Abstract
Mutations in human TBX22 cause X-linked cleft palate with ankyloglossia syndrome (CPX; OMIM 303400). Since the secondary palate was an adaptation to breathing on land, we characterized zebrafish tbx22 to study molecular mechanisms regulating early vertebrate craniofacial patterning. Rapid Amplification of cDNA Ends (RACE) analyses revealed two zebrafish tbx22 splice isoforms, tbx22-1 and tbx22-2, encoding proteins of 444 and 400 amino acids, respectively. tbx22-1 resembles canonical Tbx22 orthologs, while tbx22-2 lacks conserved N-terminal sequence. Developmental RT-PCR revealed that tbx22-1 is maternally and zygotically expressed, while tbx22-2 is expressed zygotically. WISH analyses revealed strong tbx22 mRNA expression in ectomesenchyme underlying the stomodeum, a bilaminar epithelial structure demarcating early mouth formation, and in early presumptive jaw joints. Zebrafish tbx22 expression mirrored some aspects of mammalian Tbx22, consistent with roles in early vertebrate face patterning. These studies identify an early transcription factor governing vertebrate facial development, which may underlie common craniofacial birth disorders.
Keywords: zebrafish, craniofacial development
INTRODUCTION
Human craniofacial dysmorphologies such as cleft lip and/or palate, in either the syndromic or nonsyndromic forms, exact a great cost in morbidity and mortality (Wyszynski, 2002). Mutations found within the human TBX22 gene are the cause of syndromic, X-linked, Cleft-Palate/Ankyloglossia (CPX) (Braybrook et al., 2001). Human TBX22 maps to the Xq21.1 locus, which was implicated by linkage analysis as a major locus in human nonsyndromic cleft lip and/or palate (Prescott et al., 2000), the more common form of orofacial clefting. Mutations within the TBX22 gene have also been implicated as a frequent cause of cleft palate only (Marcano et al., 2004).
Multiple subfamilies of T-box genes have been identified in all metazoans, with five major groups and eight subgroups present in the chordate lineage (Takatori et al., 2004; see Supplemental Data File 1). Pharyngeal arch expression has been especially noted for the Tbx1/10, Tbx20 and Tbx15/18/22 subgroups (Ahn et al., 2000; Begemann et al., 2002; Kochilas et al., 2003; Mahadevan et al., 2004; Piotrowski et al., 2003), implicating these genes in craniofacial development. Tbx15 has been shown to influence the formation of the vertebral column and head skeletal elements, while Tbx18 is expressed in the unsegmented paraxial mesoderm of the head (Haenig and Kispert, 2004; Singh et al., 2005). In the mouse, Tbx22 is expressed at the base of the tongue, in mesenchyme of the nasal septum and eye, and in the posterior palatal shelves prior to fusion (Bush et al., 2002; Herr et al., 2003).
To examine evolutionary roles for TBX22 in craniofacial development, zebrafish tbx22 was cloned and sequenced. Two alternatively spliced tbx22 transcripts were identified. The tbx22-1 spliced isoform encodes a protein of 444 amino acids, while tbx22-2 encodes a protein of 400 amino acids. Gene expression analyses by whole mount in situ hybridization (WISH) revealed discrete tbx22 expression in pharyngula stage zebrafish. Distinct expression domains were observed in ectomesenchymal cells underlying two bilateral sets of epithelial sheets that will eventually invaginate to form the zebrafish mouth. The expression of zebrafish tbx22 resembles that of higher vertebrates (Figure 6), consistent with conserved roles for tbx22 in early vertebrate craniofacial patterning.
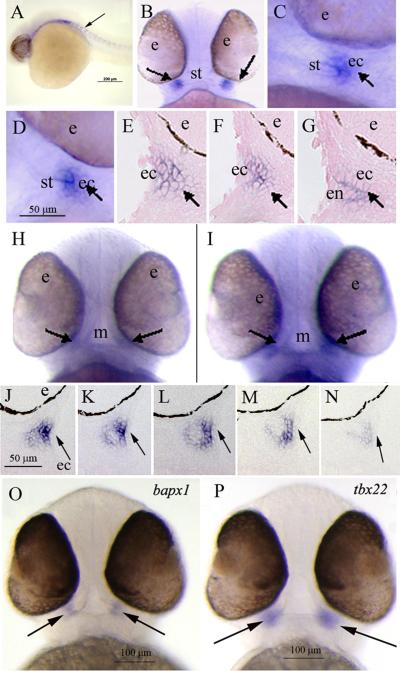
Figure 6. WISH analysis of zebrafish tbx22 expression in 38 - 42 hpf pharyngeal arch structures.
