of killer immunoglobulin-like receptors (KIRs), CD94/NKG2C, NKG2D, 2B4, NKp30, NKp44, and NKp46 (Figure 1). They signal through association with either DAP10 which allows PI3-kinase activation or DAP12, CD3ζ, or FcεRIγ, which all contain immunotyrosine-based activation motifs (ITAM). In addition, adhesion molecules, such as LFA-1 and DNAM-1, are also important for the lytic function of NK cells, but they do not associate directly with the known ITAM-containing molecules [5]. The inhibitory NK receptors include the long-tail members of the KIR family, CD94/NKG2A, leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1), and killer cell lectin-like receptor subfamily G member 1 (KLRG-1) which signal via cytoplasmic immunotyrosine-based inhibitory motifs (ITIM). From a structural perspective, all known NK receptors adopt either an immunoglobulin- (Ig-) like or a C-type lectin-like receptor fold with both folds present in the activating and inhibitory receptor families. In the last decade, a large amount of structural information for many of these receptors has become available, and this review focuses on the structures of human NK cell receptors and their ligand recognition (Table 1).
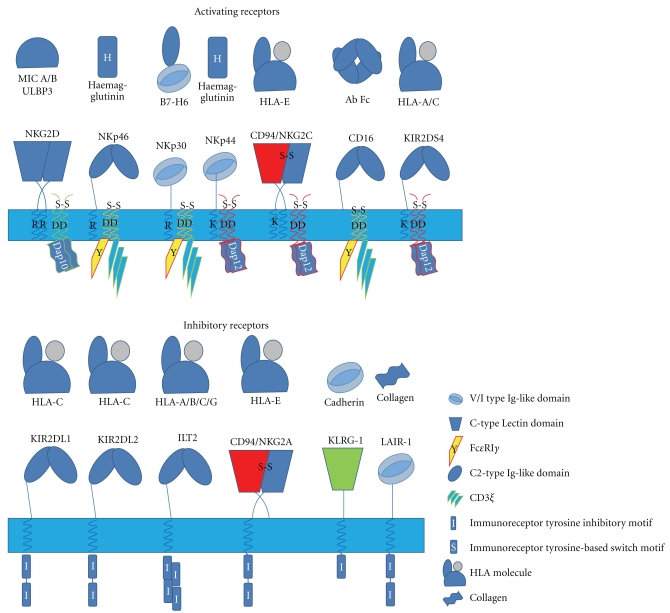
Figure 1.
Overview of the structurally characterized activating and inhibitory human NK receptors and their ligands. The upper panel displays the chief activating receptors found on human NK cells including the homodimer NKG2D, the natural cytotoxicity receptors, NKp46, NKp44, and NKp30, the c-type lectin heterodimer CD94 : NKG2C, the Fc receptor CD16, and also the KIR activating receptors. The associated cytoplasmic signaling molecules for these activating receptors which include DAP12, DAP10, and CD3ζ/FcεRIγ and the specific transmembrane residues which facilitate the interaction are also shown. The lower panel illustrates the structurally characterized inhibitory receptors found on NK cells. These include the KIR receptors KIR 2DL1 and KIR 2DL2 which have been characterized in complex with peptide bound MHC class I molecules, the heterodimeric CD94 : NKG2A which binds to peptide bound HLA-E. The C-type lectin receptor KLRG-1 which binds to cadherin and the NK cell receptor LAIR-1 molecule which binds to collagen are also shown. The known ligands for both the activating and inhibitory receptors are illustrated above their respective NK cell receptor.
Table 1.
List of human NK receptor structures.
| Structure | Resolution (Å) | PDB entry | Reference |
|---|---|---|---|
| KIR2DL1 | 1.7 | 1NKR | Fan et al. [6] |
| KIR2DL2 | 2.9 | 2DLI | Snyder et al. [10] |
| 2DL2 | |||
| KIR2DL3 | 3.0 | 1B6U | Maenaka et al. [8] |
| KIR2DS2 | 2.3 | 1M4K | Saulquin et al. [9] |
| KIR2DS4 | 2.5 | 3H8N | Graef et al. [7] |
| KIR2DL2/HLA-Cw3 | 3.0 | 1EFX | Boyington et al. [11] |
| KIR2DL1/HLA-Cw4 | 2.8 | 1IM9 | Fan et al. [12] |
| NKp46 | 2.2 | 1P6F | Foster et al. [13] |
| 1.9 | 1OLL | Ponassi et al. [14] | |
| NKp44 | 2.2 | 1HKF | Cantoni et al. [15] |
| NKp30 | 1.8 | 3NOI | Joyce et al. [16] |
| LAIR-1 | 1.8 | 3KGR | Brondijk et al., 2010 |
| CD94 | 2.6 | 1B6E | Boyington et al. [17] |
| CD94/NKG2A | 2.5 | 3BDW | Sullivan et al. [18] |
| CD94/NKG2A/HLA-E | 2.5 | 3CDG | Petrie et al. [19] |
| 3.4 | 3CII | Kaiser et al. [20] | |
| NKG2D | 1.95 | 1HQ8 | Wolan et al. [21] |
| NKG2D/MICA | 2.7 | 1HYR | Li et al. [22] |
| NKG2D/ULBP3 | 2.6 | 1KCG | Radaev et al. [23] |
| NKG2D/Rae-1β | 3.5 | 1JSK | Li et al. [24] |
| KLRG1/E-cadherin | 1.8 | 3FF7 | Li et al. [25] |
| DAP12/NKG2C | NMR | 2L35 | Call et al. [26] |
2. Immunoglobulin-Like NK Receptors
2.1. Structure of KIR
Shortly after the identification of inhibitory NK cell receptors and their MHC ligands, questions regarding the mechanisms of ligand recognition were raised. Prior to the NK cell receptor ligand findings, T cell receptors were the only MHC binding molecules. Characterization of the TCR-MHC recognition mechanism was a central tenet in molecular immunology. The fundamental principle of TCR-MHC recognition is based on a number of factors which include the presentation of peptides by a MHC molecule and the polymorphic MHC residues which together interact with the TCR. In addition, the TCR uses complementarity determining regions (CDR) to recognize MHC molecules, in a similar manner to that seen with antibody ligand interactions. These CDR regions are the result of gene rearrangement and recombination, thus essentially allowing an unlimited number of TCR specificities. This diverse receptor repertoire is necessary to recognize variable pathogenic peptides and the presenting polymorphic MHC molecules which altogether enable exquisite TCR-MHC allele specificities.
The identification of NK receptors which could bind to MHC molecules immediately challenged our understanding of MHC recognition. While the potential T cell receptor repertoire can be greater than 106 molecules, NK receptors are germ-line encoded and there are less than a dozen KIR molecules in a given individual. A number of questions arose such as how can such a small number of KIRs recognize a much larger number of peptide-MHC molecules? While the KIR genes are also members of the Ig super family, could they form TCR-like structures and recognize MHC molecules in a similar fashion to the TCRs even though the putative CDRs are nonvariable?
