Abstract
(GIMAPs) GTPase of the immunity associated protein family are a novel protein family of putative small GTPases. GIMAPs are mainly expressed in the cells of the immune system and have been associated with immunological functions, such as thymocyte development, apoptosis of peripheral lymphocytes and T helper cell differentiation. GIMAPs have also been linked to immunological diseases, such as T cell lymphopenia, leukemia and autoimmune diseases. In this review we examine the role of GIMAP proteins in T-lymphocyte biology.
1. GTPase of the Immunity-Associated Protein Family (GIMAP)
GTPase of the immunity-associated protein family (GIMAPs), also termed immune-associated nucleotide-binding proteins (IANs), are a relatively recently described, uncharacterized protein family of putative small GTPases conserved among vertebrates and higher plants [1–4]. The original publications describing the different human, mouse, and rat GIMAP family members used a variety of different names for the genes generating confusion. Thus, the GIMAP nomenclature followed also in this minireview was introduced by the HUGO Gene Nomenclature Committee and includes the human, mouse, and rat orthologs [5] (Table 1).
Table 1.
Nomenclature of the human, mouse, and rat GIMAP genes and gene expression sites.
| Gene | Synonyms, human | Synonyms, mouse | Synonyms, rat | Size kDa (human) | Expression | Reference |
|---|---|---|---|---|---|---|
| GIMAP1 | GIMAP1, IMAP1, HIMAP1, IMAP38, hIan2 | Gimap1, imap, IAP38, Imap38, mIan2 | Gimap1, rIan2, Imap38, MGC156493 | 34.4 | thymocytes, spleen, lymphocytes (T, B, NK), macrophages, cell lines (LTR6, C1498, TK-1, A20, P815) | [6–10] |
| GIMAP2 | GIMAP2, IMAP2, HIMAP2, hIan12, MGC24275, DKFZp586D0824 | no mouse ortholog | no rat ortholog | 38.0 | spleen, lymph nodes, PBL, thymus | [3] |
| GIMAP3 | GIMAP3P (pseudogene) | Gimap3, mIan4, Gimap5, 2010110D23Rik | not annotated | spleen, cell lines expressing BCR/ABL | [11] | |
| GIMAP4 | GIMAP4, IAN1, IMAP4, hIan1, HIMAP4, MSTP062, FLJ11110 | Gimap4, mIan1, IMAP4, mIAN1, AU019574, MGC11734, E430007K16Rik | Gimap4, rIan1 | 37.5 | splenocytes, lymphocytes (T and B), thymocytes | [12–16] |
| GIMAP5 | GIMAP5, IAN4, IAN5, IMAP3, hIan5, HIMAP3, IAN4L1, FLJ11296, Irod | Gimap5, mIan5, D630024P16, E230026N22Rik | Gimap5, IAN4, rIan5, Ian4l1 | 34.8 | wide tissue distribution outside the central nervous system | [17–24] |
| GIMAP6 | GIMAP6, hIan6, hIAN2, FLJ22690, DKFZp686A01175 | Gimap6, mIan6, FLJ00102; MGC41522, mFLJ00102, 4833419H03Rik | Gimap6, rIan6, MGC108948 | 32.9 | spleen, lymph nodes, lungs, placenta | [3] |
| GIMAP7 | GIMAP7, IAN7, hIan7, MGC27027 | Gimap7, mIan7, mIan3, MGC41480 | Gimap7, rIan3, MGC108919 | 34.5 | spleen, thymus, lymph nodes, PBL | [3] |
| GIMAP8 | GIMAP8, IANT, hIAN6, MGC129545, DKFZp667I133, hIan11/10/9, hIan4/3/6 | Gimap8, IAN9, Gm457, IMAP8, mIan9 mIan11/10/9 | Gimap8, rIan11/10/9, IanT, MGC116406 | 74.9 | thymus, spleen | [3, 12, 13, 25] |
| GIMAP9 | orthologous to human GIMAP7 | Gimap9, BB145400, A630002K24, mIan3 mIan7 | Gimap9, MGC124918, rIan3 | thymus, lymph nodes | [3, 13, 20] | |
| aGIMAP10P | no human ortholog | Gimap10-ps (pseudogene), mIan8 | no rat ortholog | [3] |
Small GTP (guanosine triphosphate) binding proteins, also known as small GTPases, Ras-like GTPases, or Ras superfamily of GTP binding proteins, regulate key cellular functions in virtually all living organisms. They are involved in signal transduction events and regulation of gene expression in almost all cell types, including the cells of the immune system [26–28]. The Ras superfamily can be subclassified into Ras, Rho, Rab, and Arf families, and the closely related Gα family of the heterotrimeric G proteins, which sometimes are excluded from the RAS superfamily [29]. The Ras proteins induce signaling pathways that include a variety of second messengers, such as calcium and cAMP. The Ras superfamily proteins play key roles in a variety of cellular functions in the immune system, such as cell migration [30], T-cell anergy [31, 32], antigen presentation, [33] and radical formation [34].
The GIMAP family members have unique primary structures and, thus, they define a new family of G proteins distinct from the Ras superfamily and the heterotrimeric G proteins [1]. The expression of GIMAPs in vertebrates has been shown to be highest in the cells of the immune system, although a more ubiquitous expression has also been suggested. Several studies have associated GIMAPs with immunological functions, such as thymocyte development and apoptosis regulation in lymphocytes. These are discussed in what follows.