Panels B, H, I, O, P display ventral views, and all other panels present sagittal views. A) At 28 hpf, tbx22 is expressed in paraxial mesodermal tissues in a segmental pattern suggestive of vertebral pre-patterning (arrow). B, C) In 38 hpf embryos, tbx22 is first detectable as discrete, bilateral domains at either corner of the forming mouth (arrows). D) At 40 hpf, tbx22 is expressed in similar bilateral domains (arrow). E, F, G) Consecutive sagittal sections illustrate tbx22 expression underlying the bilaminar epithelium of the forming stomodeum, which bifurcates into dorsal and ventral elements (arrows). At 42 hpf, faint tbx22 expression is observed in tissues surrounding the mouth (H, arrows), and at each corner of the mouth opening (I, arrows), as shown in consecutive serial sections (J-N, arrows). (O, P) At 50 hpf, bapx1 and tbx22 exhibit similar expression patterns in the developing jaw joint (arrows). Abbreviations: e, eye; ec, ectoderm; m, mouth; s, stomodeum.
RESULTS AND DISCUSSION
Syntenic analysis of the zebrafish tbx22 gene
To identify the zebrafish tbx22 gene, homology was first established between the human TBX22 mRNA and Fugu tbx22 genomic nucleotide sequences. The Fugu tbx22 sequence was then used as BLAST bait to screen the Sanger Institute Danio rerio (Dr) database using Ensembl software. A putative zebrafish tbx22 ortholog was initially identified within contig ctg10123.1 of the Zv4 assembly, and a zebrafish tbx22 partial transcript was identified as ENSDART00000020757, found at Scaffold_Zv5_NA5339: 72.86k. Identification of limited synteny between the human TBX22 and zebrafish tbx22 locus, consisting of an adjacent FAM46 group transcript, ENSDARESTT00000032536, convinced us to proceed to clone the zebrafish tbx22 gene from this site.
Cloning, sequencing, and in vitro translation of full-length zebrafish tbx22-1 and tbx22-2 splice variants
Primers chosen from the bioinformatically identified partial tbx22 coding sequence were used in 5′ and 3′ RACE reactions to generate a series of overlapping PCR products (Figure 1A, PCR fragments ‘a-c’), which were subsequently used to generate full length cDNAs (Figure 1A, PCR fragments d-1 and d-2). Nucleotide sequence analysis of 5′ RACE products revealed two alternatively spliced zebrafish tbx22 transcripts, which were named tbx22-1 and tbx22-2 (Figure 1B). The 1,856 bp full length tbx22-1 cDNA spans 8 exons of the zebrafish tbx22 locus. The initiation codon prediction program ATGpr (Salamov et al., 1998) identified a start codon located at nucleotide 174 of the tbx22-1 isoform clone with a reliability score of 0.47, which predicts a Tbx22-1 protein of 444 amino acids. An additional in frame start codon located 54 nucleotides downstream of the first putative ATG start codon, and predicting a protein of 426 amino acids, had a much lower ATGpr reliability score (0.14), indicating that the first start codon is most likely the correct start site. The 1,969 bp full length tbx22-2 cDNA start codon has an ATGpr reliability score of 0.43, and spans 7 exons. The tbx22-2 transcript retains an intron between exons 1-1 and 1-2 that is spliced out of tbx22-1, along with an additional upstream, in frame, stop codon (Figure 1A). Zebrafish tbx22-2 encodes a predicted protein of 400 amino acids. Note that the tbx22-2 start codon is spliced out of the tbx22-1 transcript, along with an additional upstream, in frame, stop codon. In tbx22-2, the upstream ATG start codon for tbx22-1 is eliminated as a possible start codon by an in-frame stop codon located 147 nucleotides downstream. Nucleotide sequence comparison of tbx22-1 and tbx22-2 reveals the alternative splicing and ATG start codons, with otherwise identical nucleotide sequence (Figure 2A).
Figure 1. Alternatively spliced zebrafish tbx22 transcripts, tbx22-1 and tbx22-2.
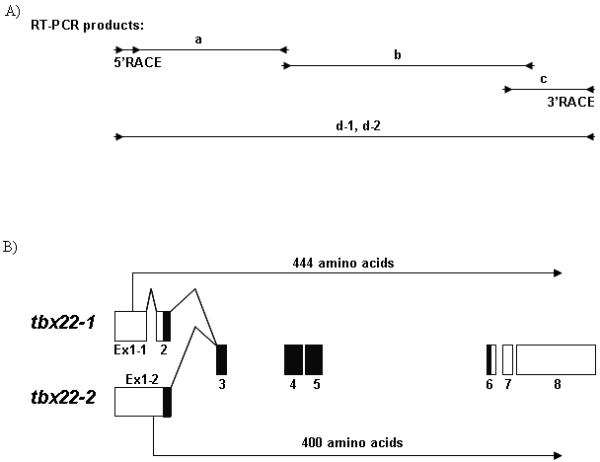
A) Intron and exon maps of identified overlapping zebrafish tbx22 RT-PCR products are shown. The tbx22 RT-PCR product ‘a’ spans exons 1- 4, fragment ‘b’ spans exons 4 - 8, and fragment ‘c’ spans exons 7 - 8. RT-PCR products ‘d-1’ and ‘d-2’ represent the two full length clones, tbx22-1 and tbx22-2 respectively, generated from 5′ and 3′ RACE products. Internal primers for fragment ‘a’ were used in developmental RT-PCR analyses presented in Figure 5. B) The transcript map depicts the two alternatively spliced zebrafish tbx22 transcripts, tbx22-1 and tbx22-2. Exons 1-8 are illustrated by boxes, open reading frames are indicated by arrows, and T-box domains are indicated in black.
