Abstract
To compare the efficiency of novel mitogenic agents and traditional mitosis inductors, 18 patients with splenic marginal zone lymphoma (SMZL) were studied. Three cultures using oligodeoxynucleotide (ODN) plus interleukin-2 (IL-2), or TPA, or LPS were setup in each patient. Seventeen/18 cases with ODN + IL2 had moderate/good proliferation (94, 4%) as compared with 10/18 cases with TPA and LPS (55%) (P = .015); 14/18 (77, 7%) cases with ODN + IL2 had sufficient good quality of banding as compared with 8/18 cases (44, 4%) with TPA and LPS. The karyotype could be defined from ODN + IL2-stimulated cultures in all 18 patients, 14 of whom (77, 7%) had a cytogenetic aberration, whereas clonal aberrations could be documented in 9 and in 3 cases by stimulation with LPS and TPA, respectively. Recurrent chromosome aberrations in our series were represented by aberrations of chromosome 14q in 5 patients, by trisomy 12 and 7q deletion in 4 cases each, and by abnormalities involving 11q and 13q in two cases each. These findings show that stimulation with ODN + IL2 offers more mitotic figures of better quality and results in an increased rate of clonal aberrations in SMZL, making this method ideal for prospective studies aiming at the definition of the prognostic impact of cytogenetic aberrations in this disorder.
1. Introduction
Splenic marginal zone lymphoma (SMZL) is an indolent disease, representing <2% of the lymphoid neoplasms, which was recognized as separate clinicopathological entity in the World Health Organization 2008 (WHO) classification [1]. Studies using conventional cytogenetic analysis and molecular cytogenetic techniques disclosed a number of recurrent chromosome and genetic lesions in this disorder [2]. Due to low spontaneous mitotic activity, stimulation using 12-O-tetradecanoylphorbol 12-myristate 13-acetate (TPA), or a combination of TPA and lipopolysaccharide (LPS) in parallel cultures, was employed in previous studies [3–8] revealing abnormal karyotypes in 43–72% of the cases [5, 9]. Indeed, the combination of data derived from conventional cytogenetic analysis, along with data derived from the application of fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH) [10, 11], allowed for the definition of a cytogenetic profile of SMZL. Deletion of the long arm of chromosome 7 is regarded as the most characteristic anomaly of SMZL [12]; other recurrent aberrations are represented by total/partial trisomy 3q in 20–40% of the cases and by 14q32/IgH translocations, +12, 17p/TP53 deletion, 6q−, +12, and +18 in 5–10% of the cases, as documented in a recent large multicentre study using the traditional TPA mitogen stimulation [13].
Because conventional karyotype analysis is the only method allowing for the visualization in a single experiment of any type of chromosomal defect, including gains, losses, and balanced translocations, attention was recently devoted to the development of efficient mitogenic stimulation. In chronic lymphocytic leukemia (CLL), the introduction of stimulation by CpG oligodeoxynucleotide (ODN-DSP30) combined with IL2 allowed for the identification of more cases with clonal aberrations than in previous analyses using other mitogens and showed that a fraction of cases with apparently normal FISH results may carry chromosome lesions in regions not covered by conventional probe panels [14, 15]. Because the identification of an optimal mitogen stimulation can be expected to reduce the rate of normal results in low-grade lymphoproliferative disorder, we designed this study which aimed at analyzing the impact of the innovative combination of immunostimulatory ODN + IL2 in cell culture in 18 cases of well-documented SMZL.
2. Design and Methods
2.1. Patients and Samples
Eighteen cases of SMZL seen at our Institution between 2008 and June 2010 were included in the present analysis (Table 1). All the patients were studied by cytogenetic analysis as part of diagnostic workup. Samples from 13 patients were obtained at disease presentation, whereas 5 patients were sent for cytogenetic analysis 5–12 months after initial presentation.
Table 1.