2. Genomic Organization of GIMAP Genes
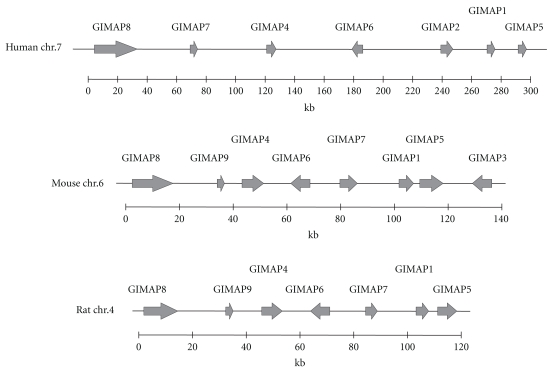
All vertebrate species examined so far have GIMAP genes in tight clusters in their genome [3, 4, 12]. The seven functional human GIMAP genes and one pseudogene are clustered on chromosome 7q36.1 [3] and there are eight functional mouse Gimaps clustered on chromosome 6 and seven functional genes in rat chromosome 4 [13, 25]. The ongoing sequencing project of the genome of Danio rerio (zebrafish) has revealed the existence of Gimap orthologs also in a lower vertebrate. The genomic organization of human, mouse and rat GIMAP genes is depicted in Figure 1.
Figure 1.
GIMAP gene clusters in human, mouse, and rat chromosomes. The GIMAP genes are clustered in human chromosome 7q36.1, mouse chromosome 6, and rat chromosome 4.
Homolog searches in available corn, soybean, and tobacco genomes by Liu et al. [4] came up with one to two homologs of GIMAP/IAN genes in each genome. However, searches within the well-characterized genomes of the unicellular organisms Saccharomyces cerevisiae (Baker's yeast) and Schizosaccharomyces pombe (fission yeast), or invertebrates, such as Caenorhabditis elegans (free-living roundworm) and Drosophila melanogaster (fruit fly) did not reveal any homologs of the GIMAP gene family [4]. Thus, GIMAP genes exist only in vertebrates and angiosperm (i.e., flowering) plants and the yet poorly characterized cellular functions of the GIMAP proteins are specific for vertebrates and higher plants.
GIMAP/IAN proteins emerged before plants and animals split into their own evolutionary paths [4]. Phylogenetic analyses of both protein and genomic sequences [3, 4] showed that human and mouse GIMAPs 1, 4, 5, 6, 7, and 8 form highly orthologous pairs, and, thus, suggest that a gene duplication event in a common ancestor of rodents and primates gave rise to these genes. The phylogenetic analyses by Liu et al. [4] place the Arabidopsis and rice IANs to a clade distinct from the mouse and human GIMAP proteins, thus indicating that the gene duplication events have taken place after the divergence of vertebrates and plants.
3. Features of GIMAP Proteins
Human GIMAP proteins are relatively small proteins with one GTPase domain. Their molecular sizes range from 34 kDa to 38 kDa. GIMAP8 makes an exception by having three GTPase domains, which is extremely unusual not only for GIMAPs, but for small GTPases in general, too. Thus, its molecular size is 74.9 kDa, making it by far the largest GIMAP protein.
The GTPase domain with the five motifs G1-G5 characteristic for all small GTPases is included in the AIG1 domain, named after the prototype gene AIG1 found in Arabidopsis thaliana (avrRpt2-induced gene) [35]. The AIG1 domain is found in all GIMAP and IAN proteins and besides the GTPase motifs, it contains a conserved box, which is characteristic for all AIG1 domain GTPases [3]. All human GIMAPs also contain putative coiled coil domains which suggest protein-protein interactions. Some GIMAPs, namely, GIMAP1, 2, 4, and 5, contain putative transmembrane domains in their COOH-terminal ends and GIMAP7, GIMAP6, GIMAP1, and GIMAP2 have basic amino acids in their NH2- or COOH-terminus with weak similarity to endoplasmic reticulum- (ER)-localization signals [3]. However, localization studies found GIMAP4 mainly in cytosol [12, 14] but also in ER and Golgi [3] and in membrane fraction of fractionated CD4+ T-lymphocytes [36]. GIMAP1 has been reported to localize in ER [6] and GIMAP7 in ER and Golgi [3].
Human GIMAP4 was shown to be able to bind guanosine nucleotides GTP and GDP but not GMP or other nucleotide triphosphates ATP, CTP or TTP [15]. GIMAP4 showed also intrinsic GTPase activity [15]. Besides the GTP, binding domain, GIMAP4 protein has other interesting motifs. It has a carboxy-terminal IQ domain, which is known to bind calmodulin, a second messenger molecule involved in, among other cellular processes, TCR signaling, and activation of NFAT in T-cells [14, 37]. The IQ motif usually consists of ~23 residues and contains the consensus sequence hydrophobQxxxRxxxxRxxxR/K, where hydrophob is a hydrophobic residue [14]. This motif was proven to be functional in murine GIMAP4 and, furthermore, seems to be absent from other GIMAP protein family members [14]. Also the putative phosphoylation sites of murine GIMAP4 were shown to be functional, since mouse GIMAP4 was phosphrorylated rapidly after 10 minutes of PMA/ionomycin stimulation in primary splenocytes [14]. The phosphorylation was mediated by PKC since it was inhibited by a PKC inhibitor, Rottlerin. Interestingly, this phosphorylation was diminished completely after 40–60 minutes of stimulation. Four of the six putative phosphorylation sites of mouse GIMAP4 are also found in human GIMAP4. Thus, GIMAP4 is a lymphocytic signaling molecule.