Figure 2. Detailed genomic organization of zebrafish tbx22-1 and tbx22-2.
A) Nucleotide sequence comparison of the first three exons for tbx22-1 and the first two exons for tbx22-2 cDNA sequences distinguish the alternatively spliced isoforms. A Clustal W 1.83 alignment of the zebrafish tbx22-1 and tbx22-2 nucleotide sequences is shown. Coding sequence is shown in upper case, and noncoding is shown in lower case font. Asterisks indicate nucleotide identity, and dashes indicate gaps in the alignment.
B) Multisequence alignments of the predicted amino acid sequences for zebrafish tbx22-1 and tbx22-2, as compared to human and mouse Tbx22 proteins, are shown in nexus format. The T-box domain (as defined in Muller and Herrmann, 1997) is indicated in bold type. Functionally conserved amino acids within the T-box domain are indicated as follows: amino acids involved in Tbx22 dimerization are underlined, and amino acids involved in DNA binding are italicized. Dots represent identity, and dashes represent gaps in the alignment. Human TBX22, ENSP00000362393; Mouse Tbx22, ENSMUSP00000033593.
To determine which zebrafish tbx22 isoform gene product represents the more evolutionarily conserved canonical Tbx22, both isoform gene products were compared to the human TBX22 sequence by BLAST at NCBI, and found to have 68% amino acid identity (156/228) and 83% amino acid similarity (190/228) to human TBX22 (Figure 2B). However, a multi-sequence alignment was found to be more informative. The predicted amino acid sequences of zebrafish tbx22-1 and tbx22-2 cDNAs were compared to human and mouse Tbx22 amino acid sequences (Figure 2B). This comparison clearly shows that zebrafish Tbx22-1 and Tbx22-2 differ only at the amino terminal portion of the protein, and that zebrafish Tbx22-1 most closely resembles human and mouse Tbx22, as it aligns with a stretch of conserved positively and negatively charged amino acids just upstream of the T-Box that are missing from zebrafish Tbx22-2. Curiously, both human and mouse Tbx22 have a conserved Groucho repression motif, ‘FSVEAL’, located just upstream of the conserved hydrophilic region, which is not present in zebrafish Tbx22-1. An earlier report identified this motif in the closely related human TBX 15, −18, −22, −20, −2 and −3 paralog groups (Copley, 2005), suggesting that the Groucho repression domain was present in the common ancestors of Tbx22 paralogs, but was subsequently lost within the zebrafish lineage.
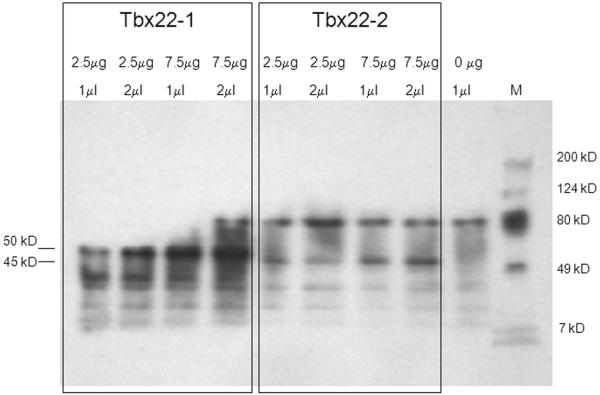
Coupled in vitro transcription and translation was used to confirm the predicted translational start sites for tbx22-1 and tbx22-2. Full-length cDNAs for both splice variants were cloned into expression vectors designed for the high protein yield in coupled transcription and translation reactions. Biotin-conjugated lysine tRNAs were used to label the 24 and 21 lysine residues predicted to be present in Tbx22-1 and Tbx22-2, respectively. Two amounts of plasmid template (2.5 μg and 7.5 μg), and two loading volumes (1μl and 2μl), were used to reveal all translation products using gel electrophoresis (Figure 3). The predicted molecular weights for the two possible translation start sites for Tbx22-1 are 50.3 kD, and 48.3 kD, respectively, and that of Tbx22-2 is 45.2 kD. The biotinylated translation products are expected to migrate at slighter higher molecular weights than non-biotinylated products. Translation products corresponding to the biotinylated 50.3 kD Tbx22-1 and 45.2 kD Tbx22-2 were observed. A putative 48.3 kD Tbx22-1 translation product was not observed, neither was there a small 5-6 kD Tbx22-2 translation product observed, which could have been generated from a tbx22-2 transcript using the same start codon as Tbx22-1. These results confirmed the location of the ATG start codon at bp 174 of the tbx22-1 cDNA, as predicted by the strong Kozak consensus sequence (Kozak, 1996).