Broadly speaking, there are overall large differences between the size and structures of TCR molecules compared to the KIR molecules. TCR molecules are made up of a heterodimer of an alpha and beta chain each constituting ~200 amino acids in length and peptide-MHC recognition is dictated by both the alpha and beta chains while in contrast KIR molecules are monomeric consisting of ~200 amino acids in length. To date, there are five crystal structures available for KIR family members, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DS2, and KIR2DS4 [6–10]. All structures display essentially the same fold with two C2-type Ig-like domains, each made up of a β-sandwich with strands ABE packed against C′CFGA′ (Figure 2; note that in contrast to C2-type Ig-domains, a C1-type Ig-like domain is made up of ABED strands stacking against CFG strands). The two Ig-like domains, named D1 and D2, exhibit a tilted side by side arrangement with a major hinge region, which results in a substantial surface buried area between the D1 and D2 domains. The hinge angle observed in the structure of KIR2DL1 was about 65°, substantially smaller than those in the other four structures, which were about 80°, which indicated that the hinge angle might be important for ligand binding. However, the hinge angle of KIR2DL1 in the presence of bound HLA molecule was found to be similar to that of KIR2DL2, suggesting that the smaller hinge angle observed in the ligand-free structure of KIR 2DL1 is likely influenced by crystal packing. Many of the hinge residues are conserved among members of the KIR family, including the three domain KIRs, suggesting they all share the same relative domain arrangement. It is worth noting, however, that there is no structure available at present for any member of the KIR3D receptors which differ from the KIR2D molecules by the presence of an additional D0 domain.
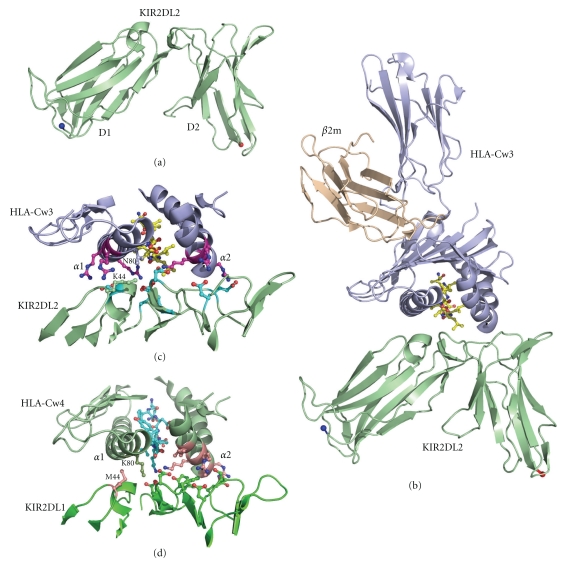
Figure 2.
Structure of inhibitory KIR molecules. (a) Structure of KIR2DL2 which is made up of two C2-like domains is shown in cartoon representation with the N-terminus and C-terminus described by a blue and red sphere, respectively. (b) Structure of KIR2DL2 in complex with the MHC class I molecule HLA-Cw3 and β2m shown in cartoon representation with bound peptide shown in ball and stick representation with yellow carbon atoms. (c) Closeup of the HLA-Cw3 : KIR2DL2 interface with interacting residues displayed in ball and stick representation. The bound peptide is shown with yellow colored carbon atoms while the interacting residues from HLA-Cw3 are shown in purple and those from the KIR molecule are shown in off-white coloring. The critical residues for HLA-KIR specificity, K44 from KIR 2DL2 and N80 from HLA Cw3 are shown in green and light-blue, respectively. (d) Closeup of the HLA-Cw4 : KIR2DL1 interface with interacting residues displayed in ball and stick representation. The bound peptide is shown with white carbon atoms while the interacting residues from HLA-Cw4 have green carbon atoms and those from KIR 2DL1 have yellow carbon atoms. The critical residues for HLA-KIR specificity, M44 from KIR 2DL1, and K80 from HLA Cw4 are shown in dark green and light green, respectively.
2.2. KIR Recognition of MHC Molecules
Understanding KIR recognition of MHC molecules through the mapping of mutational data on the KIR structure suggested that critical MHC contact regions were not at the tip of the KIR D1 domain as is the case with TCR receptor binding, rather they were located near the receptor hinge region [6, 10]. Furthermore, while KIRs were known to recognize degenerate MHC ligands, the ligand recognition was not entirely promiscuous, but in fact rather specific. That is, KIR2DL2 has C1-type specificity (KIR molecule contains Lys-44 and binds to Asn-80 residue in HLA-molecule) recognizing HLA-Cw1, 3, 7, and 8. In contrast, KIR2DL1 has C2-type specificity (KIR molecule contains Met-44 and binds to Lys-80 residue in HLA molecule) binding to the ligands HLA-Cw2, 4, 5, 6, and 15. How can MHC molecules with their polymorphic heavy chains which are necessary for presenting variable peptides, and whose recognition requires variable regions of TCR, serve as ligands to nonvariable KIR receptors? These principle defining questions prompted an intense effort to resolve the structure of KIR in complex with HLA ligands.
The first KIR-MHC complex structure was determined between KIR2DL2 and HLA-Cw3 bound to a nonamer self-peptide from human importin-α1 [11]. Subsequently, the structure of KIR2DL1 in complex with HLA-Cw4 was also determined [12]. Since the two KIR receptors recognize different class I MHC allotypes, the structure solution of both KIR-MHC complexes provided a rather complete and complementary structural view of both the overall KIR-MHC recognition and the specific interactions critical to their class I MHC specificities. The overall KIR binding mode used for HLA recognition is very similar between the two complexes. Both KIR2DL1 and KIR2DL2 bind to the peptide-binding groove of the MHC molecules using the receptor D1 and D2 domains which interact with the α1- and α2-helices of the MHC heavy chain, respectively (Figures 2(c) and 2(d)). Although many detailed interface interactions differ between the two KIR-MHC complexes, both KIRs recognize their MHC ligands through primarily salt bridges and charge complementarity rather than hydrophobic interactions and shape complementarity. KIR receptors contact primarily nonpolymorphic HLA residues on both the α1- and α2-helices. The molecular mechanism of the C1- and C2 allotype specificities of KIR also became apparent. KIR2DL2 can be switched to recognize C2 ligands by just one amino acid mutation at residue 44 [27]. Similarly, the C1 and C2 type class I ligand engagement appeared to be controlled by residue 80 on the MHC heavy chain [28]. The structures suggest that this specificity switch is due to primarily the compatibility of one hydrogen bond between Lys 44 of KIR 2DL2 and Asn 80 of HLA-Cw3 [11].
The overall KIR docking orientation on the MHC molecule is very similar to that of TCR with the D1 and D2 domains of KIRs occupying equivalent positions to the Vα and Vβ of a TCR. However, KIR receptor binding to the peptide is altered in comparison to TCR binding. Typically peptides which are bound to the MHC class I molecules are 8–10 amino acids in size and they are numbered from N-terminus to C-terminus with the first peptide amino acid named P1 and so on. TCR binding interactions with the peptide are primarily centered on the P4-P5 amino acids and are highly specific while KIR binding is shifted towards the C-terminus of the bound peptide interacting with the P7 and P8 residues of the peptide. This results in a mutually exclusive region of peptide binding between KIR and TCR but also some level of overlap. This partial overlap between KIR and TCR is potentially important to avoid conflicting simultaneous recognition by both the activating T and inhibitory NK cell receptors. The nonoverlapping peptide regions used in KIR and TCR binding mechanisms allows a TCR to evolve and mature against the variable parts of presented peptides and also the polymorphic MHC residues while KIR maintains a dual recognition mechanism of both the nonpolymorphic MHC residues and bound self-peptides via its specific binding mechanism ultimately resulting in inhibition of NK cell activation pathways.