4. GIMAPs in T-lymphocyte Development and Function
4.1. GIMAPs in Thymocyte Development
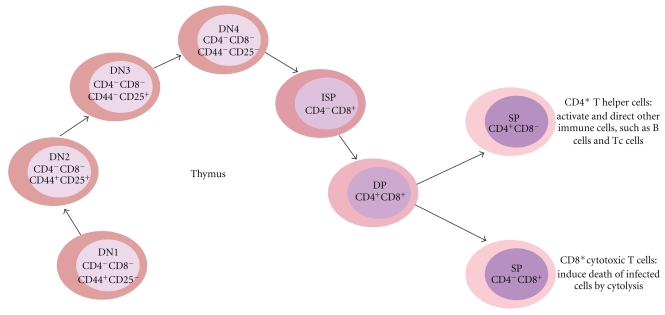
Cell surface expression of leukocyte markers CD4 and CD8 defines the stages of T-cell development in the thymus. During the development process, thymocytes mature from CD4−CD8− double negative (DN) cells into CD4+CD8+ double positive (DP) cells and finally into single positive (SP) CD4+ or CD8+ cells which exit the thymus and enter the periphery (Figure 2). The DN cells are further categorized according to their CD44 and CD25 cell surface expression: (1) DN1 (CD44+CD25−), (2) DN2 (CD44+CD25+), (3) DN3 (CD44−CD25+), and (4) DN4 (CD44−CD25−). Furthermore, the DN4 cells pass through a CD4−CD8+ immature single positive (ISP) stage before developing into DP cells. The different maturation stages are phenotypically and functionally distinct.
Figure 2.
Thymocyte development. T-cells are developed in the thymus. The development can be defined by cell surface expression of CD4 and CD8. During the process, the thymosytes mature from CD4−CD8− double negative (DN) cells into CD4+CD8+ double positive (DP) cells and finally into single positive (SP) CD4+ or CD8+ cells. The DN cells can be further categorized into DN1, DN2, DN3 and DN4 cells. ISP: immature single positive.
The expression of GIMAP genes is strictly regulated during T-cell development in the thymus. Dion et al. [13] studied the expression of rat Gimaps in different thymic subpopulations and observed a significant elevation in expression of all rat Gimaps but Gimap 4 genes between the DN and DP stages. The expression patterns for Gimap1, 6, 8, and 9 during thymocyte development were very similar showing the highest expression in the DP stage. Mouse Gimap 3, 4, 5, and 7 expression was increased during the transition from CD4+CD8+ DP thymocytes into CD4+ or CD8+ SP thymocytes [12]. While shRNA mediated knockdown of mouse GIMAP4 hade no effect on thymocyte development in fetal thymic organ culture model shRNA mediated knockdown of mouse GIMAP3 and GIMAP5 disturbed thymocyte development [12]. Nitta et al. [12] showed that GIMAP3 was needed for optimal positive selection of SP CD4+ and CD8+ T-cells and Gimap5 was important for optimal development of DP thymocytes. It has also been shown that the proportion of CD4−CD8+ SP thymocytes was somewhat reduced in Gimap5 −/− mice [38].
Expression of mouse GIMAP4 protein during thymocyte development has been studied in more detail by Poirier et al. [1] and Schnell et al. [14]. It was not detectable in DN1, DN2, DN3, or DP thymocytes whereas high levels of GIMAP4 were expressed in SP CD4+ cells and DN4 cells [1, 14]. The development of thymocytes in Rag−/− mice is blocked at the DN3 stage but in vivo administration of anti-CD3 antibody allows a low number of thymocytes to proceed up to the DP-stage. This system allows for the enrichment of the low-frequency cell subsets from the different developmental stages, namely, DN3, DN4, and ISP, as well as DP cells. Using the Rag2−/− model, Schnell et al. [14] demonstrated that mouse Gimap4 transcript was absent in differentiation arrested Rag2−/− DN3 thymocytes while it was present in high amounts in DN4, in lower amounts in ISP, and absent in DP cell subsets of anti-CD3 treated Rag2−/− mice. Similar results were obtained by Poirier et al. [1] with a Rag−/− fetal thymic organ culture (FTOC) model where thymocytes from Rag−/− mice blocked at the DN3 stage were cultured in vitro with anti-CD3 antibodies. The antibody treatment resulted in the transient expression of GIMAP4 protein between the DN3 and DP developmental stages. The transitions both from DN3 to DN4 stages and from DP to SP stages require TCR-mediated signals, and, thus, the results described above indicate that GIMAP4 expression is directly or indirectly induced by TCR-mediated signals during thymocyte development. GIMAP4 was also shown to be upregulated by IL-7 signaling in mouse DN thymocytes [39]. However, GIMAP4−/− mice showed no significant role for GIMAP4 in T-cell development in thymus [14]. Thus, GIMAP4 seems to be dispensable for T-cell development, although its expression is tightly regulated during the development process.
4.2. GIMAPs in Peripheral T-cell Functions
Studies have indicated several roles for GIMAP proteins in peripheral T-lymphocyte functions. GIMAPs have been studied in most detail in apoptotic functions and in T-helper (Th) cell differentiation and these processes are depicted in Figures 3 and 4 and discussed below.
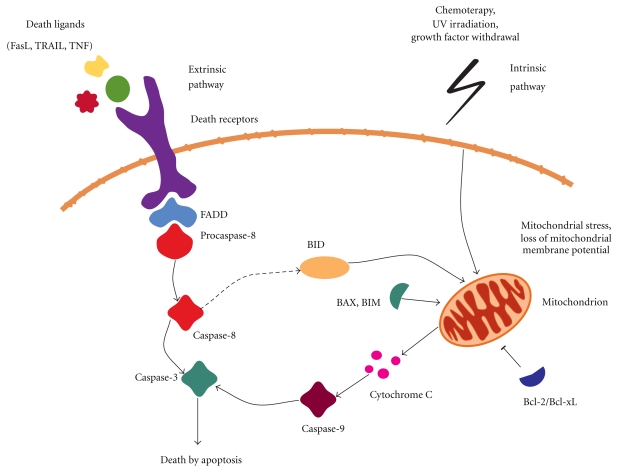
Figure 3.