Figure 3. In vitro transcription and translation of tbx22 splice variants.
Western blot analysis revealed distinct biotinylated Tbx22-1 and Tbx22-2 in vitro translation products corresponding to the predicted 50.3 and 45.2 kD Tbx22 isoforms, respectively. No putative 48.3 kD Tbx22-1 translation product was observed, consistent with the location of the Tbx22-1 start codon at bp 147. Plasmid concentrations used in the in vitro transcription reactions, and the amount of translation product loaded in each well, are as indicated. A no template negative control is also shown. M indicates molecular weight protein standard markers.
Phylogenetic analysis of the zebrafish tbx22 gene
The predicted amino acid sequences of the zebrafish tbx22-1 and tbx22-2 cDNAs were used to confirm the identity of the zebrafish tbx22 transcripts, and to establish that no additional tbx22 paralogs were present in the Zv7, August 2007 zebrafish genomic assembly. Closely related human T-box containing proteins with known craniofacial expression domains, and several other T-box proteins representing other paralogous groups, were used in phylogenetic analyses to help assign zebrafish T-box genes to the previously identified eight T-box paralog subfamily groups (Takatori et al., 2004). The abbreviated gene and protein sequence identifiers used in these comparisons are listed in FASTA format in Supplemental Data File 1.
Phylogenetic analyses of the predicted amino acid sequences of the 19 identified zebrafish T-box genes (Supplemental Data File 1) used only the 125 highly conserved amino acid positions previously used to define the major vertebrate T-box paralog groups (Ruvinsky et al., 2000). MEGA 3.1 software was used to generate the Maximum Parsimony (MP) phylogenetic tree shown below (Figure 4). Neighbor-Joining (NJ) analyses of these same sequences produced a nearly identical tree topology (data not shown). The MP tree illustrates the strong separation of the Tbx15/18/22 T-box subfamily from the other paralogs (bootstrap value = 88). In addition, the zebrafish tbx22 sequence separation from the Tbx15 and 18 paralogs is also well supported (bootstrap value = 82). These analyses helped us to assign previously ill-defined zebrafish T-box gene sequences to their appropriate orthologous families. In conclusion, our phylogenetic analyses placed the predicted amino acid sequence for the newly identified zebrafish tbx22 into closest relationship to the human TBX22 sequence, with high bootstrap values ranging from 82 to 88. No other Tbx22 coding ESTs were identified in an extensive search of the ENSEMBL and NCBI databases, performed by BLAST search to identify all putative Tbox loci within the zebrafish genome and determining the predicted amino acid sequence for each, followed by global phylogenetic analysis to assign each to its appropriate orthologous group. In this way, our phylogenetic analyses indicated that the bioinformatically identified and cloned zebrafish tbx22 cDNAs likely represent the only zebrafish ortholog of the vertebrate Tbx22.
Figure 4. Maximum parsimony tree analysis of zebrafish Tbx22 within the eight major paralog subfamilies of T-box proteins.
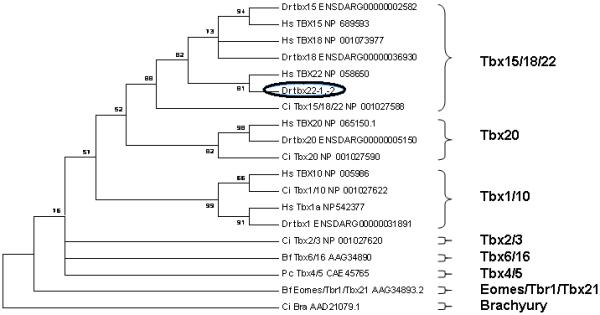
Paralogous zebrafish T-Box subfamily members found in the zebrafish ENSEMBL Zv7 assembly, release 46, were analyzed along with representative members of each paralog subfamily (see Supplemental Data File 1). Human TBX15, −18, −22, −1, −10, and −20 were included to provide more robust paralog family definitions. The maximum parsimony (MP) tree was constructed from the multi-sequence alignment of the 125 most conserved amino acids within the protein sequence (See Supplementary Data). This data is presented as a condensed, topology only maximum parsimony tree, rooted on the Ciona Brachyury T paralog group. Only bootstrap values above 50% are shown. The position of the zebrafish tbx22 is highlighted by a black oval.
