Abstract
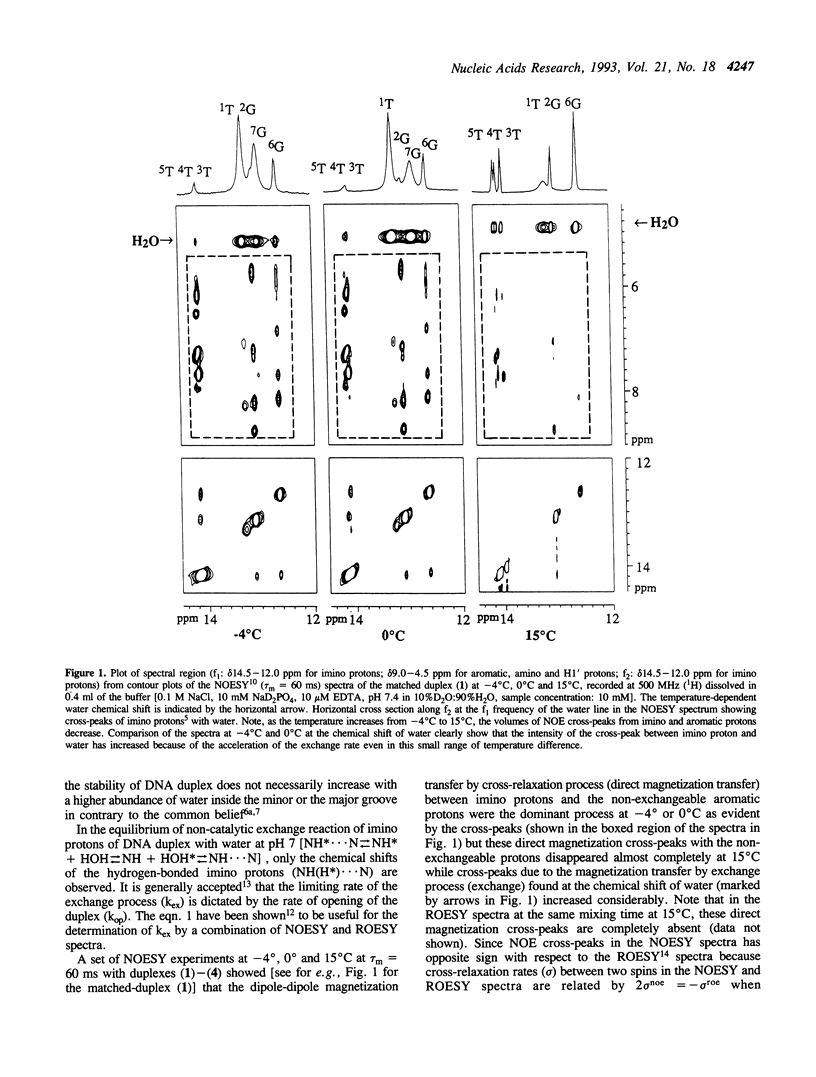
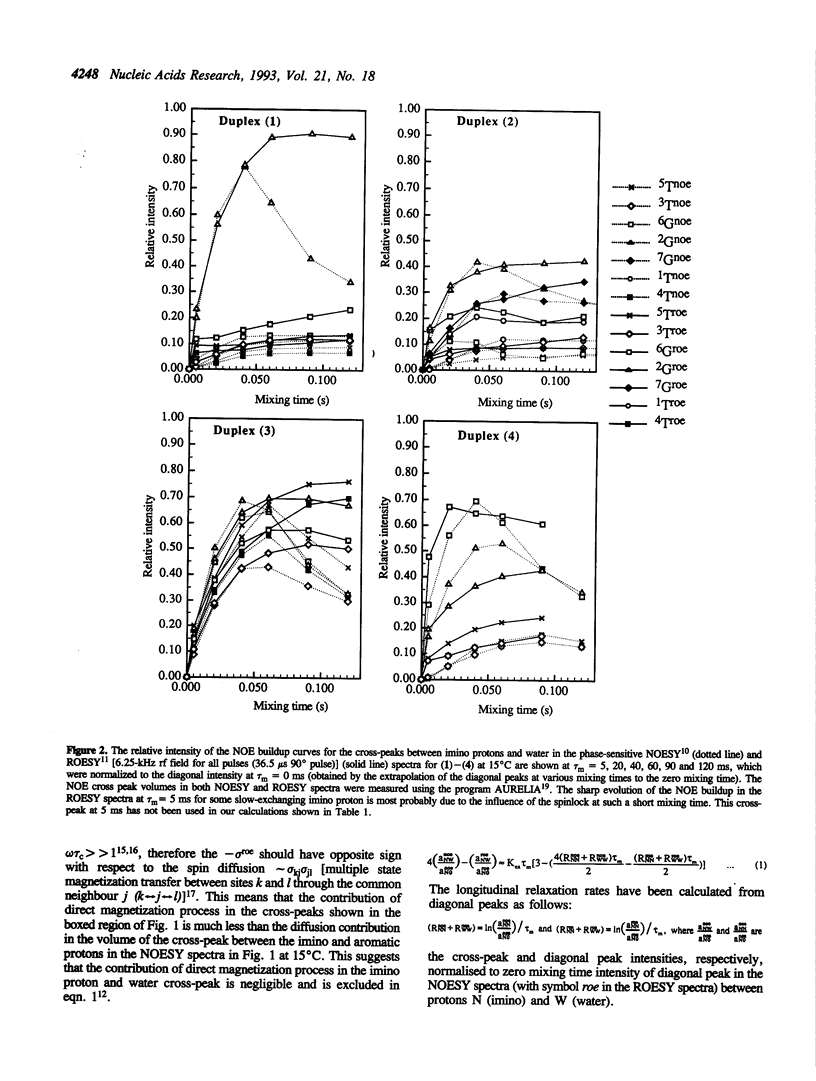
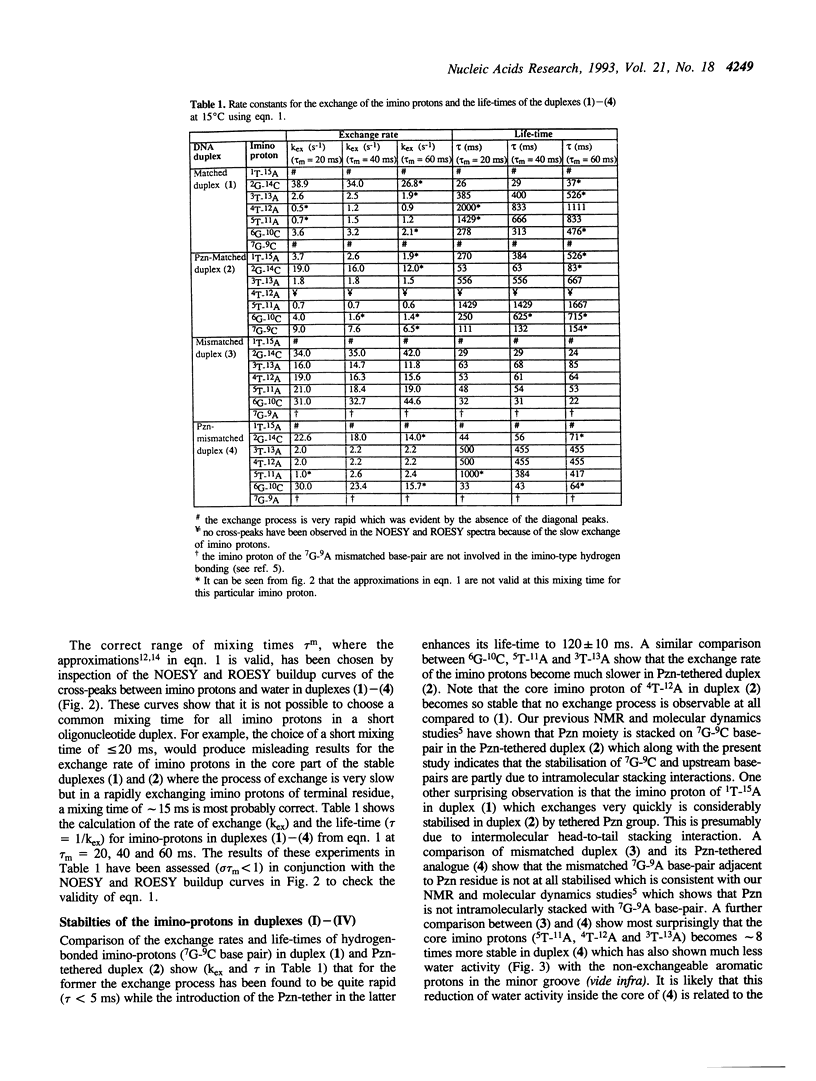
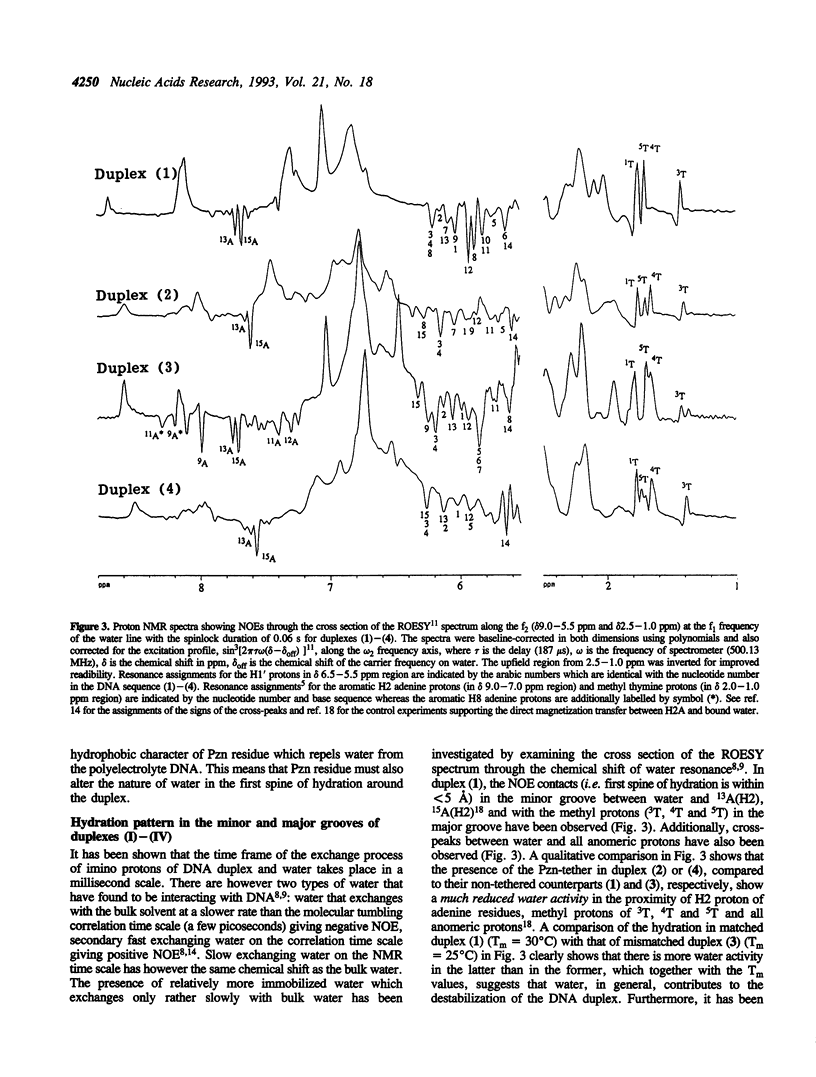
A combination of NOESY and ROESY experiments show that the higher stabilities (Tm) of phenazine tethered matched (2) and G-A mismatched (4) DNA duplexes are due to the decrease of the exchange-rate (i.e. increase of the life-time) of the imino-protons and the reduced water activity in their minor grooves compared to their non-tethered counterparts (1) and (3).
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asseline U., Toulme F., Thuong N. T., Delarue M., Montenay-Garestier T., Hélène C. Oligodeoxynucleotides covalently linked to intercalating dyes as base sequence-specific ligands. Influence of dye attachment site. EMBO J. 1984 Apr;3(4):795–800. doi: 10.1002/j.1460-2075.1984.tb01887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Brown D. G., Spink N., Skelly J. V., Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution. J Mol Biol. 1992 Aug 20;226(4):1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- Liepinsh E., Otting