Apoptosis. Apoptosis can be initiated by the extrinsic or intrinsic pathways. The extrinsic pathway involves binding of ligands, such as tumor necrosis factor (TNF) or Fas ligand (FasL) to the death receptor. Subsequently, FAS-associating death domain (FADD) adapter protein is recruited to the receptor which leads to activation of caspase-8. The intrinsic pathway is initiated by cellular stress which induces loss of mitochondrial membrane potential, release of cytochrome C, and activation of caspase-9. The mitochondrial apoptotic pathway is controlled by the proapoptotic and antiapoptotic members of the Bcl-2 family. Cytochrome C release is inhibited by the prosurvival members Bcl-2/Bcl-xL and promoted by proapoptotic BAX/BIM. Both extrinsic and intrinsic pathways lead to activation of caspase-3 and, ultimately, cell death by apoptosis [40].
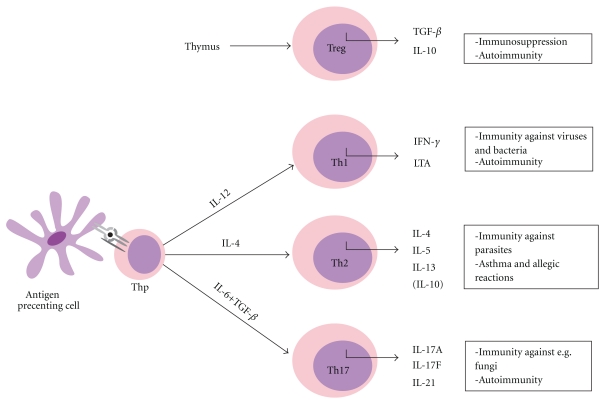
Figure 4.
T-helper cell differentiation. Naïve Thp cells are able to differentiate into functional effector cell subsets according to the cytokine milieu. The differentiation process is initiated by antigen encounter. Naturally occurring regulatory T-cells are produced in the thymus. The different roles of the effector cells in the periphery are indicated in the figure.
4.3. GIMAP1
Mouse Gimap1 was first identified by Krücken et al. [7] as a gene expressed in the spleens of mice infected with Plasmodium chabaudi malaria. C57BL/10 mice are capable of self-healing the P. chabaudi malaria infection and this immunity is suppressed by testosterone. In the absence of testosterone the immunity progresses to a long-lasting protective immunity upon rechallenge. Once acquired, the immunity is testosterone independent and mediated by spleen cells [41]. The high expression of Gimap1 in the spleen cells of P. chabaudi infected mice was strongly associated with the acquisition of the testosterone-resistant, immunity-mediating phenotype and was observed to be highest in macrophages with lower expression in B cells and T-cells [7]. Later it was shown that the expression of Gimap1 in the spleens of the immune mice remains constitutively high for at least 13 weeks postinfection [8]. Surprisingly, however, a later study detected no upregulation of mouse GIMAP1 mRNA or protein in spleen cell lysates from mice infected with P. chabaudi [9] and this phenomenon could not be explained. Mouse Gimap1 has also been reported to be upregulated by p53 during apoptosis in a mouse myeloid leukemia cell line LTR6 [10]. A conditional knockout of Gimap1 in murine lymphocytes led to severe reduction of mature T and B cells but there was little effect on immature thymocytes [42]. Thus, GIMAP1 is critical for peripheral lymphocyte survival.
Both human and mouse GIMAP1 orthologs are preferentially expressed in the spleen with some expression in the lymph nodes [6]. More detailed analysis of GIMAP1 protein expression in mouse splenic, thymic, and bone marrow cell populations revealed GIMAP1 expression in T (CD3+) and B (B220+) cells, DN, DP, and CD4+ and CD8+ SP thymocytes, as well as in NK/NKT (NK1.1+) cells [9]. Interestingly, GIMAP1 was expressed at significant levels in 5 out of 11 tested lymphoid cancer cell lines, namely C1498, TK-1, A20 and P815 [9].
It was shown that GIMAP1 was differentially expressed during human Th1/Th2 differentiation [36]. Its expression was induced by Th1 promoting cytokine IL-12 whereas its expression was downregulated by IL-4, a Th2-inducing cytokine. Promoter analysis of human GIMAP1 gene revealed the existence of multiple silencer elements in the promoter [6]. Stamm et al. [6] studied a large 6.2 kb fragment containing also the first intron (from −3760 to +2419). Deletion of sequences from the first intron and from the 5′ flanking area resulted in inducible promoter activity indicating the removal of silencers. The smallest promoter clone of 0.8 kb (from −760 to +76) was highly active in Jurkat cells thus suggesting that it contained all the elements necessary for active transcription.
4.4. GIMAP3
GIMAP3-ps is a pseudogene in humans [3] and Gimap3 is not annotated in the rat genome. Mouse Gimap3 gene expression was detected at low levels in spleen, but no expression was detected in thymus, liver, or kidney [11]. However, high expression was detected in cell lines expressing BCR/ABL. BCR/ABL is a chimeric oncogenic protein generated from translocation between chromosomes 9 and 22 resulting in the so-called Philadelphia chromosome. BCR/ABL has constitutive tyrosine kinase activity and has been shown to activate several signal transduction pathways, such as RAS, RAF, MYC, and STAT [11]. Mouse GIMAP3 was also shown to bind GTP and localize in mitochondrial membranes [11].
4.5. GIMAP4
GIMAP4 protein expression is highly regulated at the post-transcriptional level [14, 15]. During T- and B-cell activation by anti-CD3/anti-CD28 and CD40-ligand/IL-4 stimulation, respectively, the mRNA level expression of GIMAP4 was shown to be quite stable [15]. However, GIMAP4 protein level started to decrease after 4 days of T-cell activation and was undetectable at day 6. Similarly, yet more rapidly, B-cell activation resulted in the reduction of GIMAP4 protein already after two days of activation. The rapid activation induced phosphorylation/dephosphorylation/degradation of mouse GIMAP4 observed by Schnell et al. [14] also suggest a tight post-transcriptional regulation.