Clinical features at presentation in 18 cases of SMZL.
| Median age, y (range) | 74 (56–85) |
| Sex, male/female | 13/5 |
| Splenomegaly yes/no | 11/7 |
| Lymphadenopathy (yes/no) | 1/14 |
| >40% lymphs in the BM aspirate | 11/4 |
| Lymphocytosis ≥5 × 109/L yes/no | 11/7 |
| Absolute lymphocyte count (×109/L) | 0.68–31.49 (median 6,63) |
| Villous lymphocytes yes/no | 6/9 |
| Hb < 12 g/dL yes/no | 5/13 (8.6–15.2) |
| Platelet count ≤100 × 109/L yes/no | 4/14 (52–239) |
| CD5 expression yes/no | 5/13 |
Diagnosis was made according to the WHO [1] histopathologic criteria in one patient in whom splenectomy was performed for diagnostic purposes; in the remaining 17 patients, diagnosis was based on the combination of presentation features, morphologic and immunologic features, and bone marrow findings [16]. Minimal requirements were represented by (a) peripheral and bone marrow lymphocytosis (i.e., >5 × 109/L B-lymphocytes in the PB and/or >40% lymphocytes in the BM aspiration or lymphoid infiltrate on biopsy sections), with or without splenomegaly and minimal adenopathy, (b) morphology consistent with SMZL (i.e., small-to-medium-sized lymphocytes, with or without villous lymphocytes and/or plasmacytoid features, and (c) immunophenotype consistent with chronic B-cell proliferation with a Matutes score ≤3 [17].
2.2. Conventional Cytogenetic Analysis
Conventional cytogenetic analysis was performed on cells obtained from peripheral blood (PB) in 14 cases, from BM aspirate in 3 cases, and from a spleen sample in 1 case (Table 2). Methods for cytogenetic analysis used in our laboratories were previously published [18]. Spleen samples were minced with a scalpel to obtain a single cell suspension. After separation by centrifugation over Ficoll-Hypaque, PB, BM, and splenic cells were cultured for 72 h in 10 ml RPMI 1640 (Gibco-Invitrogen) supplemented with 20% fetal calf serum (FCS-Gibco-Invitrogen), 2 mmol/L GlutaMAX (Gibco-Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco-Invitrogen). Three separate cell cultures were setup in all patients, using the 3 different mitogens: (i) 12-O-tetradecanoylphorbol 12-myristate 13-acetate (TPA; 50 ng/mL–Sigma-Aldrich), (ii) lipopolysaccharide (LPS; 40 μg/mL–Sigma-Aldrich), and (iii) immunostimulatory CpG-oligonucleotide DSP30 plus IL2 (2 μmol/L GpC-ODN-TCGTCGCTGTCTCCGCTTCTTCTTGCC) (TibMolBiol, Berlin, Germany/IL2 100 U/mL Stem Cell Technologies Inc) according to the method described by Dicker et al. [14]. Whenever possible, an additional 72 h unstimulated control culture was setup (6 cases). All cultures were setup with a cell concentration of 2 × 106/mL and incubated at 37°C in a 5% CO2 fully humidified atmosphere under standard conditions which have remained unchanged at our laboratories during the study period. Colcemid (Kario Max Colcemid Solution 0,05 μg/mL Gibco, Invitrogen) was added for four hours before harvest. Harvesting and slide preparation were performed by the same technician (ET) throughout the study period using hypotonic treatment (20 minutes incubation in 0,075 mol/L potassium chloride); a classical 3 : 1 methanol/acetic acid solution was used as fixative. Slides were prepared using a predetermined volume (i.e., 20 μl) of fixed cell suspension, and metaphases were G-banded with Wright's stain [19]. Whenever possible, 20 or more metaphases were analyzed from each culture, and karyotypes were described according to the International System for Human Cytogenetics Nomenclature (ISCN 2005) [20]. Complex karyotype was defined by the presence of 3 or more cytogenetic aberrations in the same clone. To compare the efficiency of the 3 different mitogens, the following cytogenetic features were assessed in the different culture types by visualization at the microscope of the metaphases present on one slide.
Table 2.
Efficiency of different mitogens and karyotypes in 18 patients with SMZL.