Developmental expression of zebrafish tbx22 isoform mRNAs
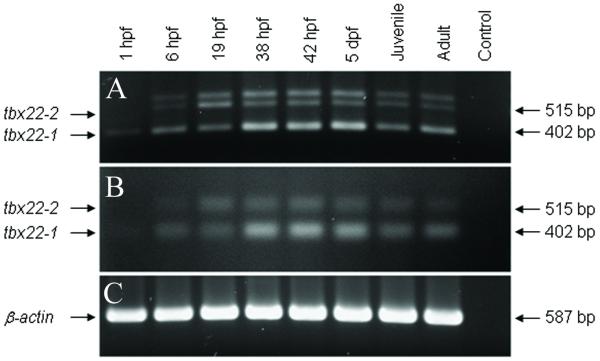
Based on the alternative splice sites for tbx22-1 and tbx22-2, PCR primers were designed to generate isoform specific, 402 bp tbx22-1, and 515 bp tbx22-2 RT-PCR products. The developmental expression of tbx22 isoform mRNAs was examined by RT-PCR, using total RNA isolated from staged zebrafish at 1, 6, 19, 38 and 48 hours post fertilization (hpf), 5 and 44 days post fertilization (dpf), and 6 month old adult zebrafish. Unexpectedly, three distinct PCR products of approximately 545, 515, and 402 bp were consistently generated (Figure 5A). To confirm the identities of the PCR products, each band was isolated, cloned and nucleotide sequenced. As anticipated, the 515 and 402 bp PCR products were found to be specific for the tbx22-1 and tbx22-2 isoform sequences, respectively. Nucleotide sequence analysis of 46 clones generated from the 545 bp PCR products revealed 20 tbx22-1 clones, 26 tbx22-2 colonies, suggesting that the larger sized RT-PCR product was in fact an incompletely denatured heteroduplex of the two zebrafish tbx22 isoform products. Analysis of the RT-PCR products in denaturing gels revealed only the 515 and 402 bp (Figure 5B), consistent with this interpretation. Developmental RT-PCR analysis revealed that tbx22-1 was expressed at all developmental stages examined, from maternal to adult, while tbx22-2 was not maternally expressed, but exhibited zygotic expression at all stages examined. The expression of zebrafish β-actin was used as an internal control (Figure 5C).
Figure 5. Developmental RT-PCR analysis of tbx22-1 and tbx22-2 mRNAs.
A) The developmental expression of zebrafish tbx22-1 and tbx22-2 RT-PCR products, 402 and 515 bp respectively, size fractionated in a non-denaturing TAE agarose gel. B) tbx22-1 and tbx22-2 RT-PCR products size fractionated in a denaturing, alkaline agarose gel. The non-denatured, heteroduplex, high molecular weight product present in Panel A is eliminated in the alkaline denaturing gel. C) β-actin control RT-PCR products.
WISH analyses were performed using the full-length tbx22 probe to characterize tbx22 expression in developing branchial arch and craniofacial structures. tbx22 was first detected in 28 hpf embryos, where discrete expression was observed in segmental paraxial mesodermal tissues (Figure 6A, arrow), in a manner suggestive of pre-patterning of the vertebrae. This expression was quite transient - no tbx22 expression was detected in 26 or 30 hpf embryos. Distinct, bilateral tbx22 craniofacial expression domains were first detectable at 38 hpf, coincident with the onset of pectoral fin bud growth (Figure 6B, C, arrows). Analyses of whole mount and sectioned 40hpf embryos revealed bilateral tbx22 expression domains (Figure 6D, E-G, respectively), localized to mesenchymal cells underlying the oral epithelial bilayer. These tissues develop into bifurcated upper and lower elements prior to the formation of mouth opening at 42 hpf (Figure 6H, I). Medial to lateral serial sections of 42 hpf WISH stained specimens revealed discrete tbx22 expression at each mouth corner (Figure 6, J-N). tbx22 was also detected in the jaw joint of 51 and 55 hpf embryos, in a similar pattern as bapx1 (Miller et al., 2003) (Figure 5O, P, arrows), consistent with roles for tbx22 in primitive jaw joint formation.