The expression of rat GIMAP4 protein was approximately 10-fold greater in wild type lymph node (LN) T-cells compared to LN T-cells from lyp/lyp (Gimap5 −/−) rat [13]. It is not known whether this is due to GIMAP4 and GIMAP5 interaction in some shared biochemical pathway or a result of the altered activation status of T-cells from lyp/lyp animals. The latter seems plausible in the light of the findings by Cambot et al. [15] and Schnell et al. [14] indicating that both human and mouse GIMAP4 protein expression decreased in response to activation. While the majority of the peripheral T-cells of wt animals are resting G0 cells, the T-cells in lyp/lyp rats are a mixture of semiactivated cells and recent thymic emigrants (RTE) [43, 44].
Although Gimap4 knockout seemed to be redundant for T-cell development as well as selection and activation of T-cells in vivo, the knockout had a significant effect on T-cell apoptosis [14]. Apoptosis induces alterations in the plasma membrane composition and permeabilisation. A convenient method for measuring apoptosis is the staining of exposed phosphatidylserine (PS) on plasma membrane with fluorochrome labeled Annexin V and measuring the accumulation of propidium iodide (PI) in the nucleus after alterations in plasma membrane permeability. Apoptotic cells are, thus, PS positive and PI negative. When mature splenic T-cells from wild-type and Gimap4−/− mice were induced to apoptosis, the number of apoptotic (PS+/PI−) cells was greater among the Gimap4−/− T-cells than the wt T-cells and the number of dead cells were reduced accordingly, that is, the cells executed apoptosis with slower kinetics [14]. Furthermore, the accumulation of apoptotic cells could be inhibited by caspase inhibitors, and there were no changes in caspase-3 activation between apoptotic ko and wt T-cells. This indicated that GIMAP4 acts downstream of caspase-3 and plays a role rather in the execution than the induction phase of apoptosis. A similar, but less marked effect was found in T-cells of the inbred Brown Norway (BN) rat, which carries a natural hypomorphic variant of the Gimap4 gene [16]. Further support for the proapoptotic function of GIMAP4 came also from the findings of Nitta et al. [12], who showed that ectopic expression of GIMAP4 in immature mouse thymocytes led to increased apoptosis. Furthermore, GIMAP4 protein was shown to associate with the proapoptotic Bcl-2 family protein Bax but not other Bcl-2 family members [12, 45].
Although Schnell et al. [14] and Carter et al. [16] showed that GIMAP4 is largely a cytosolic protein Filén et al. [36] found it to be expressed in the membrane fraction of human CD4+ T-helper cells. GIMAP4 expression was furthermore shown to be tightly regulated during early human CD4+ T-helper cell differentiation towards Th1 and Th2 [36]. Proteomic analysis of IL-4 regulated membrane proteins revealed downregulation of GIMAP4 during early Th2 differentiation and it was further shown that IL-12 upregulated GIMAP4 expression during Th1 differentiation. The differential expression of GIMAP4 was detected also on mRNA level. The expression of two distinct isoforms of GIMAP4 in response to IL-12 and IL-4 was also studied and it was shown that they followed a similar expression pattern in response to the studied cytokines, although the short isoform was more highly abundant. RNAi studies further showed that GIMAP4 protein expression was negatively regulated by STAT6 in response to IL-4R signaling during Th2 differentiation [36].
4.6. GIMAP5
Human GIMAP5 mRNA expression showed very wide tissue distribution among most human tissues outside the central nervous system [17, 18]. Its expression was upregulated during human peripheral T-cell activation [19]. It seemed to localize to the mitochondrial membrane [18] although localization to the centrosome/ER/plasma membrane [19] or to a distinct sedimentable subcellular fraction outside ER and mitochondria [46] has also been detected. The regulation of GIMAP5 gene expression remains largely unknown but it has been shown that GIMAP5 is a transcriptional target of Notch signaling in Jurkat cells [47].
An extensively studied genetic defect of lymphopenia (i.e., lyp, also called Iddm2) in diabetes-prone BioBreeding rat (BBDP) was shown to be caused by a frameshift mutation in the Gimap5 gene [20–23]. This frameshift deletion of 1 bp introduces a premature STOP codon leading to a truncated protein product. This deletion was absent in the diabetes resistant BB (BBDR) rat [20, 21]. The BBDP spontaneously develop insulin-dependent diabetes mellitus (IDDM). It closely resembles human type 1 diabetes and is a consequence of Th1-mediated destruction of pancreatic islet β cells characterized by high levels of IFN-γ and IL-12 [48, 49]. The BBDP rat has severe, life-long T-cell lymphopenia [43, 50] caused by increased apoptosis and shortened life-span of recent thymic emigrants and peripheral T-cells [51–53]. However, the RTE can be rescued from apoptosis by activation through TCR [51]. Furthermore, peripheral lyp/lyp T-cells display normal Ca2+ signaling and proliferation in response to TCR crosslinking [52]. Interestingly, the in vitro life-span of lyp/lyp B cells is not affected by the mutation [52].