| Patient | Age | Sample (*) | Score (**) | Karyotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPA | LPS | ODN + IL2 | ||||||||||
| Quality | Proliferation | Efficiency | Quality | Proliferation | Efficiency | Quality | Proliferation | Efficiency | ||||
| 1 | 70 | Pb (21.9) | 3 | 3 | N | 3 | 2 | N | 3 | 3 | N | 46, XY [20] |
| 2 | 78 | BM (38%) | 2 | 3 | N | 2 | 3 | A | 3 | 3 | A | 47, XX, +12 [2]/46,XX [18] |
| 3 | 77 | Pb (7.28) | 0 | 1 | F | 1 | 3 | A | 3 | 3 | A | 47, XY, add(5)(p15),+22 [15]/47, XY, add (5)(p15), del (7)(q22), i(8)(p10), +del (14)(q24) [5] |
| 4 | 75 | BM (35%) | 3 | 3 | N | 3 | 4 | A | 3 | 2 | A | Near tetraploid: 91, XXY, idic (1)(p11), del (11)(q21), add(14)(q32), +mar1, +mar2 [3]/46, XY [17] |
| 5 | 76 | Pb (8.63) | 0 | 1 | F | 0 | 1 | F | 2 | 3 | A | 46, XY, der (3)(p26), del (7)(q32), del (13)(q22) [4]/46, XY [21] |
| 6 | 69 | Pb (34.8) | 0 | 1 | F | 3 | 3 | N | 3 | 3 | N | 46, XY [20] |
| 7 | 55 | Pb (16.1) | 3 | 3 | N | 3 | 2 | A | 3 | 3 | A | 46, XY, t(3;13)(q21;q13), del (11)(q12) [13]/46, XY [7] |
| 8 | 84 | Pb (20) | 0 | 1 | F | 3 | 3 | A | 4 | 3 | A | 46, XX, -9, t(14;19)(q32;q13), +add (14)(q32) [19]/46, XX [1] |
| 9 | 76 | Pb (4.5) | 3 | 3 | A | 3 | 3 | A | 4 | 4 | A | 47, XY, +12 [15]/46, XY [5] |
| 10 | 63 | Pb (NA) | 2 | 4 | N | 2 | 3 | N | 2 | 4 | N | 46, XY [20] |
| 11 | 86 | Pb (6.1) | 3 | 3 | A | 1 | 2 | A | 3 | 3 | A | 46,XX,del(14) (q22) [16]/46,XX [5] |
| 12 | 76 | Spleen | 0 | 1 | F | 0 | 1 | F | 2 | 3 | N | 46, XX [20] |
| 13 | 75 | Pb (23.1) | 3 | 3 | N | 1 | 2 | N | 3 | 4 | A | 48, XY, +12, +15 [5]/46, XY [15] |
| 14 | 83 | BM (30%) | 2 | 3 | N | 2 | 3 | N | 2 | 4 | A | 46, XY, del (3)(p13) [3]/46, XY [17] |
| 15 | 85 | Pb (4.2) | 0 | 1 | F | 3 | 3 | A | 3 | 3 | A | 47, XY, +12, del (14)(q24) [12]/46, XY [8] |
| 16 | 72 | Pb (2.9) | 0 | 1 | F | 0 | 1 | F | 4 | 3 | A | 46, XY, dup (1)(q21q32), del (7)(q32), del (13)(q14q22) [2]/47, XY, del (7)(q32), der (12) t(3;12)(p11;p11), |
| del (13)(q14q22), +mar1 +mar2 [1]/46, XY [17] | ||||||||||||
| 17 | 56 | Pb (4.9) | 3 | 3 | A | 3 | 4 | A | 3 | 3 | A | 46, XY, del (7)(q32) [8]/46, XY [12] |
| 18 | 82 | Pb (7.1) | 3 | 2 | N | 0 | 1 | F | 3 | 3 | A | 47, XX, +3 [4]/46, XX [16] |
*Absolute lymphocyte count × 109/L at the time of cytogenetic investigation, or % BM infiltration, as appropriate. NA: not available.
**See materials and methods for details; A: abnormal, N: normal, and F: failure.
(a) Proliferation —
Based on the number of mitotic figures, the following score was adopted.
- Score 1:
failure, defined by the presence of 0-1 mitotic figures.
- Score 2:
poor proliferation, defined by the presence of 2–10 mitotic figures.
- Score 3:
moderate proliferation, defined by the presence of 11–19 mitotic figures.
- Score 4:
good proliferation, defined by the presence of ≥20 mitotic figures.
(b) Quality of Banding —
The number of chromosomal bands per haploid set of chromosomes was counted referring to the ideograms of banding patterns present in the guidelines of ISCN 2005 [20]. The following score was adopted.
- Score 1:
insufficient quality for karyotyping (<100 visible chromosome bands).
- Score 2:
poor quality (<200 visible chromosome bands).
- Score 3:
sufficient quality (200–300 visible chromosome bands).
- Score 4:
good quality (>300 visible chromosome bands).
(c) Stimulation Efficiency —
The number of metaphases with clonal abnormalities in each culture system was evaluated, and the karyotypes were divided in three groups:.