The expression patterns of zebrafish tbx22 mirror some aspects of Tbx22 expression in the mouse (Bush et al., 2002; Herr et al., 2003), where tbx22 mRNAs were found to be expressed in mesenchymal tissue underlying the inferior nasal septum epithelium, which eventually fuses with the right and left palatal shelves to form the secondary palate. Mouse tbx22 mRNAs were also detected in caudal regions of the palatal shelves prior to fusion, and in basal anterior tongue mesenchyme (Bush et al., 2002; Herr et al., 2003). The expression pattern of zebrafish tbx22 is notable for its association with the stomodeum, the depression marking the future location of the mouth opening, and in the developing jaw joint. Expression of tbx22 in the developing mouse joint has not been reported. It is interesting that tbx18, a very close paralog of tbx22, is also expressed around the zebrafish mouth at 40 hpf, and in the palate of 3 dpf embryos (Begemann et al., 2002). In Xenopus, stomodeal ectoderm and endodermal layers progressively thin via apoptosis to form the primary mouth opening (Dickinson and Sive, 2006). Although the zebrafish mouth opening is thought to form by a different mechanism, where changes in intercellular junctions and cell orientations occur prior to rupture of the oral membrane (Hamlett et al., 1996; Waterman and Kao, 1982), tbx22 expression in the relatively simple zebrafish mouth formation correlates well with that of the comparatively complex palatal shelf development in the mouse and human.
SUMMARY
Our phylogenetic analysis of all known zebrafish T-box sequences containing genes identified a single zebrafish tbx22 ortholog. Two alternatively spliced tbx22 transcripts were identified, tbx22-1 and tbx22-2, which are predicted to encode proteins of 444 and 400 amino acids, respectively. Developmental RT-PCR analyses revealed that tbx22-1 was maternally expressed, while tbx22-2 was not. Both zygotically expressed tbx22 transcripts were present at all developmental stages examined. WISH analyses revealed discrete tbx22 expression in pharyngula staged embryos within ectomesenchymal tissue underlying the posterior and lateral aspects of the bilateral, bilaminar, stomodeal epithelial sheets. Tbx22 was also detected in the presumptive jaw joint, and may play a role in the formation of this structure. The relatively simple craniofacial expression pattern of tbx22 in developing zebrafish resembles the more complex Tbx22 expression patterns observed in mammalian craniofacial development, and provides insight into ancestral vertebrate facial patterning mechanisms. We anticipate that further studies of tbx22, and of other genes expressed in early zebrafish craniofacial development, will provide valuable insight into the etiology, and molecular based therapies, for the prevention and/or correction of a variety of human craniofacial dysmorphologies.
EXPERIMENTAL PROCEDURES
Zebrafish husbandry
Danio rerio were bred and maintained at 28°C on a 14 hr light/10 hr dark cycle, as described (Westerfield, 1995). Embryos were collected from natural matings, grown in system water and staged as described (Westerfield, 1995).
Reverse transcription polymerase chain reaction (RT-PCR) and 5′/3′ Rapid Amplification of cDNA Ends (RACE)
Total RNA was isolated from 19 hpf nacre and AB* zebrafish using TRIzol reagent (Invitrogen, Carlsbad, CA), and 1μg RNA was used as template for RT-PCR and RACE analyses, performed using the SMART RACE cDNA Amplification Kit (BD BioSciences, Palo Alto, CA). PCR amplification of RACE products was performed using Advantage 2 Polymerase (BD BioSciences, Palo Alto, CA), the forward primer 5′-GGGGAAGGAGAAGGTGAACACACCA-3′, and the reverse primer 5′-TGGATCTTCAGCGAGTGGTA-3′. Internal primers used to generate tbx22-1 and tbx22-2 isoform specific RT-PCR products were as follows: fragment ‘a’, forward primer t22-1F2, 5′-TCACGCGACAAGTGGATCATACTTCAGG-3′ and reverse primer t22-4R, 5′- CCCACTGAGAGCTGTGATAAACATACCTG - 3′; fragment ‘b’, forward primer t22-4F, 5′-GCTCTCAGTGGGTGATTGCTGGAAA-3′ and reverse primer t22-8R, 5′-TATGCGAGGAAACAGCTTGA-3′; fragment ‘c’, forward primer t22-8F, 5′- TTGGATCTTCAGCGAGTGGTA - 3′ and reverse nested universal primer (NUP) 5′-AAGCAGTGGTATCAACGCAGAGT-3′ (BD SMART RACE cDNA Amplification Kit; BD Biosciences/ Clontech, Mountain View, CA). Full-length zebrafish tbx22-1 and tbx22-2 cDNAs (fragments d-1 and d-2, respectively) were generated using primer T22-1F1, 5′-CCCATTTGCAAAGCGATGAGTTCG-3′, and reverse primer 5′-TGTGCCAAATGGCTCACAGAA-3′, and cloned into the TOPO-TA, pCRII cloning vector (Invitrogen, Carlesbad, CA). All clones were confirmed by double stranded nucleotide sequence analyses.