The lymphopenic phenotype of the BBDP rat argues for anti-apoptotic function for GIMAP5. The studies with the mutated GIMAP5 in BBDP rat by Pandarpurkar et al. [24], Keita et al. [46], and Pino et al. [54] and with human GIMAP5 by Sandal et al. [17] support this finding. Mitochondrial membrane potential was found to be lower in BBDP T-cells than in BBDR T-cells. The lack of full-length GIMAP5 in BBDP rat T-cells lead to ER stress signaling and C/EBP homologous protein-(CHOP)-mediated apoptosis [54]. Thus, GIMAP5 is a regulator of ER homeostasis in rat T-cells. The BBDP rat T-cells also display an impaired TCR-stimulated Ca2+ response which suggests that GIMAP5 is a regulator of calcium responses in T-lymphocytes [55]. Moreover, knockdown of Gimap5 by RNAi in Jurkat T-cells resulted in increased apoptosis [24]. Gimap5 −/− mice exhibit peripheral T-cell lymphopenia, especially of CD8+ T-cells [38]. The Gimap5 −/−CD8+ splenocytes furthermore showed increased apoptosis [38]. Overexpression of human GIMAP5 in Jurkat T-cell line resulted in increased resistance to okadaic acid or γ-irradiation induced apoptosis. The anti-apoptotic function of GIMAP5 was shown to be upstream of caspase-3 [17]. However, more recent studies by Dalberg et al. [19] were contradictory to the earlier findings and showed a proapoptotic function for human GIMAP5. They showed that GIMAP5 knockdown by RNAi did not affect the number of apoptotic cells but overexpression of GIMAP5 in both Jurkat T-cells and naïve T-cells led to increased apoptosis [19]. Furthermore, they showed that while expression of the wt rat GIMAP5 led to increased apoptosis of the cells, the expression of the truncated GIMAP5-LYP in C58, a rat T-cell line, led to a very rapid death of the transfected cells [19]. This indicated a function for the truncated GIMAP5-LYP. Albeit the results obtained by different research groups seem contradictory, they may only indicate that the role of GIMAP5 in apoptosis is dependent on the activation status of the cells and the availability of growth factors.
When the rat Gimap5-Lyp mutation was backcrossed into PVG-RT1u rat strain the lyp/lyp rats developed a spontaneous, progressive, inflammatory bowel disease with Th2 characteristics [56]. Indeed, the purified lyp/lyp CD4+CD45RClo cells produced increased amounts of IL-4 but similar amounts of IFN-γ compared to control wt cells. Also the expression of IL-4 and IL-13 was higher in the lyp/lyp cells compared to wt [56]. Accordingly, chemical mutation of mouse Gimap5 led to severe intestinal inflammation and wasting disease [57].
A common polyadenylation polymorphism in the human GIMAP5 gene was associated with the risk to autoimmune systemic lupus erythematosus (SLE) [58] and with the increased prevalence of IA-2 autoantibodies in patients with type I diabetes [59]. This polymorphism produces an inefficient polyadenylation signal to the 3′ part of the GIMAP5 mRNA and leads to increased proportion of nonterminated mRNA [58].
5. Concluding Remarks
There are a growing number of reports describing a functional role for GIMAP family proteins in lymphocyte biology and we have discussed the role of GIMAPs in T-lymphocyte functions. More investigation is warranted to reveal the detailed molecular mechanism of GIMAP protein function. Determining, for example, the interaction partners, site of action, and signaling pathways will strengthen our knowledge of the function of this highly interesting novel family of GTPases.
References
- 1.Poirier GMC, Anderson G, Huvar A, et al. Immune-associated nucleotide-1 (IAN-1) is a thymic selection marker and defines a novel gene family conserved in plants. Journal of Immunology. 1999;163(9):4960–4969. [PubMed] [Google Scholar]
- 2.Nitta T, Takahama Y. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends in Immunology. 2007;28(2):58–65. doi: 10.1016/j.it.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Krücken J, Schroetel RMU, Müller IU, et al. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene. 2004;341(1-2):291–304. doi: 10.1016/j.gene.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Wang T, Zhang W, Li X. Computational identification and analysis of immune-associated nucleotide gene family in Arabidopsis thaliana. Journal of Plant Physiology. 2008;165(7):777–787. doi: 10.1016/j.jplph.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79(4):464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- 6.Stamm O, Krücken J, Schmitt-Wrede H-P, Benten WPM, Wunderlich F. Human ortholog to mouse gene imap38 encoding an ER-localizable G-protein belongs to a gene family clustered on chromosome 7q32-36. Gene. 2002;282(1-2):159–167. doi: 10.1016/s0378-1119(01)00837-x. [DOI] [PubMed] [Google Scholar]
- 7.Krücken J, Schmitt-Wrede H-P, Markmann-Mulisch U, Wunderlich F. Novel gene expressed in spleen cells mediating acquired testosterone-resistant immunity to Plasmodium chabaudi malaria. Biochemical and Biophysical Research Communications. 1997;230(1):167–170. doi: 10.1006/bbrc.1996.5876. [DOI] [PubMed] [Google Scholar]
- 8.Krücken J, Stamm O, Schmitt-Wrede H-P, Mincheva A, Lichter P, Wunderlich F. Spleen-specific expression of the malaria-inducible intronless mouse gene imap38. Journal of Biological Chemistry. 1999;274(34):24383–24391. doi: 10.1074/jbc.274.34.24383. [DOI] [PubMed] [Google Scholar]
- 9.Saunders A, Lamb T, Pascall J, et al. Expression of GIMAP1, a GTPase of the immunity-associated protein family, is not up-regulated in malaria. Malaria Journal. 2009;8(1, article 53) doi: 10.1186/1475-2875-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannan K, Kaminski N, Rechavi G, Jakob-Hirsch J, Amariglio N, Givol D. DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene. 2001;20(26):3449–3455. doi: 10.1038/sj.onc.1204446. [DOI] [PubMed] [Google Scholar]