The peptide sensitivity of KIR-MHC recognition remains an important issue. While the structures show that KIR receptors either directly or through water molecules interact with the MHC bound peptide, the KIR-peptide interactions only serve to constrain the type of residues placed at the P7 and P8 position of the peptide instead of formally defining specific contacts. In the case of KIR2DL2, the receptor and peptide contact requires a hydrophobic residue at the P7 and the presence of a small amino acid, such as an Ala or Ser residue at the P8 position [11]. Whether such constraints contribute to KIR differentiation of self versus pathogenic peptides remains an open question. Since the peptide positions recognized by KIR are not involved in TCR binding, it is thus likely that a self-peptide presenting an MHC ligand of KIR can also present pathogenic peptides for TCR. In other words, MHC molecules can evolve simultaneously with abilities both to rapidly mutate their polymorphic regions to match and present pathogenic antigens for T cell-mediated immunity and also to maintain the nonvariant regions for self peptide binding and recognition by KIR for NK cell mediated immune protection.
2.3. Activating Killer Ig-Like Receptors
While the inhibitory group of KIR molecules has been extensively studied, the function and ligands of the activating KIR molecules are less well understood. Activating KIR molecules occur less commonly than the inhibitory KIR receptors and are comprised of the two domain KIR2DS family and the three domain KIR3DS family. KIR2DS1 has similar binding specificity to KIR2DL1 in that both bind to HLA-C2 but KIR2DS1 has a much lower affinity for this receptor. Illustrating the lack of understanding of these receptors is the fact that the ligands for KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1 are currently unknown.
Sequence comparison between activating and inhibitory ligands indicates that the activating and inhibitory molecules can be shockingly close in sequence identity to each other but the most minimal changes still result in differences in ligand binding specificity and affinity. One example is the case of KIR2DS2 in which the ligand specificity is unknown while this activating molecule has been structurally characterized [9]. The sequence difference between KIR2DS2 and KIR2DL3 is only two amino acid changes with Tyr45 and Glu35 found in KIR2DS2 as opposed to Phe45 and Gln35 in KIR2DL3. The ligand for KIR2DL3 is known to be HLA-C molecules but KIR2DS2 has undetectable binding affinity for these molecules. KIR2DS2 differs in sequence from KIR2DL2 by four amino acids as follows, P16R, Y45F, R148C, and T200I. The F45Y change in KIR2DS2 does not lead to any steric clashes based on modeling of bound HLA molecule but it will clearly disrupt extensive hydrophobic interactions which are seen in the 2DL2 structure perhaps explaining the absence of HLA-C binding (Figures 3(b) and 3(f)).
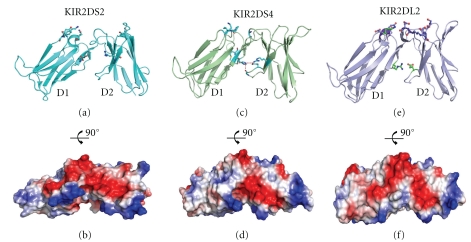
Figure 3.
Structure of activating KIR receptors. (a) KIR2DS2 shown in cartoon representation with residues likely important for ligand binding and also the hinge region residues shown in ball and stick representation. (b) Charged surface representation of the membrane distal face (binding face) of KIR2DS2. (c) KIR2DS4 shown in cartoon representation with ligand binding and hinge region residues shown in ball and stick representation. (d) Charged surface representation of the membrane distal face (binding face) of KIR2DS4. (e) For comparative purposes, the inhibitory receptor KIR2DL2 is shown in cartoon representation with ligand binding and hinge region residues shown in ball and stick representation. (f) Charged surface representation of the membrane distal face (binding face) of the inhibitory receptor KIR2DL2.
A second activating KIR, KIR2DS4, has recently been characterized as an HLA class I receptor which binds specifically to subsets of C1+ and C2+ HLA-C and also to HLA-A*11 molecules [7]. The presence of activating KIR molecules appears to occur late in evolution and only KIR2DS4 has a homologue in any other species [29]. The structure of this activating KIR molecule KIR2DS4 has also been determined [7]; (Figure 3). Overall, the structure of KIR2DS4 is very similar to other KIR molecules as judged by very low rmsd values (KIR2DL1 bound structure: rmsd 1.04 Å; KIR2DL2 bound structure: rmsd 1.12 Å; KIR2DS2: rmsd 2.37 Å). KIR2DS4 D1/D2 domains form an angle of 69° which is similar to the unbound KIR2DL1 structure (66°) but contrasts with the KIR2DL2 (81°) and KIR2DL3 (78°) structures. Further analysis of the hinge region residues illustrates a core hydrophobic region which is made up of 13 amino acids and is very similar to the inhibitory KIR molecules. Of these residues, only residue 102 shows some variability amongst the KIR family, and this has been shown to lead to changes in HLA specificity implying the hinge region hydrophobic core can indirectly impact ligand specificity and binding. Comparison of KIR2DS4 with other KIR molecules indicates that the main structural changes include the altered orientation of residues 43–45 away from the ligand binding area which results in the critical Lys44 of inhibitory KIRs lying ~2.5 Å away from the location observed in KIR2DL2/3. A second critical region is the Pro71-Val72 region which differs from the KIR2DL2 Gln71-Asp72 residues and is likely one of the main specificity components for KIR2DS4.
The presence of activating receptors with such close ligand specificity to the inhibitory KIR molecules is somewhat perplexing, but analysis of the presence of these genes in different populations indicates that KIR2DS4 may play a role in EBV control within Southeast Asian populations [30]. There is also support for the role of activating receptors in the control of cytomegalovirus infection following kidney transplantation [31]. The presence of paired or balancing KIR receptors may exist to prevent viral mutations which would result in uncontrolled infections. An additional role of activating KIR receptors in embryo implantation and the initial stages of pregnancy has been illuminated by Moffett and coworkers [32]. Their studies indicate that the presence of the activating KIR2DS1 receptor may influence embryo implantation and prevent reproductive failure.
2.4. Natural Cytotoxicity Receptors
The natural cytotoxicity receptors have been identified as the effector molecules responsible for the majority of NK driven cytotoxicity against tumor cells or virally infected cells with blocking of these receptors resulting in significantly decreased NK cell killing [33]. The NCRs are made up of three Ig-like proteins named NKp46, NKp30, and NKp44. NKp46 and NKp30 are constitutively expressed on NK cells while NKp44 is expressed upon IL-2 driven activation of NK cells. To date, all three proteins have been structurally characterized (Figure 4) but some controversy still surrounds their ligands due to the absence of ligand bound structures.
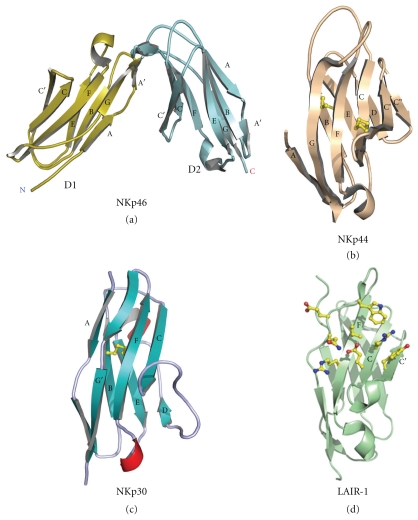
Figure 4.
Structures of the natural cytotoxicity Receptors in cartoon representation; (a) NKp46, (b) NKp30 (c) NKp44. (d) Structure of human LAIR-1 with collagen binding residues shown in ball and stick representation.