- Score 1:
failure, less than 10 analyzable metaphases.
- Score 2:
normal, absence of clonal abnormalities.
- Score 3:
abnormal I, karyotype with clonal chromosomal abnormalities, that is, the same structural rearrangement or chromosome gain in at least two mitotic figures, or chromosome loss in at least three mitoses.
3. Results
3.1. Hematologic and Clinical Features
All 18 patients had an unequivocal diagnosis of SMZL with PB and/or BM involvement by a clonal expansion of B-lymphocytes consistent with a marginal zone phenotype as assessed by immunophenotyping. The patients had 2,9–34,8 × 109/L PB lymphocytes at time of sampling for cytogenetic analysis, and no patient had cytologic and/or histologic features suggestive of transformation into high-grade lymphoma. BM involvement with >40% lymphocytes was detected in 11/15 cases, splenomegaly was present in 11/18 cases. A minority of patients had anemia or thrombocytopenia. Demographics and hematologic data in our patients are presented in Table 1.
3.2. Outcome of Cytogenetic Investigations
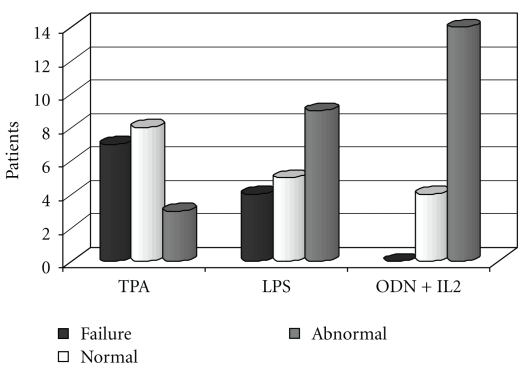
The karyotype could be defined in all 18 cases. No analyzable mitoses were obtained from 72 h unstimulated parallel culture in 6 cases. The outcome of cytogenetic investigations using different mitogens is shown in Figures 1–5.
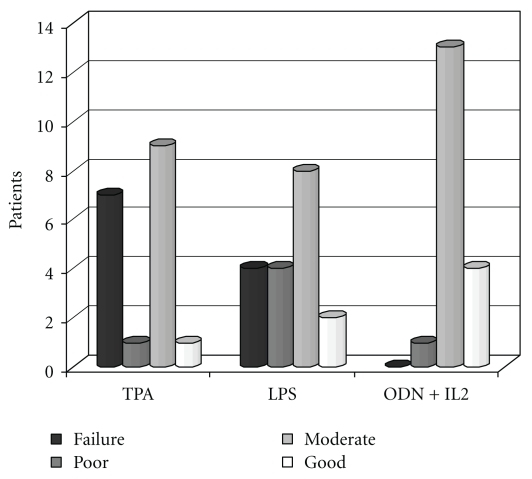
Figure 1.
No. of patients with failure, poor, moderate, and good proliferation following stimulation by different mitogens.
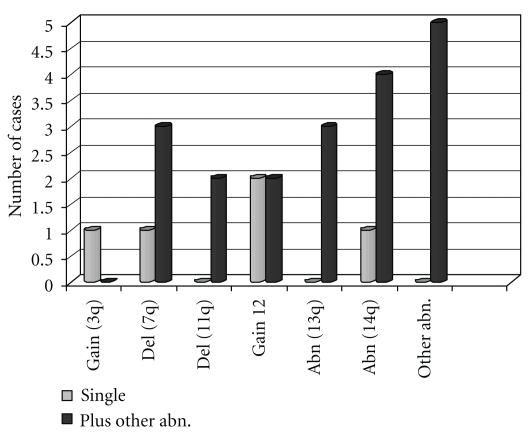
Figure 5.
Total clonal chromosome abnormalities: distribution of recurrent abnormalities if they were found as single aberration or in association with other abnormalities.
3.3. Proliferation
Proliferation with at least 1 mitogen was assessable in all 18 cases. The number of cases with failure, low, moderate, and good proliferation is shown in Figure 1. More cases with score 3-4 were seen in ODN + IL2-stimulated cultures (17 cases = 94,4%) as compared with TPA and LPS (10 cases each; 55,5%) (P = .015).
Seven/18 patients (38,8%) with TPA and 4/18 patients (22,2%) with LPS had score 1 (failure), whereas no failure was observed in ODN + IL2-stimulated culture.