- 11.Dahéron L, Zenz T, Siracusa LD, Brenner C, Calabretta B. Molecular cloning of Ian4: a BCR/ABL-induced gene that encodes an outer membrane mitochondrial protein with GTP-binding activity. Nucleic Acids Research. 2001;29(6):1308–1316. doi: 10.1093/nar/29.6.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nitta T, Nasreen M, Seike T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biology. 2006;4(4, article e103) doi: 10.1371/journal.pbio.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dion C, Carter C, Hepburn L, et al. Expression of the Ian family of putative GTPases during T cell development and description of an Ian with three sets of GTP/GDP-binding motifs. International Immunology. 2005;17(9):1257–1268. doi: 10.1093/intimm/dxh302. [DOI] [PubMed] [Google Scholar]
- 14.Schnell S, Démollière C, van den Berk P, Jacobs H. Gimap4 accelerates T-cell death. Blood. 2006;108(2):591–599. doi: 10.1182/blood-2005-11-4616. [DOI] [PubMed] [Google Scholar]
- 15.Cambot M, Aresta S, Kahn-Perlès B, de Gunzburg J, Roméo P-H. Human immune associated nucleotide 1: a member of a new guanosine triphosphatase family expressed in resting T and B cells. Blood. 2002;99(9):3293–3301. doi: 10.1182/blood.v99.9.3293. [DOI] [PubMed] [Google Scholar]
- 16.Carter C, Dion C, Schnell S, et al. A natural hypomorphic variant of the apoptosis regulator Gimap4/IAN1. Journal of Immunology. 2007;179(3):1784–1795. doi: 10.4049/jimmunol.179.3.1784. [DOI] [PubMed] [Google Scholar]
- 17.Sandal T, Aumo L, Hedin L, Gjertsen BT, Døskeland SO. Irod/Ian5: an inhibitor of γ-radiation- and okadaic acid-induced apoptosis. Molecular Biology of the Cell. 2003;14(8):3292–3304. doi: 10.1091/mbc.E02-10-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zenz T, Roessner A, Thomas A, et al. hlan5: the human ortholog to the rat lan4/lddm1/lyp is a new member of the Ian family that is overexpressed in B-cell lymphoid malignancies. Genes and Immunity. 2004;5(2):109–116. doi: 10.1038/sj.gene.6364044. [DOI] [PubMed] [Google Scholar]
- 19.Dalberg U, Markholst H, Hornum L. Both Gimap5 and the diabetogenic BBDP allele of Gimap5 induce apoptosis in T cells. International Immunology. 2007;19(4):447–453. doi: 10.1093/intimm/dxm009. [DOI] [PubMed] [Google Scholar]
- 20.MacMurray AJ, Moralejo DH, Kwitek AE, et al. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Research. 2002;12(7):1029–1039. doi: 10.1101/gr.412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornum L, Rmer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes. 2002;51(6):1972–1979. doi: 10.2337/diabetes.51.6.1972. [DOI] [PubMed] [Google Scholar]
- 22.Michalkiewicz M, Michalkiewicz T, Ettinger RA, et al. Transgenic rescue demonstrates involvement of the Ian5 gene in T cell development in the rat. Physiological Genomics. 2005;19:228–232. doi: 10.1152/physiolgenomics.00126.2004. [DOI] [PubMed] [Google Scholar]
- 23.Rutledge EA, Fuller JM, Van Yserloo B, et al. Sequence variation and expression of the Gimap gene family in the BB rat. Experimental Diabetes Research. 2009;2009:10 pages. doi: 10.1155/2009/835650. Article ID 835650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandarpurkar M, Wilson-Fritch L, Corvera S, et al. Ian4 is required for mitochondrial integrity and T cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(18):10382–10387. doi: 10.1073/pnas.1832170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krücken J, Epe M, Benten WPM, Falkenroth N, Wunderlich F. Malaria-suppressible expression of the anti-apoptotic triple GTPase mGIMAP8. Journal of Cellular Biochemistry. 2005;96(2):339–348. doi: 10.1002/jcb.20552. [DOI] [PubMed] [Google Scholar]
- 26.Scheele JS, Marks RE, Boss GR. Signaling by small GTPases in the immune system. Immunological Reviews. 2007;218(1):92–101. doi: 10.1111/j.1600-065X.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- 27.Genot E, Cantrell DA. Ras regulation and function in lymphocytes. Current Opinion in Immunology. 2000;12(3):289–294. doi: 10.1016/s0952-7915(00)00089-3. [DOI] [PubMed] [Google Scholar]
- 28.Rajalingam K, Schreck R, Rapp UR, Albert Š. Ras oncogenes and their downstream targets. Biochimica et Biophysica Acta. 2007;1773(8):1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Colicelli J. Human RAS superfamily proteins and related GTPases. Science’s STKE. 2004;2004(250):p. RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLeod SJ, Gold MR. Activation and function of the Rap1 GTPase in B lymphocytes. International Reviews of Immunology. 2001;20(6):763–789. doi: 10.3109/08830180109045589. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, Zha Y, Gajewski TF. Molecular regulation of T-cell anergy. EMBO Reports. 2008;9(1):50–55. doi: 10.1038/sj.embor.7401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz RH. T cell anergy. Annual Review of Immunology. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 33.Shurin GV, Tourkova IL, Chatta GS, et al. Small Rho GTPases regulate antigen presentation in dendritic cells. Journal of Immunology. 2005;174(6):3394–3400. doi: 10.4049/jimmunol.174.6.3394. [DOI] [PubMed] [Google Scholar]
- 34.Finkel T. Intracellular redox regulation by the family of small GTPases. Antioxidants and Redox Signaling. 2006;8(9-10):1857–1863. doi: 10.1089/ars.2006.8.1857. [DOI] [PubMed] [Google Scholar]