NKp46 was first identified by Moretta and coworkers in 1997 as a cell surface receptor expressed on freshly isolated and activated NK cells and which plays a pivotal role in natural cytotoxicity and targeting of transformed cells [34]. NKp46 is critical to NK cell activation in response to Newcastle disease virus [35], primary tumor melanocytes [36], urothelial cancers [37], medulloblastoma [38], myeloma cancer [39], filovirus infected dendritic cells [40], Herpes simplex virus [41], vaccinia virus [42], influenza virus, parainfluenza and sendai viruses [43], and the response to mycobacterium tuberculosis infected monocytes [44]. NKp46 is a type I membrane protein made up of two N-terminal Ig-like extracellular domains, a 40 amino acid linker region, single transmembrane domain, and a short highly charged 25 amino acid cytoplasmic region. NKp46 contains three glycosylation sites, two which are located in the 40 amino acid stalk region and one in the D2 domain at Thr 225. NKp46 lacks an activating cytoplasmic component but through a transmembrane Arg residue associates with CD3ζ and FcεRIγ which can transmit a cell activating signal via their multiple ITAM containing domains. A number of viral hemagglutinin, neuraminidase-hemagglutinin proteins which become surface expressed following viral infection have been proposed as ligands. In addition, Vimentin binding by NKp46 following mycobacteria infection of monocytes has been described [44]. NKp46 binding of specific heparan sulfate proteoglycans has also been proposed as a ligand and may be critical for tumor cell killing but its relevance to virally infected cell killing is unclear [45, 46].
The structure of NKp46 was determined by X-ray crystallography in 2003 [13, 14] revealing a structure made up of two C2-type Ig-like domains (Figure 4). Each domain is made up of eight β-strands which form two β-sheets with a typical disulphide bond formed between each sheet maintaining the Ig-domain fold. The two domains are maintained at an angle of 85° respective to each other by a large number of hydrophobic interactions and extensive hydrogen bonding. An area of 1021 Å is buried between the two NKp46 domains illustrating the extensive interactions and within the core region of the domain interactions, 11 of 18 amino acids are conserved in NKp46 sequences from diverse species. Although associated by function with NKp30 and NKp44, NKp46 shows very low homology to the other natural cytotoxicity receptors (Figure 4). The most structurally similar molecules to NKp46 include the KIR family of proteins, ILT2 ([47] rmsd: 2.2 Å, hinge angle: 86°), glycoprotein VI ([48] rmsd: 1.6 Å, hinge angle: 90°), and also FcαRI ([49] CD89; rmsd: 2.4 Å; hinge angle: 92°). The majority of these homologous molecules have been structurally defined in complex with their respective ligands. All of these molecules have a very similar overall fold made up of two C2 type Ig-like domains and a hinge angle comparable to that observed in NKp46 (Figure 5). Altogether, the description of the binding sites of these homologous proteins suggests that NKp46 would also utilize the hinge region for ligand binding and the description of a ligand binding site as determined from structural studies or large-scale mutagenesis is eagerly anticipated.
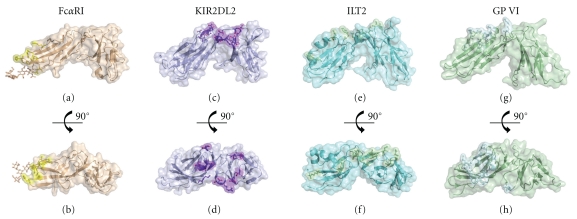
Figure 5.
Structures of NKp46 homologous proteins ((a), (b)) FcαRI, ((c), (d)) KIR2DL2, ((e), (f)) ILT2, and ((g), (h)) GP VI shown in cartoon representation with transparent surface also shown. The known ligand binding residues of these molecules are given in ball and stick representation with the surface of these amino acids also highlighted. KIR binding to MHC molecules utilizes a number of residues found in the AB, CC′, EF loop regions of D1, the GA hinge region between D1 and D2, and the BC loop and FG loop regions of D2. ILT-2 uses residues located in the C strand, the CE loop region, EF loop region, and G strand of D1 while also using residues located in the BC loop region of D2. GP VI utilizes residues in the CC′ loop, C strand, CE loop, E strand, EF loop, and F strand of D1 while also using the FG loop region of D2. CD89 uses residues from the BC loop, C′ strand, C′E loop, and the FG loop of D1 in a side-on mechanism of binding in contrast to the other homologous proteins which all utilize the membrane distal face of the receptor for ligand binding. The lower panel shows the respective molecules in an “above-cell view” orientation which also highlights their ligand binding sites.
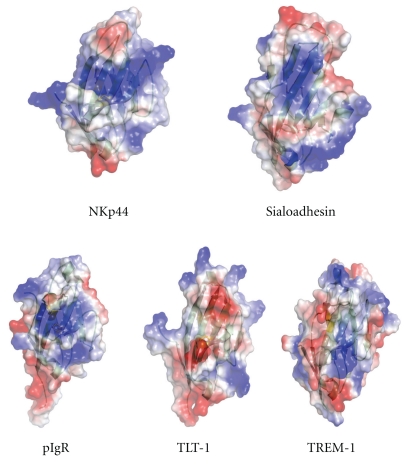
NKp44 is a natural cytotoxicity receptor which is found on activated NK cells and leads to enhanced killing of both tumor cells and virally infected cells [50–52]. NKp44 is made up of a single extracellular IgV domain with a 64 amino acid stalk region which contains a number of glycosylation sites, a transmembrane region containing a lys residue, and a short cytoplasmic domain. Following NKp44 ligand binding, NK cell activation signals are transduced via DAP12 which associates with NKp44 through its transmembrane domain. The crystal structure of the extracellular domain of NKp44 displays a compact V domain structure [15]. It is made up of two typical β-sheets constructing the Ig-V domain structure. Unique features of this structure include the presence of a second disulphide bond which in combination with the atypical orientation of the equivalent CDR3 loop region creates a large grooved area on one face of the protein. This groove is also positively charged and has been proposed as a ligand binding site. The NKp44 structure is homologous to a number of other Ig-like structures including TREM-1 (triggering receptor expressed on myeloid cells; rmsd: 2.0 Å) [53, 54], TREM-like transcript-1 (rmsd: 1.7 Å) [55], poly Ig receptor (rmsd: 2.1 Å) [56], sialoadhesin (rmsd: 2.4 Å) [57], and IREM-1 (inhibitory receptor expressed on myeloid cells, rmsd: 2.58 Å) [58] (Figure 6).
Figure 6.
Charged surface representation of NKp44 and closely related homologues sialoadhesin, pIgR, TLT-1, and TREM-1. The orientation of each molecule is identical to that of NKp44 shown in Figure 4 and allows a view of the G, F, C, and C′ sheets of each molecule.
NKp44 binding to influenza hemagglutinin and other viral hemagglutinin-neuraminidase proteins due to the presence of sialic acid in the NKp44 stalk region have been presented in the literature [35, 50]. Since NKp44 is also involved in the increased lysis of tumor cells, it will likely bind to a cellular ligand utilizing its Ig-like domain. In addition to transformed cells, NKp44 has also been shown to bind to the cell surface of mycobacteria and other bacteria which may contain a similar or homologous ligand to that found on human cells [59].