In vitro coupled transcription and translation
The complete cDNA sequences for tbx22-1 and tbx22-2 were subcloned into the EcoRI site of the pTnT expression vector (Promega Corporation, Madison, WI), and used in in vitro coupled transcription and translation reactions, using the TnT SP6 High-Yield Protein Expression System (Promega, Madison, WI), and biotin-conjugated lysine tRNAs. The resulting biotinylated translation products and Prestained Kaleidoscope Standard Molecular Weight Markers (Bio-Rad, Richmond, CA) were size fractionated on 4-20% Tris-HCL acrylamide gels (Bio-Rad, Richmond, CA), and transferred to nitrocellulose. Western blotting was performed via chemiluminescence using the Transcend, Non-Radioactive Translation Detection System (Promega, Madison, WI).
Phylogenetic analyses
Zebrafish tbx22 isoform ATG initiation codons were identified using the start codon prediction program, ATGpr (Salamov et al., 1998, http://www.hri.co.jp/atgpr). Phylogenetic analyses were performed on 19 T-box sequences chosen to represent the major subgroups of the T-box gene family (Agulnik et al., 1996; Ruvinsky et al., 200; and Takatori et al., 2004), using only the most highly conserved orthologous amino acids within the T-box DNA binding domain. The actual amino acid sequences used in our phylogenetic analysis are listed in Supplemental Data File 1. Zebrafish T-box amino acid sequences were compared to those of higher taxa to confirm the identification of each T-box paralog family (Takatori et al., 2004). Neighbor joining and maximum parsimony trees were constructed using the MEGA 3.1 software (http://www.megasoftware.net). The maximum parsimony tree was inferred with 1,000 bootstrap replicates, searched by close-neighbor-interchange (CNI) settings, using all 125 orthologous sites.
RT-PCR analysis of tbx22-1 and tbx22-2 isoform developmental expression
Developmentally staged zebrafish were collected, frozen in liquid nitrogen, and stored at −80°C. Total RNA was isolated using Trizol as described above, and 4.5 μg of each total RNA sample was used in RT reactions using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). The resulting RT products were used as template in 50-μl PCR reactions, performed with Platinum PCR SuperMix High Fidelity (Invitrogen, Carlsbad, CA), using the following reaction conditions: denaturation at 95°C for 5 minutes, followed by 35 cycles of 95°C for 40 seconds, 60°C for 1 minute, and 72°C for 2 minutes, followed by a final 7 minute extension at 72°C. The internal primers for fragment ‘a’ identified above, were used in this reaction. Twelve and 15 microliter aliquots of each PCR reaction were resolved by regular or alkaline agarose gel electrophoresis, respectively, stained with ethidium bromide, and digitally photographed. Control PCR primers for the zebrafish β-actin gene were βaF, 5′-GGAGAAGATCTGGCATCACACCTTCTAC-3′ and βaR, 5′-TGGTCTCGTGGATACCGCAAGATTCCAT-3′.
Whole mount and sectioned in situ hybridization analyses
Digoxigenin labeled sense and anti-sense riboprobes were generated from linearized zebrafish tbx22 and bapx1 (Miller et al., 2003) cDNAs using the DIG RNA Labeling Kit (SP6/T7) (Roche Applied Science, Indianapolis, IN). WISH was performed as previously described by us (Albertson et al., 2005), using a modification of published protocols (Thisse et al., 1993). Embryos were cleared, and photographed using Zeiss Stemi SV11, Leica DMRE, and Leica MZ12 microscopes, and Zeiss AxioCam HRc digital camera with Axiovision 3.1 software. Selected WISH labeled embryos were embedded in JB-4, and sectioned at 6 micron intervals.
Supplementary Material
Supplemental Data File 1. FASTA files of curated T-box coding sequences and identifiers for the genes and taxa used in the phylogenetic analysis.
ACKNOWLEDGMENTS
We wish to thank all of the members of the Yelick lab for helpful discussions, advice and expertise, and Loic Fabricant and Seija Cope for expert zebrafish husbandry. We thank Andrea Moreira and Angela Allard for their technical assistance, and The Forsyth Institute DNA Sequencing Core staff for their expertise. This research was supported in part by NIH/NIDCR grants DE14528 (PAJ), DE14683 (TLF), and DE12076, DE018043 (PCY).
Grant Sponsor: NIH/NICDR
Grant Numbers: DE14528 (PAJ), DE14683 (TLF), and DE018043, DE12076 (PCY).