- 35.Reuber TL, Ausubel FM. Isolation of arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell. 1996;8(2):241–249. doi: 10.1105/tpc.8.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filén J-J, FiLén S, Moulder R, et al. Quantitative proteomics reveals GIMAP family proteins 1 and 4 to be differentially regulated during human T helper cell differentiation. Molecular and Cellular Proteomics. 2009;8(1):32–44. doi: 10.1074/mcp.M800139-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugiura R, Sio SO, Shuntoh H, Kuno T. Molecular genetic analysis of the calcineurin signaling pathways. Cellular and Molecular Life Sciences. 2001;58(2):278–288. doi: 10.1007/PL00000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulteis RD, Chu H, Dai X, et al. Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5. Blood. 2008;112(13):4905–4914. doi: 10.1182/blood-2008-03-146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duthie KA, Osborne LC, Foster LJ, Abraham N. Proteomics analysis of interleukin (IL)-7-induced signaling effectors shows selective changes in IL-7Rα449F knock-in T cell progenitors. Molecular and Cellular Proteomics. 2007;6(10):1700–1710. doi: 10.1074/mcp.M600468-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Mechanisms of disease: cell death. The New England Journal of Medicine. 2009;361(16):1570–1583. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wunderlich F, Peter W, Benten M, Bettenhaeuser U, Schmitt-Wrede H-P, Mossmann H. Testosterone-unresponsiveness of existing immunity against Plasmodium chabaudi malaria. Parasite Immunology. 1992;14(3):307–320. doi: 10.1111/j.1365-3024.1992.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 42.Saunders A, Webb LMC, Janas ML, et al. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 2010;115(16):3249–3257. doi: 10.1182/blood-2009-08-237586. [DOI] [PubMed] [Google Scholar]
- 43.Ramanathan S, Poussier P. BB rat lyp mutation and type 1 diabetes. Immunological Reviews. 2001;184:161–171. doi: 10.1034/j.1600-065x.2001.1840115.x. [DOI] [PubMed] [Google Scholar]
- 44.Groen H, van der Berk JMMM, Nieuwenhuis P, Kampinga J. Peripheral T cells in diabetes prone (DP) BB rats are CD45R-negative. Thymus. 1989;14(1–3):145–150. [PubMed] [Google Scholar]
- 45.Häcker G, Bauer A, Villunger A. Apoptosis in activated T cells: what are the triggers, and what the signal transducers? Cell Cycle. 2006;5(21):2421–2424. doi: 10.4161/cc.5.21.3397. [DOI] [PubMed] [Google Scholar]
- 46.Keita M, Leblanc C, Andrews D, Ramanathan S. GIMAP5 regulates mitochondrial integrity from a distinct subcellular compartment. Biochemical and Biophysical Research Communications. 2007;361(2):481–486. doi: 10.1016/j.bbrc.2007.07.048. [DOI] [PubMed] [Google Scholar]
- 47.Chadwick N, Zeef L, Portillo V, et al. Identification of novel Notch target genes in T cell leukaemia. Molecular Cancer. 2009;8, article 35 doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zipris D, Greiner DL, Malkani S, Whalen B, Mordes JP, Rossini AA. Cytokine gene expression in islets and thyroids of BB rats: IFN-γ and IL-12p40 mRNA increase with age in both diabetic and insulin-treated nondiabetic BB rats. Journal of Immunology. 1996;156(3):1315–1321. [PubMed] [Google Scholar]
- 49.Rabinovitch A, Suarez-Pinzon W, El-Sheikh A, Sorensen O, Power RF. Cytokine gene expression in pancreatic islet-infiltrating leukocytes of BB rats: expression of Th1 cytokines correlates with β-cell destructive insulitis and IDDM. Diabetes. 1996;45(6):749–754. doi: 10.2337/diab.45.6.749. [DOI] [PubMed] [Google Scholar]
- 50.Poussier P, Nakhooda AF, Falk JA, Lee C, Marliss EB. Lymphopenia and abnormal lymphocyte subsets in the “BB” rat: relationship to the diabetic syndrome. Endocrinology. 1982;110(5):1825–1827. doi: 10.1210/endo-110-5-1825. [DOI] [PubMed] [Google Scholar]
- 51.Ramanathan S, Norwich K, Poussier P. Antigen activation rescues recent thymic emigrants from programmed cell death in the BB rat. Journal of Immunology. 1998;160(12):5757–5764. [PubMed] [Google Scholar]
- 52.Hernández-Hoyos G, Joseph S, Miller NGA, Butcher GW. The lymphopenia mutation of the BB rat causes inappropriate apoptosis of mature thymocytes. European Journal of Immunology. 1999;29(6):1832–1841. doi: 10.1002/(SICI)1521-4141(199906)29:06<1832::AID-IMMU1832>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 53.Whalen BJ, Weiser P, Marounek J, Rossini AA, Mordes JP, Greiner DL. Recapitulation of normal and abnormal biobreeding rat T cell development in adult thymus organ culture. Journal of Immunology. 1999;162(7):4003–4012. [PubMed] [Google Scholar]
- 54.Pino SC, O’Sullivan-Murphy B, Lidstone EA, et al. CHOP mediates endoplasmic reticulum stress-induced apoptosis in Gimap5-deficient T cells. PLoS ONE. 2009;4(5, article e5468) doi: 10.1371/journal.pone.0005468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ilangumaran S, Forand-Boulerice M, Bousquet SM, et al. Loss of GIMAP5 (GTPase of immunity-associated nucleotide binding protein 5) impairs calcium signaling in rat T lymphocytes. Molecular Immunology. 2009;46(6):1256–1259. doi: 10.1016/j.molimm.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 56.Cousins L, Graham M, Tooze R, et al. Eosinophilic bowel disease controlled by the BB rat-derived lymphopenia/Gimap5 gene. Gastroenterology. 2006;131(5):1475–1485. doi: 10.1053/j.gastro.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 57.Barnes MJ, Aksoylar H, Krebs P, et al. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. Journal of Immunology. 2010;184(7):3743–3754. doi: 10.4049/jimmunol.0903164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hellquist A, Zucchelli M, Kivinen K, et al. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. Journal of Medical Genetics. 2007;44(5):314–321. doi: 10.1136/jmg.2006.046185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin J-H, Janer M, McNeney B, et al. IA-2 autoantibodies in incident type I diabetes patients are associated with a polyadenylation signal polymorphism in GIMAP5. Genes and Immunity. 2007;8(6):503–512. doi: 10.1038/sj.gene.6364413. [DOI] [PubMed] [Google Scholar]