The most recently discovered NCR is NKp30 which was identified by Moretta and coworkers [60]. NKp30 has been shown to be the dominant activating receptor responsible for the lysis of a number of tumor cell types [60]. In addition, NKp30 has been shown to cause the activation and expansion of resting NK cells upon interaction with DCs and to cause the death of imDCs [61]. NKp30 has a single extracellular Ig-like domain with a short stalk region (~5 aa), and a transmembrane domain which associates with CD3ζ homodimers through a charged transmembrane interaction. The physiological ligands for NKp30 remain a controversial issue as a large number of NKp30 interacting molecules have been proposed. These include a human cytomegalovirus tegument protein pp65 [62], duffy-binding-like- (DBL-) 1α of Plasmodium falciparum erythrocyte membrane protein-1 (PfEMP-1) [63], leukocyte antigen-B-associated transcript 3 (BAT3) [64, 65], and a group of heparan sulfate/heparin molecules [45]. Most recently, NKp30 was shown to recognize a B7 family homolog (B7-H6) as its ligand [66]. Unlike the other protein ligands proposed, B7-H6 is expressed on a number of tumor cell lines, such as K562 and Raji, as well as on primary cancer cells. However, the lack of structural information for NKp30 makes it difficult to parse out the most important ligand involved in cancer destruction from those proposed. The structure of NKp30 was recently, determined within our group and is shown to be a single I-type Ig-like domain [16]. The structure displays strong structural similarity to the CD28 family of receptors. Mutagenesis work carried out on NKp30 and binding studies on the B7H6 ligand indicates that binding is very similar to that observed for PD-1/PD-L1 interactions and involves residues found on the upper portion of the F and C strands.
2.5. The Structure of LAIR-1
The inhibitory receptor LAIR-1 is found on a large number of immune cells including NK cells and is specifically involved in collagen recognition [67] and increasing the threshold for NK cell activation. In contrast to LAIR-1, glycoprotein VI which is found on platelets also binds to collagen and through its association with the FcRγ chain, an activation signal is induced which leads to increased thrombosis. Recently the crystal structure of LAIR-1 was determined and the collagen binding site was mapped (Brondijk et al., 2010). LAIR-1 is made up of a single E-type Ig-like fold made up of two β-sheets consisting of β-strands ABE and A′GFCC′ with high structural homology to the D1 domain of KIR2DL2 (rmsd: 1.34 Å), ILT11 (rmsd: 1.36 Å), and GP VI (rmsd: 1.09 Å). Using mutagenesis and binding experiments in addition to NMR residue assignment signal shifts, it was possible to map the collagen binding site onto the A′FGCC′β-sheet specifically involving residues Arg59 and Glu61 from the LAIR-1 F strand and Trp109 and Glu111 from the G strand (Figure 4(d)). The collagen binding site of LAIR-1 is quite different to the site proposed for GP VI, and this may allow the design of molecules which can act as immune modulators by blocking LAIR-1 or affecting thrombosis via GP VI collagen binding.
3. C-Type Lectin-Like NK Receptors
Many NK receptor structures exhibit a C-type lectin fold, similar to that found among carbohydrate binding animal lectins and whose function requires bound calcium ions. Most C-type lectin-like NK receptors recognize membrane-bound protein ligands independent of carbohydrates and do not require calcium for their ligand recognition. Examples include CD69, CD94/NKG2 receptors which recognize HLA-E, NKG2D which recognizes stress-induced and tumor ligands such as MICA/B and ULBPs. In mouse, the classical class I MHC recognizing receptors, the Ly49 family receptors as well as Nkrp1 receptors, are also C-type lectin-like molecules. Within this review, we shall focus on the structures of human CD94/NKG2, NKG2D and their mechanisms of ligand recognition.
3.1. The Structure of the CD94/NKG2 Receptor and Its Recognition of HLA-E
CD94 exists primarily in a heterodimeric form with NKG2A, C, and E on the cell surface. Depending on the associated NKG2 subunit, CD94/NKG2 can function as either an activating (NKG2C, and E) or an inhibitory (NKG2A) receptor. The inhibitory form has intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIM), while the activating forms contain a positive transmembrane charged residue which facilitates interaction and signaling through the ITAM-containing DAP12 molecule. HLA-E was identified as the ligand for CD94/NKG2 [68–70]. Since C-type lectins are historically associated with carbohydrate binding, it was not clear how CD94/NKG2 would recognize HLA-E and whether the recognition involved the glycosylation of the class I MHC. In addition, the only class I binding receptors known at the time were T cell receptors, which are members of the immunoglobulin superfamily. Thus, part of the need to solve the CD94 structure was to understand how a C-type lectin fold receptor could recognize an MHC ligand. The crystal structure of a homodimeric human CD94 showed that the receptor maintained a canonical C-type lectin fold except that one of the two α-helices in the canonical C-type lectin fold (helix 2) was missing and replaced with a loop in the structure of CD94 (Figure 7(a)) [17]. More importantly, the CD94 structure showed that the receptor is missing four of the five calcium binding ligands and does not have a bound calcium ion in its putative calcium binding site. Thus, the structural identification suggested that the C-type lectin-like NK receptors functioned differently from a typical lectin calcium and carbohydrate binding molecule.
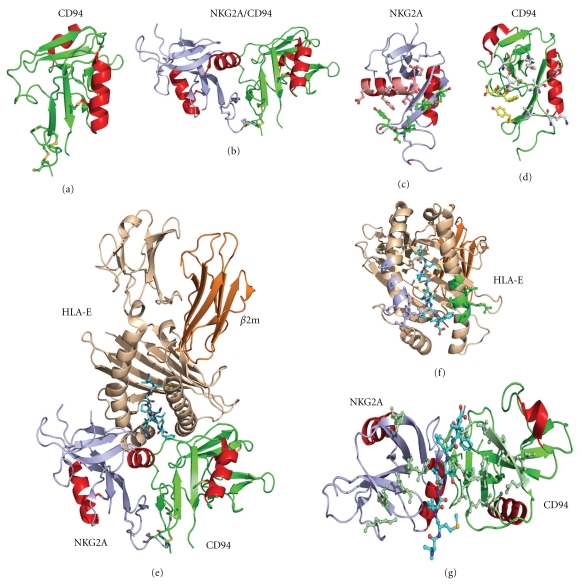
Figure 7.
Structure of CD94 and associated complexes. (a) CD94 monomer shown in cartoon representation. (b) NKG2A/CD94 heterodimer structure with NKG2A shown in light blue and CD94 shown in green cartoon representation with each α-helix from both molecules colored red. (c) NKG2A shown in cartoon representation rotated 90° in relation to (7B) displaying interacting residues in ball and stick representation. Residues colored in green are part of interaction region 1 described in the text while residues colored salmon are part of region 2 also described in the text. (d) CD94 in cartoon representation rotated 90° in relation to (B) with residues colored yellow part of interaction region 1 while residues colored in grey are part of interaction region 2. (e) Cartoon representation of NKG2A/CD94 in complex with peptide bound HLA-E molecule. The bound peptide is shown in ball and stick representation with off-white carbon atoms (f) HLA-E molecule rotated 90° from (7E) illustrating the HLA-E interaction surface with residues which form bonds with NKG2A and CD94 shown in light blue coloring, and green coloring, respectively. Bound peptide is also shown in ball and stick representation. (g) NKG2A/CD94 molecule rotated 90° from (7E) showing the HLA-E interaction surface with the bound peptide shown for reference, residues which interact with HLA-E and the peptide are shown colored green in ball and stick representation.
CD94/NKG2A binding of HLA-E by NK cells is critical for the careful monitoring of MHC class I expression on healthy cells. HLA-E molecules present peptides which have been generated from digested MHC class I molecules and the CD94/NKG2A complex detects this HLA-E/MHC class I peptide complex. This mechanism serves as a double-check to ensure that MHC class I molecules are being produced by a cell in a normal manner. Upon NKG2A/CD94 ligand binding of the HLA-E/peptide complex an inhibitory signal is transduced via cytoplasmic ITIM motifs found on NKG2A. The crystal structure of CD94/NKG2A first showed us how this heterodimeric complex is constructed [18]. The dimer interface between NKG2A and CD94 is extensive (1500 Å) and is made up of largely polar interactions with salt bridge formation observed at the edge of the dimer interface and a further central hydrophobic area maintaining the dimer (Figures 7(b), 7(c), and 7(d)). CD94/NKG2A dimerization is also driven by extensive main chain interactions between the β1 strands of both molecules, thus leading to the formation of a β-sheet made up of three β-strands from one molecule and three from the adjacent molecule.