3.4. Quality of Banding
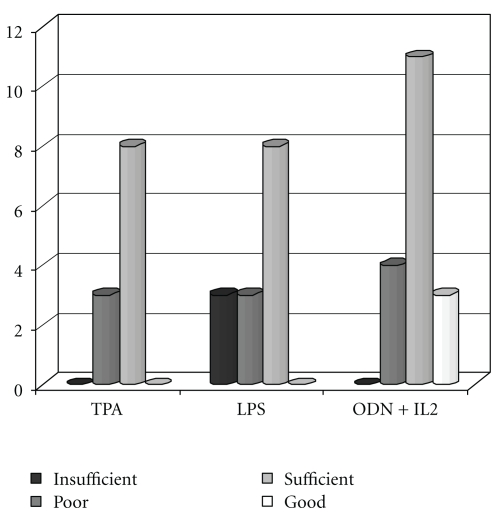
The quality of banding expressed as number of bands in mitotic figures from the different cell cultures is shown in Figure 2. A good quality of banding (score 4) was observed in 3/18 cases with ODN + IL2 and in no case with TPA or LPS. Overall, 14/18 cases with ODN + IL2 had score 3-4 (77,7%) as compared with 8/18 cases (44,4%) with TPA and LPS (P = .067). An example of the quality of chromosome banding is shown in Figure 3.
Figure 2.
Quality of banding: black-coloured column corresponds to insufficient chromosome quality, grey-coloured column corresponds to poor quality, light grey and white correspond to sufficient and good quality, respectively, in every stimulation procedure.
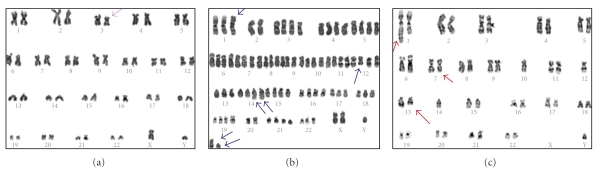
Figure 3.
G-banding karyotypes showing some examples of poor quality (score2—(a), patient 14), sufficient quality (score3—(b), patient 4), and good quality (score4—(c), patient 16).
3.5. Stimulation Efficiency
The karyotypes are described in Table 2, along with outcome measures (i.e., quality of banding and proliferation score) using different mitogens. The karyotype could be defined from ODN + IL2-stimulated cultures in all 18 patients, 14 of whom (77,7%) had a cytogenetic aberration.
Clonal aberrations could be documented in 9 cases (50%) and in 3 cases (16,6%) by stimulation with LPS and TPA, respectively, whereas in the remaining cases, normal karyotype or failure was observed with these mitogens as shown in Figure 4.
Figure 4.
Stimulation efficiency: grey colour column represents abnormal cells, and dark and white colour columns represent failure and normal, respectively, in every stimulation procedure.
Five patients had a complex karyotype (pat. 3, 4, 5, 8, and 16 in Table 2), with numerical gains and structural abnormalities. One case was in the near-tetraploid range (pat. 4). The most frequent abnormalities (see Figure 5) were represented by aberrations of chromosome 14q in 5 patients, 3 of whom had a 14q interstitial deletion (nos. 3, 11, and 15). In patient 8 a t(14; 19)(q32; q13) translocation was detected. Trisomy 12 and 7q deletion were observed in 4 cases each. Abnormalities involving 11q and 13q were observed in two cases each.
4. Discussion
Cytogenetic analysis has an established role in the diagnostic workup [21] and risk assessment of chronic lymphoproliferative disorders [13, 22, 23]. Because the mitotic index in these indolent disorders is low, stimulation with LPS and TP was widely employed [3, 24]. Evidence was recently provided that conventional karyotyping may allow for the detection of aberrations, especially translocations, not detectable by molecular cytogenetic methods [25] and that ODN + IL2 stimulation may disclose more cytogenetically abnormal cases than was previously thought in CLL [14, 26]. In particular, in a CLL Research Consortium study, more clonal abnormalities were observed after culture of CLL cells with ODN than with the traditional pokeweed mitogen (PWM) plus TPA [27]. All clonal abnormalities in PWM + TPA cultures were observed in ODN cultures, whereas ODN identified some clones not found by PWM + TPA. These results were reproducible in five different laboratories, and all abnormalities were concordant with FISH.