G., Wüthrich K. NMR observation of individual molecules of hydration water bound to DNA duplexes: direct evidence for a spine of hydration water present in aqueous solution. Nucleic Acids Res. 1992 Dec 25;20(24):6549–6553. doi: 10.1093/nar/20.24.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycksell P. O., Gräslund A., Claesens F., McLaughlin L. W., Larsson U., Rigler R. Base pair opening dynamics of a 2-aminopurine substituted Eco RI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res. 1987 Nov 11;15(21):9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltseva T., Sandström A., Ivanova I. M., Sergeyev D. S., Zarytova V. F., Chattopadhyaya J. Structural studies of the 5'-phenazinium-tethered matched and G-A-mismatched DNA duplexes by NMR spectroscopy. J Biochem Biophys Methods. 1993 May;26(2-3):173–236. doi: 10.1016/0165-022x(93)90046-q. [DOI] [PubMed] [Google Scholar]
- Narayana N., Ginell S. L., Russu I. M., Berman H. M. Crystal and molecular structure of a DNA fragment: d(CGTGAATTCACG). Biochemistry. 1991 May 7;30(18):4449–4455. doi: 10.1021/bi00232a011. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Farmer B. T., 2nd, Wüthrich K. Protein hydration studied with homonuclear 3D 1H NMR experiments. J Biomol NMR. 1991 Jul;1(2):209–215. doi: 10.1007/BF01877232. [DOI] [PubMed] [Google Scholar]
- Sun J. S., François J. C., Montenay-Garestier T., Saison-Behmoaras T., Roig V., Thuong N. T., Hélène C. Sequence-specific intercalating agents: intercalation at specific sequences on duplex DNA via major groove recognition by oligonucleotide-intercalator conjugates. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9198–9202. doi: 10.1073/pnas.86.23.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J. C., Weber I. T. Crystal structure of a B-DNA dodecamer containing inosine, d(CGCIAATTCGCG), at 2.4 A resolution and its comparison with other B-DNA dodecamers. Nucleic Acids Res. 1992 Oct 25;20(20):5457–5464. doi: 10.1093/nar/20.20.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Quintana J., Dickerson R. E. Alternative structures for alternating poly(dA-dT) tracts: the structure of the B-DNA decamer C-G-A-T-A-T-A-T-C-G. Biochemistry. 1992 Sep 1;31(34):8009–8021. [PubMed] [Google Scholar]