Most recently, the structure of CD94/NKG2A in complex with HLA-E with a bound HLA-G peptide has been determined [19, 20]. The structure of CD94/NKG2A does not undergo any structural rearrangements upon ligand binding, indicating that a lock and key mechanism of binding is used. NKG2A and CD94 interact with the α2 and α1 helices of HLA-E, respectively, with the presence of charge complementarity between the ligand and receptor clearly evident such that 8 salt bridges and 19 H bonds are observed; there is also a small hydrophobic patch in the ligand receptor interface. The buried surface area is very large (2100 Å) with the majority of it contributed by CD94. In addition, the surface complementarity is higher for CD94 interactions with HLA-E (0.68 versus 0.31 for NKG2A). Overall, the peptide makes up 23% of the ligand receptor interface, and again CD94 is dominant in its interactions with the peptide (80%) with CD94 interacting with the P5-Arg, P6-Thr, and P8-Phe while NKG2A only interacts with the P5-Arg.
3.2. Activating Receptor NKG2D and Its Ligand Bound Structures
The first NKG2D structure reported was that of the murine NKG2D homodimer [21], and, subsequently, both human and murine NKG2D structures were reported in complexes with their respective ligands, including ULBP3, MICA, and Rae-1β [22–24]. Similar to the CD94 receptor, the second α-helix in NKG2D is also distorted into a one-turn helix (Figure 8(a)) and the receptor does not have a functional calcium binding site, which was also demonstrated for the mouse C-type lectin-like Ly49 family of receptors [71].
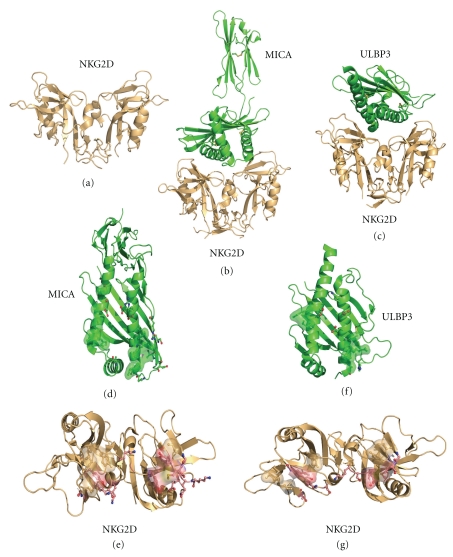
Figure 8.
Structure of NKG2D homodimer and associated complexes. (a) NKG2D homodimeric structure. (b) NKG2D in the same orientation as (a) in complex with the two domain MICA molecule. (c) NKG2D in complex with ULBP3. (d) MICA structure rotated 90° in orientation relative to (b); residues forming hydrogen bonds with NKG2D are shown in ball and stick representation with red carbon and blue nitrogen atoms while residues forming hydrophobic interactions are shown in yellow transparent surface representation. (e) NKG2D structure rotated 90° in orientation relative to (b); residues forming hydrogen bonds with MICA are shown in ball and stick representation with red carbon and blue nitrogen atoms while residues forming hydrophobic interactions are shown in green transparent surface representation. (f) ULBP3 structure rotated 90° in orientation relative to (c); residues forming hydrogen bonds with NKG2D are shown in ball and stick representation with red carbon and blue nitrogen atoms while residues forming hydrophobic interactions are shown in yellow transparent surface representation. (g) NKG2D structure rotated 90° in orientation relative to (b); residues forming hydrogen bonds with ULBP3 are shown in ball and stick representation with red carbon and blue nitrogen atoms while residues forming hydrophobic interactions are shown in green transparent surface representation.
NKG2D binds to a number of proteins which have high structural homology to the α1 and α2 domains of MHC class I molecules. Although the overall orientation of the NKG2D ligand bound structures are highly similar (Figure 8), there are very specific differences in the residues which interact with MICA compared to those seen in the ULBP3 complex. The homodimeric structure of NKG2D forms a central concave surface on its membrane distal face which allows the elevated central α-helices of the MHC class I homologues to bind NKG2D. The major binding region between the ligands MICA and ULBP3 and NKG2D receptor involves the c-terminal part of the α1 helix and the n-terminal region of the α3 helix of the ligands (Figure 8). NKG2D utilizes identical loop regions from both of its subunits to bind the ligand, and there is minimal structural reorganization following ligand binding. The binding interface between NKG2D ligands is extensive (1930 Å ULBP3 complex and 2180 Å MICA complex) and also quite specific (shape complementarity: 0.65 ULBP3 complex; 0.72 MICA complex) resulting in strong binding between the receptor and ligands. Analysis of the orientation of the two complexes indicates there is a subtle difference in the angle of orientation of ULBP3 and MICA on NKG2D resulting in a 6° difference. Interestingly, due to this slight change in the binding, it results in a difference in the number of hydrogen bonds between the complexes even though the strongest hydrogen bonds are conserved. In addition to the hydrogen bonding pattern, there is also an extensive hydrophobic region involved in ligand binding.
3.3. Other C-Type Lectin-Like NK Cell Receptors
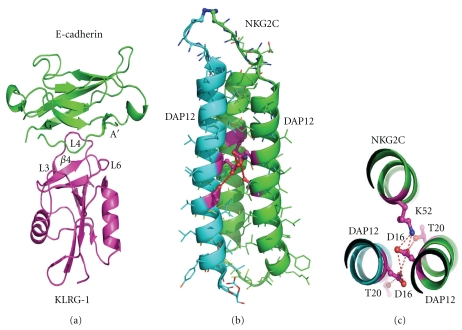
In addition to the well-studied inhibitory molecules KIR proteins and also the collagen binding inhibitory molecule LAIR-1, killer cell lectin-like receptor G1 (KLRG1) is an additional inhibitory molecule [72]. KLRG1 contains an extracellular c-type lectin-like receptor, a 19 amino acid stalk region, a single transmembrane domain, and a cytoplasmic ITIM domain allowing the elicitation of an inhibitory signal within NK cells [73]. KLRG1 is found on the vast majority of NK cells, and, interestingly, surface expression is highly upregulated following viral or parasitic infection [74]. Most recently KLRG1 has been shown to interact with E-, N, and R-cadherins [75]. E-cadherin is a critical molecule found in the junction between epithelial cells where it mediates tight contacts while N- and R-cadherins perform similar functions with different cell types. In the presence of KLRG1/E-cadherin interactions, NK cell lysis is prevented. Relevant to NK surveillance of transformed cells, epithelial cells which have become malignant typically have reduced cadherin expression which in turn promotes tumor invasiveness and subsequent metastasis [76]. The crystal structure of KLRG1 in complex with E-cadherin has recently been determined and provides a clear understanding of KLRG interaction with E-cadherin [25]. KLRG-1 binds to E-cadherin with a relatively low affinity (100–200 μM). KLRG1 structure is similar to other c-type lectin domains such as NKG2D made up of two alpha helices, two antiparallel beta sheets and the presence of intermolecular disulphide bonds.