In this study, we were able to show that improved mitotic stimulation can be obtained in SMZL, a low-grade lymphoproliferative disorder, by using ODN + IL2 in analogy with CLL. Indeed, a significantly greater number of mitotic figures could be observed in ODN + IL2-stimulated cultures, which offered chromosomes of better quality with more clonal aberrations with respect to TPA/LPS-stimulated cultures. The karyotype could be defined in 100% of ODN + IL2-stimulated cultures, 77,7% of which showed a clonal abnormality, as compared with 50% karyotypically abnormal cases obtained by a combination of results from LPS/TPA-stimulated cultures in the same patients. The percentage of cytogenetically abnormal cases in LPS/TPA-stimulated cultures was in line with previous reports [5, 9, 13]. It is worth noting that at the time of sampling for cytogenetic analysis the majority of our patients were at an initial stage of the disease with moderate lymphocytosis and splenomegaly, no lymph node involvement, and absence of anemia or thrombocytopenia in the majority of them. Interestingly, the capability to detect abnormal clones was independent of the degree of lymphocytosis and BM involvement, since all cases with <5 × 109/L lymphocytes in the PB and with <40% BM lymphocytes could be shown to have an abnormal karyotype (Table 2).
Thus, our data demonstrate that ODN + IL2 is the method of choice to enhance cell divisions of good quality for karyotyping in SMZL, in keeping with a recent analysis documenting a 97% rate of aberrant karyotypes in 29 SMZL [28].
The cytogenetic profile in our patients confirms that 7q deletion, 3q abnormalities, +12, and 14q32 translocations are frequently encountered in this disease. Interestingly, a 14q22-24 deletion occurred as single abnormality in 2 cases and as additional aberration in 1, resulting in a 16,6% incidence for this aberration. Only 3% of the SMZL studies by Salido and coworkers [13] were found to carry 14q deletions, which showed a 1,5% overall incidence in a study of 3054 mature B-cell neoplasms [29]. The majority of cases with 14q deletion in the latter analysis were represented by atypical CLL cases having a therapy-demanding disease. Our 3 SMZLs with 14q deletion also had a therapy-demanding disease, requiring treatment after 1, 8, and 13 months from cytogenetic analysis.
One case in this study with a t(14;19)(q32;q13) adds to a previous report of 4/330 cases carrying this translocation [13]. The 14;19 translocation involving IgH and BCL3 is a rare aberration usually associated with a heterogeneous group of B-cell malignancies [30], including an atypical form of CLL with aggressive clinical features [31]. The identification of this subtle rearrangement may be difficult when banding quality is suboptimal, and it is possible that improved resolution obtained in ODN + IL2-stimulated cultures made the detection of this subtle rearrangement easier. In line with previous studies [32], our case had a rapidly progressive disease requiring treatment 3 months after diagnosis due to rapid lymphocyte doubling time.
In conclusion, we have shown that stimulation of mitosis with ODN + IL2 offers more mitotic figures of better quality and results in an increased rate of clonal aberrations in SMZL, in analogy with CLL. The profile of chromosome lesions obtained with this mitogen was in line with previous data, confirming that cytogenetic findings may be useful for the diagnosis of this lymphoid neoplasia. The detection of subtle rearrangements, such as 14q deletion and 14;19 translocations, might have been facilitated by improved banding resolution. Although more sensitive molecular genetic techniques using array CGH technology [33] may be of value for the detection of subtle unbalanced genetic lesions, this cytogenetic method may be ideal for prospective studied aiming at the definition of the prognostic impact of chromosome aberrations in SMZL.