REFERENCES
- Ahn DG, Ruvinsky I, Oates AC, Silver LM, Ho RK. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–8. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Payne-Ferreira TL, Postlethwait J, Yelick PC. Zebrafish acvr2a and acvr2b exhibit distinct roles in craniofacial development. Dev Dyn. 2005;233:1405–18. doi: 10.1002/dvdy.20480. [DOI] [PubMed] [Google Scholar]
- Begemann G, Gibert Y, Meyer A, Ingham PW. Cloning of zebrafish T-box genes tbx15 and tbx18 and their expression during embryonic development. Mech Dev. 2002;114:137–41. doi: 10.1016/s0925-4773(02)00040-0. [DOI] [PubMed] [Google Scholar]
- Braybrook C, Doudney K, Marcano ACB, Arnason A, Bjornsson A, Patton MA, Goodfellow PJ, Moore GE, Stanier P. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nature Genet. 2001;29:179–183. doi: 10.1038/ng730. [DOI] [PubMed] [Google Scholar]
- Bush JO, Lan Y, Maltby KM, Jiang R. Isolation and developmental expression analysis of Tbx22, the mouse homolog of the human X-linked cleft palate gene. Dev Dyn. 2002;225:322–6. doi: 10.1002/dvdy.10154. [DOI] [PubMed] [Google Scholar]
- Copley RR. The EH1 motif in metazoan transcription factors. BMC Genomics. 2005;6:169. doi: 10.1186/1471-2164-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AJG, Sive H. Development of the primary mouth in Xenopus laevis. Developmental Biology. 2006;295:700–713. doi: 10.1016/j.ydbio.2006.03.054. [DOI] [PubMed] [Google Scholar]
- Haenig B, Kispert A. Analysis of TBX18 expression in chick embryos. Dev Genes Evol. 2004;214:407–11. doi: 10.1007/s00427-004-0415-3. Epub 2004 Jul 15. [DOI] [PubMed] [Google Scholar]
- Hamlett WC, Schwartz FJ, Schmeinda R, Cuevas E. Anatomy, histology, and development of the cardiac valvular system in elasmobranchs. Journal of Experimental Zoology. 1996;275:83–94. [PubMed] [Google Scholar]
- Herr A, Meunier D, Muller I, Rump A, Fundele R, Ropers HH, Nuber UA. Expression of mouse Tbx22 supports its role in palatogenesis and glossogenesis. Dev Dyn. 2003;226:579–86. doi: 10.1002/dvdy.10260. [DOI] [PubMed] [Google Scholar]
- Kochilas LK, Potluri V, Gitler A, Balasubramanian K, Chin AJ. Cloning and characterization of zebrafish tbx1. Gene Expr Patterns. 2003;3:645–51. doi: 10.1016/s1567-133x(03)00108-x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7(8):563–74. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- Mahadevan NR, Horton AC, Gibson-Brown JJ. Developmental expression of the amphioxus Tbx1/ 10 gene illuminates the evolution of vertebrate branchial arches and sclerotome. Dev Genes Evol. 2004;214:559–66. doi: 10.1007/s00427-004-0433-1. [DOI] [PubMed] [Google Scholar]
- Marcano ACB, Doudney K, Braybrook C, Squires R, Patton MA, Lees MM, Richieri-Costa A, Lidral AC, Murray JC, Moore GE, Stanier P. TBX22 mutations are a frequent cause of cleft palate. J. Med. Genet. 2004;41:68–74. doi: 10.1136/jmg.2003.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stanier DYR, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–65. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Müller CW, Herrmann BG. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–8. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Ahn DG, Schilling TF, Nair S, Ruvinsky I, Geisler R, Rauch GJ, Haffter P, Zon LI, Zhou Y, Foott H, Dawid IB, Ho RK. The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development. 2003;130:5043–52. doi: 10.1242/dev.00704. [DOI] [PubMed] [Google Scholar]
- Prescott NJ, Lees MM, Winter RM, Malcolm S. Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Hum Genet. 2000;106:345–50. doi: 10.1007/s004390051048. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Silver LM, Gibson-Brown JJ. Phylogenetic analysis of T-Box genes demonstrates the importance of amphioxus for understanding evolution of the vertebrate genome. Genetics. 2000;156:1249–57. doi: 10.1093/genetics/156.3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov AA, Nishikawa T, Swindells MB. Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics. 1998;14:384–90. doi: 10.1093/bioinformatics/14.5.384. [DOI] [PubMed] [Google Scholar]
- Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev. 2005;122(2):131–44. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Takatori N, Hotta K, Mochizuki Y, Satoh G, Mitani Y, Satoh N, Satou Y, Takahashi H. T-box genes in the ascidian Ciona intestinalis: characterization of cDNAs and spatial expression. Dev Dyn. 2004;230(4):743–53. doi: 10.1002/dvdy.20082. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail 1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Waterman RE, Kao R. Formation of the mouth opening in the zebrafish embryo. SEM. 1982;3:1249–1257. [Google Scholar]
- Westerfield M. The zebrafish book. University of Oregon; Eugene, OR: 1995. [Google Scholar]
- Wyszynski DF. Cleft Lip and Palate: From Origin to Treatment. Oxford University Press; New York, New York: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data File 1. FASTA files of curated T-box coding sequences and identifiers for the genes and taxa used in the phylogenetic analysis.