KLRG1 binds to E-cadherin by using three loop regions (L3, L4, and L6) and β strand 4 which interact with the cadherin short A′ and G β strands similar to other CTLD receptors such as NKG2D with the binding characterized by exemplary surface complementarity and an extensive hydrogen bonding network (Figure 9(a)). The low affinity of KLRG-1 to a single cadherin domain may be overcome by multiple copies of KLRG-1 binding to the multiple domains of E-cadherin.
Figure 9.
(a) Structure of KLRG1 in complex with E-cadherin shown in cartoon representation with the critical KLRG-1 loop regions which interact with the E-cadherin A′ and G sheet regions labeled. (b) Cartoon representation of the heterotrimeric NKG2C DAP12 transmembrane complex with interacting residues shown in ball and stick representation with bonding network shown by dashed lines. (c) View from above the membrane showing a closeup of the interacting residues and bonding interactions.
4. Transmembrane Signaling Complexes
Following ligand binding NK cell receptors initiate a signaling cascade which results in cell activation or inhibition. Critical to NK cell activation is the association of the transmembrane region of NK receptors with a signaling molecule containing an ITAM motif or association with adaptor molecules with a YXXM motif. The association of a NK activating receptor with an ITAM-containing adaptor molecule is mediated through transmembrane interactions and has recently been structurally characterized [26]. Utilizing solution NMR, the heterotrimeric structure of the transmembrane association of DAP12 with NKG2C was determined. The signaling assembly made up of a sophisticated electrostatic network involves a pair of aspartate residues from the DAP12 dimer, two adjacent threonine residues which together form the specific interaction with the lysine residue of NKG2C (Figures 9(b) and 9(c)). Mutagenesis and sequence analysis indicates that a very similar mechanism of heterotrimeric association occurs in the NKG2D-DAP10 complex formation. The interaction of CD16 with the CD3ζ and FcεRIγ as well as a number of other NK cell receptor and adaptor complexes is currently not structurally characterized, and this information will provide detailed understanding of the critical passage from ligand binding to cell activation.
This review has outlined the current state of human NK cell receptor structural biology. The structures which have been determined allow us to gain an understanding of the complex mechanisms involved in ligand recognition and NK cell activation and inhibition. Further high-resolution structure studies are likely over the next few years and remain eagerly anticipated in an effort to answer a number of burning questions within NK cell biology.
Acknowledgments
This paper is funded by the intramural research funding of National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- 1.Biassoni R, Bottino C, Cantoni C, Moretta A. Current Protocols in Immunology. chapter 14: unit 14.10. 2002. Human natural killer receptors and their ligands. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annual Review of Immunology. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nature Immunology. 2008;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama WM. Natural killer cell immune responses. Immunologic Research. 2005;32(1–3):317–325. doi: 10.1385/IR:32:1-3:317. [DOI] [PubMed] [Google Scholar]
- 5.Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunological Reviews. 2006;214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan QR, Mosyak L, Winter CC, Wagtmann N, Long EO, Wiley DC. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature. 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 7.Graef T, Moesta AK, Norman PJ, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. Journal of Experimental Medicine. 2009;206(11):2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maenaka K, Juji T, Stuart DI, Jones EY. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure. 1999;7(4):391–398. doi: 10.1016/s0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- 9.Saulquin X, Gastinel LN, Vivier E. Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j) Journal of Experimental Medicine. 2003;197(7):933–938. doi: 10.1084/jem.20021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(7):3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boylngton JC, Motykat SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405(6786):537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 12.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nature Immunology. 2001;2(5):452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 13.Foster CE, Colonna M, Sun PD. Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors. The Journal of Biological Chemistry. 2003;278(46):46081–46086. doi: 10.1074/jbc.M308491200. [DOI] [PubMed] [Google Scholar]
- 14.Ponassi M, Cantoni C, Biassoni R, et al. Structure of the human NK cell triggering receptor NKp46 ectodomain. Biochemical and Biophysical Research Communications. 2003;309(2):317–323. doi: 10.1016/j.bbrc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Cantoni C, Ponassi M, Biassoni R, et al. The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure. 2003;11(6):725–734. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 16.Joyce MG, Tran P, Zhuravleva MA, Jaw J, Sun PD. Crystal structure of human natural cytotoxicity receptor NKp30 and identification of its ligand binding site. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(15):6223–6228. doi: 10.1073/pnas.1100622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyington JC, Riaz AN, Patamawenu A, Coligan JE, Brooks AG, Sun PD. Structure of CD94 reveals novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors. Immunity. 1999;10(1):75–82. doi: 10.1016/s1074-7613(00)80008-4. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan LC, Clements CS, Beddoe T, et al. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 2007;27(6):900–911. doi: 10.1016/j.immuni.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Petrie EJ, Clements CS, Lin J, et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. Journal of Experimental Medicine. 2008;205(3):725–735. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA-E. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(18):6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolan DW, Teyton L, Rudolph MG, et al. Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A. Nature Immunology. 2001;2(3):248–254. doi: 10.1038/85311. [DOI] [PubMed] [Google Scholar]
- 22.Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nature Immunology. 2001;2(5):443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 23.Radaev S, Rostro B, Brooks AG, Colonna M, Sun PD. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class i MHC-like ligand ULBP3. Immunity. 2001;15(6):1039–1049. doi: 10.1016/s1074-7613(01)00241-2. [DOI] [PubMed] [Google Scholar]
- 24.Li P, McDermott G, Strong RK. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity. 2002;16(1):77–86. doi: 10.1016/s1074-7613(02)00258-3. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Hofmann M, Wang Q, et al. Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition. Immunity. 2009;31(1):35–46. doi: 10.1016/j.immuni.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Call ME, Wucherpfennig KW, Chou JJ. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nature Immunology. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. Journal of Immunology. 1997;158(9):4026–4028. [PubMed] [Google Scholar]
- 28.Mandelboim O, Reyburn HT, Vales-Gomez M, et al. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. Journal of Experimental Medicine. 1996;184(3):913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khakoo SI, Rajalingam R, Shum BP, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12(6):687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 30.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: www.allelefrequencies.net. Tissue Antigens. 2003;61(5):403–407. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 31.Stern M, Elsasser H, Honger G, Steiger J, Schaub S, Hess C. The number of activating KIR genes inversely correlates with the rate of CMV infection/reactivation in kidney transplant recipients. American Journal of Transplantation. 2008;8(6):1312–1317. doi: 10.1111/j.1600-6143.2008.02242.x. [DOI] [PubMed] [Google Scholar]
- 32.Hiby SE, Apps R, Sharkey AM, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. Journal of Clinical Investigation. 2010;120(11):4102–4110. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moretta A, Bottino C, Vitale M, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annual Review of Immunology. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 34.Sivori S, Vitale M, Morelli L, et al. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. Journal of Experimental Medicine. 1997;186(7):1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarahian M, Watzl C, Fournier P, et al. Activation of natural killer cells by newcastle disease virus hemagglutinin-neuraminidase. Journal of Virology. 2009;83(16):8108–8121. doi: 10.1128/JVI.00211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cagnano E, Hershkovitz O, Zilka A, et al. Expression of ligands to NKp46 in benign and malignant melanocytes. Journal of Investigative Dermatology. 2008;128(4):972–979. doi: 10.1038/sj.jid.5701111. [DOI] [PubMed] [Google Scholar]