Acknowledgments
This work was supported by MIUR PRIN, by Fondi Regionali ER, and by AIL-FE to A. Cuneo. L. Rizzotto is a fellow of AIL-FE A. Bardi and F. Cavazzini contributed equally. P. Musto and A. Cuneo shared senior authorship.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 2.Franco V, Florena AM, Iannitto E. Splenic marginal zone lymphoma. Blood. 2003;101(7):2464–2472. doi: 10.1182/blood-2002-07-2216. [DOI] [PubMed] [Google Scholar]
- 3.Oscier DG, Matutes E, Gardiner A, et al. Cytogenetic studies in splenic lymphoma with villous lymphocytes. British Journal of Haematology. 1993;85(3):487–491. doi: 10.1111/j.1365-2141.1993.tb03337.x. [DOI] [PubMed] [Google Scholar]
- 4.Dierlamm J, Michaux L, Wlodarska I, et al. Trisomy 3 in marginal zone B-cell lymphoma: a study based on cytogenetic analysis and fluorescence in situ hybridization. British Journal of Haematology. 1996;93(1):242–249. doi: 10.1046/j.1365-2141.1996.522522.x. [DOI] [PubMed] [Google Scholar]
- 5.Solé F, Woessner S, Florensa L, et al. Frequent involvement of chromosomes 1, 3, 7 and 8 in splenic marginal zone B-cell lymphoma. British Journal of Haematology. 1997;98(2):446–449. doi: 10.1046/j.1365-2141.1997.2163033.x. [DOI] [PubMed] [Google Scholar]
- 6.Solé F, Salido M, Espinet B, et al. Splenic marginal zone B-cell lymphomas: two cytogenetic subtypes, one with gain of 3q and the other with loss of 7q. Haematologica. 2001;86(1):71–77. [PubMed] [Google Scholar]
- 7.Gazzo S, Baseggio L, Coignet L, et al. Cytogenetic and molecular delineation of a region of chromosome 3q commonly gained in marginal zone B-cell lymphoma. Haematologica. 2003;88(1):31–38. [PubMed] [Google Scholar]
- 8.Cuneo A, Bigoni R, Roberti MG, et al. Molecular cytogenetic characterization of marginal zone B-cell lymphonia: correlation with clinicopathologic findings in 14 cases. Haematologica. 2001;86(1):64–70. [PubMed] [Google Scholar]
- 9.Troussard X, Mauvieux L, Radford-Weiss I, et al. Genetic analysis of splenic lymphoma with villous lymphocytes: a Groupe Francais d’Hematologie Cellulaire (GFHC) study. British Journal of Haematology. 1998;101(4):712–721. doi: 10.1046/j.1365-2141.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 10.Hernández JM, García JL, Gutiérrez NC, et al. Novel genomic imbalances in b-cell splenic marginal zone lymphomas revealed by comparative genomic hybridization and cytogenetics. American Journal of Pathology. 2001;158(5):1843–1850. doi: 10.1016/S0002-9440(10)64140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vega F, Cho-Vega JH, Lennon PA, et al. Splenic marginal zone lymphomas are characterized by loss of interstitial regions of chromosome 7q, 7q31.32 and 7q36.2 that include the protection of telomere 1 (POT1) and sonic hedgehog (SHH) genes. British Journal of Haematology. 2008;142(2):216–226. doi: 10.1111/j.1365-2141.2008.07176.x. [DOI] [PubMed] [Google Scholar]
- 12.James Watkins A, Huang Y, Ye H, et al. Splenic marginal zone lymphoma: characterization of 7q deletion and its value in diagnosis. Journal of Pathology. 2010;220(4):461–474. doi: 10.1002/path.2665. [DOI] [PubMed] [Google Scholar]
- 13.Salido M, Baró C, Oscier D, et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: a multicenter study of the Splenic B-Cell Lymphoma Group. Blood. 2010;116(9):1479–1488. doi: 10.1182/blood-2010-02-267476. [DOI] [PubMed] [Google Scholar]
- 14.Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: a study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108(9):3152–3160. doi: 10.1182/blood-2006-02-005322. [DOI] [PubMed] [Google Scholar]
- 15.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgVH status and immunophenotyping. Leukemia. 2007;21(12):2442–2451. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 16.Mollejo M, Menarguez J, Lloret E, et al. Splenic marginal zone lymphoma: a distinctive type of low-grade B-cell lymphoma: a clinicopathological study of 13 cases. American Journal of Surgical Pathology. 1995;19(10):1146–1157. [PubMed] [Google Scholar]
- 17.Matutes E, Oscier D, Montalban C, et al. Splenic marginal zone lymphoma proposals for a revision of diagnostic, staging and therapeutic criteria. Leukemia. 2008;22(3):487–495. doi: 10.1038/sj.leu.2405068. [DOI] [PubMed] [Google Scholar]
- 18.Castoldi GL, Lanza F, Cuneo A. Cytogenetic aspects of B-cell chronic lymphocytic leukemia: their correlation with clinical stage and different polyclonal mitogens. Cancer Genetics and Cytogenetics. 1987;26(1):75–84. doi: 10.1016/0165-4608(87)90135-x. [DOI] [PubMed] [Google Scholar]
- 19.Rooney DE. Human Cytogenetics: Malignancy and Acquired Abnormalities. Oxford university Press; 2001. [Google Scholar]
- 20.Karger MF. Guidelines for Cancer Cytogenetics: Supplement to an International System for Human Cytogenetics Nomenclature ISCN. Basel, Switzerland, 2005.