- 37.Yutkin V, Pode D, Pikarsky E, Mandelboim O. The expression level of ligands for natural killer cell receptors predicts response to bacillus Calmette-Guerin therapy: a pilot study. Journal of Urology. 2007;178(6):2660–2664. doi: 10.1016/j.juro.2007.07.118. [DOI] [PubMed] [Google Scholar]
- 38.Castriconi R, Dondero A, Negri F, et al. Both CD133+ and CD133- medulloblastoma cell lines express ligands for triggering NK receptors and are susceptible to NK-mediated cytotoxicity. European Journal of Immunology. 2007;37:3190–3196. doi: 10.1002/eji.200737546. [DOI] [PubMed] [Google Scholar]
- 39.El-Sherbiny YM, Meade JL, Holmes TD, et al. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Research. 2007;67:8444–8449. doi: 10.1158/0008-5472.CAN-06-4230. [DOI] [PubMed] [Google Scholar]
- 40.Fuller CL, Ruthel G, Warfield KL, et al. NKp30-dependent cytolysis of filovirus-infected human dendritic cells. Cellular Microbiology. 2007;9(4):962–976. doi: 10.1111/j.1462-5822.2006.00844.x. [DOI] [PubMed] [Google Scholar]
- 41.Chisholm SE, Howard K, Gómez MV, Reyburn HT. Expression of ICP0 is sufficient to trigger natural killer cell recognition of herpes simplex virus-infected cells by natural cytotoxicity receptors. Journal of Infectious Diseases. 2007;195(8):1160–1168. doi: 10.1086/512862. [DOI] [PubMed] [Google Scholar]
- 42.Chisholm SE, Reyburn HT. Recognition of vaccinia virus-infected cells by human natural killer cells depends on natural cytotoxicity receptors. Journal of Virology. 2006;80(5):2225–2233. doi: 10.1128/JVI.80.5.2225-2233.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandelboim O, Lieberman N, Lev M, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409(6823):1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 44.Garg A, Barnes PF, Porgador A, et al. Vimentin expressed on Mycobacterium tuberculosis-infected human monocytes is involved in binding to the NKp46 receptor. Journal of Immunology. 2006;177(9):6192–6198. doi: 10.4049/jimmunol.177.9.6192. [DOI] [PubMed] [Google Scholar]
- 45.Hecht ML, Rosental B, Horlacher T, et al. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. Journal of Proteome Research. 2009;8(2):712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 46.Zilka A, Landau G, Hershkovitz O, et al. Characterization of the heparin/heparan sulfate binding site of the natural cytotoxicity receptor NKp46. Biochemistry. 2005;44(44):14477–14485. doi: 10.1021/bi051241s. [DOI] [PubMed] [Google Scholar]
- 47.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nature Immunology. 2003;4(9):913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 48.Horii K, Kahn ML, Herr AB. Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI. Blood. 2006;108(3):936–942. doi: 10.1182/blood-2006-01-010215. [DOI] [PubMed] [Google Scholar]
- 49.Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structure of human Fc-alpha-RI and its complex with IgA1-Fc. Nature. 2003;423(6940):614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- 50.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. European Journal of Immunology. 2001;31(9):2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 51.Cantoni C, Bottino C, Vitale M, et al. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. Journal of Experimental Medicine. 1999;189(5):787–795. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vitale M, Bottino C, Sivori S, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. Journal of Experimental Medicine. 1998;187(12):2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelker MS, Debler EW, Wilson IA. Crystal structure of mouse triggering receptor expressed on myeloid cells 1 (TREM-1) at 1.76 A. Journal of Molecular Biology. 2004;344(5):1175–1181. doi: 10.1016/j.jmb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Radaev S, Kattah M, Rostro B, Colonna M, Sun PD. Crystal structure of the human myeloid cell activating receptor TREM-1. Structure. 2003;11(12):1527–1535. doi: 10.1016/j.str.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Gattis JL, Washington AV, Chisholm MM, et al. The structure of the extracellular domain of triggering receptor expressed on myeloid cells like transcript-1 and evidence for a naturally occurring soluble fragment. Journal of Biological Chemistry. 2006;281(19):13396–13403. doi: 10.1074/jbc.M600489200. [DOI] [PubMed] [Google Scholar]
- 56.Hamburger AE, West AP, Jr., Bjorkman PJ. Crystal structure of a polymeric immunoglobulin binding fragment of the human polymeric immunoglobulin receptor. Structure. 2004;12(11):1925–1935. doi: 10.1016/j.str.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Zaccai NR, May AP, Robinson RC, et al. Crystallographic and in silico analysis of the sialoside-binding characteristics of the Siglec sialoadhesin. Journal of Molecular Biology. 2007;365(5):1469–1479. doi: 10.1016/j.jmb.2006.10.084. [DOI] [PubMed] [Google Scholar]
- 58.Marquez JA, Galfre E, Dupeux F, Flot D, Moran O, Dimasi N. The crystal structure of the extracellular domain of the inhibitor receptor expressed on myeloid cells IREM-1. Journal of Molecular Biology. 2007;367(2):310–318. doi: 10.1016/j.jmb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Esin S, Batoni G, Counoupas C, et al. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surfaces of mycobacteria and other bacteria. Infection and Immunity. 2008;76(4):1719–1727. doi: 10.1128/IAI.00870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pende D, Parolini S, Pessino A, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. Journal of Experimental Medicine. 1999;190(10):1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Münz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. Journal of Experimental Medicine. 2002;195(3):343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnon TI, Achdout H, Levi O, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nature Immunology. 2005;6(5):515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 63.Mavoungou E, Held J, Mewono L, Kremsner PG. A duffy binding-like domain is involved in the NKp30-mediated recognition of Plasmodium falciparum-parasitized erythrocytes by natural killer cells. Journal of Infectious Diseases. 2007;195(10):1521–1531. doi: 10.1086/515579. [DOI] [PubMed] [Google Scholar]
- 64.Pogge von Strandmann E, Simhadri VR, von Tresckow B, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27(6):965–974. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Simhadri VR, Reiners KS, Hansen HP, et al. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS ONE. 2008;3(10, article e3377) doi: 10.1371/journal.pone.0003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brandt CS, Baratin M, Yi EC, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. Journal of Experimental Medicine. 2009;206(7):1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lebbink RJ, De Ruiter T, Adelmeijer J, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. Journal of Experimental Medicine. 2006;203(6):1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. Journal of Experimental Medicine. 1998;187(5):813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braud VM, Allan DS, O’Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 70.Lee N, Llano M, Carretero M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tormo J, Natarajan K, Margulles DH, Mariuzza RA. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature. 1999;402(6762):623–631. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- 72.Bryceson YT, Long EO. Line of attack: NK cell specificity and integration of signals. Current Opinion in Immunology. 2008;20(3):344–352. doi: 10.1016/j.coi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guthmann MD, Tal M, Pecht I. A secretion inhibitory signal transduction molecule on mast cells is another C-type lectin. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(20):9397–9401. doi: 10.1073/pnas.92.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voehringer D, Kaufmann M, Pircher H. Genomic structure, alternative splicing, and physical mapping of the killer cell lectin-like receptor G1 gene (KLRG1), the mouse homologue of MAFA. Immunogenetics. 2001;52(3-4):206–211. doi: 10.1007/s002510000282. [DOI] [PubMed] [Google Scholar]
- 75.Ito M, Maruyama T, Saito N, Koganei S, Yamamoto K, Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. Journal of Experimental Medicine. 2006;203(2):289–295. doi: 10.1084/jem.20051986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colonna M. Cytolytic responses: cadherins put out the fire. Journal of Experimental Medicine. 2006;203(2):261–264. doi: 10.1084/jem.20052559. [DOI] [PMC free article] [PubMed] [Google Scholar]