- 21.Hernandez JM, Mecucci C, Criel A, et al. Cytogenetic analysis of B cell chronic lymphoid leukemias classified according to morphologic and immunophenotypic (FAB) criteria. Leukemia. 1995;9(12):2140–2146. [PubMed] [Google Scholar]
- 22.Gruszka-Westwood AM, Hamoudi RA, Matutes E, Tuset E, Catovsky D. p53 abnormalities in splenic lymphoma with villous lymphocytes. Blood. 2001;97(11):3552–3558. doi: 10.1182/blood.v97.11.3552. [DOI] [PubMed] [Google Scholar]
- 23.Zenz T, Döhner H, Stilgenbauer S. Genetics and risk-stratified approach to therapy in chronic lymphocytic leukemia. Best Practice and Research. 2007;20(3):439–453. doi: 10.1016/j.beha.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Juliusson G, Oscier DG, Fitchett M, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. New England Journal of Medicine. 1990;323(11):720–724. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 25.Haferlach C, Dicker F, Weiss T, et al. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes and Cancer. 2010;49(9):851–859. doi: 10.1002/gcc.20794. [DOI] [PubMed] [Google Scholar]
- 26.Buhmann R, Kurzeder C, Rehklau J, et al. CD40L stimulation enhances the ability of conventional metaphase cytogenetics to detect chromosome aberrations in B-cell chronic lymphocytic leukaemia cells. British Journal of Haematology. 2002;118(4):968–975. doi: 10.1046/j.1365-2141.2002.03719.x. [DOI] [PubMed] [Google Scholar]
- 27.Heerema NA, Byrd JC, Dal Cin PS, et al. Stimulation of chronic lymphocytic leukemia cells with CpG oligodeoxynucleotide gives consistent karyotypic results among laboratories: a CLL Research Consortium (CRC) Study. Cancer Genetics and Cytogenetics. 2010;203(2):134–140. doi: 10.1016/j.cancergencyto.2010.07.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meloni-Ehrig A, Meck J, Christacos N, Kelly J, Matyakhina L, Schonberg S. Stimulation of B-cell mature malignancies with the CpG-oligonucleotide DSP30 and interleukin-2 for improved detection of chromosome abnormalities. Blood. 2009;114 ASH Annual Meeting Abstracts. Abs no. 1955. [Google Scholar]
- 29.Reindl L, Bacher U, Dicker F, et al. Biological and clinical characterization of recurrent 14q deletions in CLL and other mature B-cell neoplasms. British Journal of Haematology. 2010;151(1):25–36. doi: 10.1111/j.1365-2141.2010.08299.x. [DOI] [PubMed] [Google Scholar]
- 30.Martín-Subero JI, Ibbotson R, Klapper W, et al. A comprehensive genetic and histopathologic analysis identifies two subgroups of B-cell malignancies carrying a t(14;19)(q32;q13) or variant BCL3-translocation. Leukemia. 2007;21(7):1532–1544. doi: 10.1038/sj.leu.2404695. [DOI] [PubMed] [Google Scholar]
- 31.Huh YO, Abruzzo LV, Rassidakis GZ, et al. The t(14;19)(q32;q13)-positive small B-cell leukaemia: a clinicopathologic and cytogenetic study of seven cases. British Journal of Haematology. 2007;136(2):220–228. doi: 10.1111/j.1365-2141.2006.06416.x. [DOI] [PubMed] [Google Scholar]
- 32.Michaux L, Dierlamm J, Wlodarska I, Bours V, Van Den Berghe H, Hagemeijer A. t(14;19)/BCL3 rearrangements in lymphoproliferative disorders: a review of 23 cases. Cancer Genetics and Cytogenetics. 1997;94(1):36–43. doi: 10.1016/s0165-4608(96)00247-6. [DOI] [PubMed] [Google Scholar]
- 33.Robledo C, García JL, Caballero D, et al. Array comparative genomic hybridization identifies genetic regions associated with outcome in aggressive diffuse large B-cell lymphomas. Cancer. 2009;115(16):3728–3737. doi: 10.1002/cncr.24430. [DOI] [PubMed] [Google Scholar]