Maternal high-fat diet results in increased body size in third-generation female offspring through the paternal line that may be due to programming of paternally expressed genes.
Abstract
The health consequences of in utero exposure to maternal obesity on future generations are concerning because they contribute to increased rates of diabetes, cardiovascular disease, and metabolic syndrome. We previously reported that maternal high-fat diet exposure in mice resulted in an increase in body size and reduced insulin sensitivity that persisted across two generations via both maternal and paternal lineages. However, because the first generation's primordial germ cells may be affected by gestational exposure, analysis of phenotype transmission into a third generation (F3) is necessary to determine whether stable epigenetic programming has occurred. Therefore, we have examined the body size and insulin sensitivity of male and female F3 offspring. We found that only females displayed the increased body size phenotype, and this effect was only passed on via the paternal lineage. The finding of a paternally transmitted phenotype to F3 female offspring supports a stable germline-based transgenerational mode of inheritance; thus we hypothesized that imprinted genes may be involved in this epigenetic programming. Using a quantitative TaqMan Array for imprinted genes to examine paternally or maternally expressed loci in F3 female livers, we detected a potential dynamic pattern of paternally expressed genes from the paternal lineage that was not noted in the maternal lineage. These findings suggest that the environmental influence on developmental regulation of growth and body size may be the result of broad programming events at imprinted loci, thereby providing sex specificity to both the transmission and inheritance of traits related to disease predisposition.
Transgenerational epigenetic transmission of traits allows future generations to be maximally competitive in their environments. Adaptive gene programs acquired during the parental lifespan perpetuate into the next generation, enabling future generations to better survive in a challenging habitat. However, epidemiological studies suggest that exposure to environmental challenges such as stress, toxins, or unhealthy diet results in maladaptive parental responses that can be passed to offspring, predisposing them to disease (1–9). These epigenetic traits can spread quickly, resulting in a population-wide manifestation of a phenotype over several generations. Transgenerational transmission could exacerbate the rapid onset of phenotypes such as increased height, obesity, or diabetes in human populations over the last century (10–13).
High-fat diet (HFD) consumption during pregnancy increases the rates of obesity and metabolic syndrome by predisposing offspring toward an obesogenic phenotype (14). There is substantial evidence in humans and model organisms demonstrating that modulating maternal diet through either caloric restriction or high-fat feeding results in a predisposition to obesity and metabolic syndrome in the first-generation (F1) offspring (7, 15–23). Furthermore, recent data indicate that F1 can pass similar phenotypes to the second generation (F2) (6, 8, 9, 24–26), and even into the third (F3) (27). We have previously shown that maternal high-fat diet (mHFD) results in increased body length and insulin insensitivity over two generations of offspring that can be perpetuated through both maternal and paternal lineages (28). These studies suggest that maternal diet and its transgenerational consequences on offspring accelerate body length and insulin insensitivity phenotypes by hardwiring future generations toward a predisposition to these traits. Thus, an examination of mechanisms required to epigenetically program an organism is critical in furthering our understanding of how population-wide epidemics occur, and ultimately how they can be intervened upon.
Mechanistically, little is known about the transgenerational epigenetic perpetuation of traits such as obesity or metabolic syndrome. Because mHFD programs a complex array of phenotypes, it is important to determine those that result from stable germline-based epigenetic marks, and those that require active reinforcement by maternal behavior, physiology, or further environmental cues to persist. Demonstration of a germline-based mechanism requires two concurrent strategies: 1) that transmission of phenotypes is followed through the paternal lineage to eliminate complicating maternal behavioral and physiological factors; and 2) that the paternal lineage be followed to the F3 of offspring (29). Because a clear examination of germline-based transgenerational inheritance requires this specific analysis, few studies have provided direct evidence that the programmed germline is capable of transmitting information across generations after mHFD exposure.
In this study, we hypothesized that mHFD would result in a germline-based epigenetic program, increasing body length across three generations of offspring. Somatic growth is a quantitative trait controlled by many loci; thus we further hypothesized that a broad program of epigenetic gene regulation would control the body length phenotype. Because imprinted genes are important regulators of growth and are epigenetically regulated in the germline, we also performed a quantitative PCR-based array on imprinted genes to determine their potential mechanistic role.
Materials and Methods
Animals
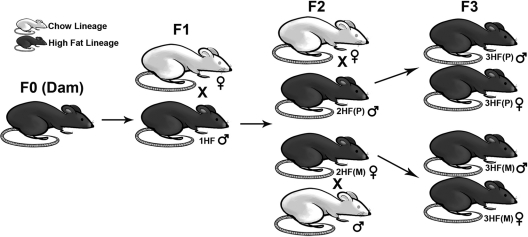
C57Bl/6:129 hybrid mice were bred in our colony and sustained on a 12-h light, 12-h dark cycle (lights on at 0700 h). Starting at wk 8, adult female mice were either given a breeder chow (Chow) or HFD for 6 wk before breeding at 14 wk as previously described (28). In total, nine maternal chow (mCh) and six maternal high fat (mHF) dams were bred with control males to produce 73 F1 offspring of the Chow lineage, and 45 F1 offspring of the High-Fat lineage (1HF) (Fig. 1). Two breeding combinations were used to produce the F2: the F2 Chow lineage (produced from Chow females crossed with Chow males, six litters producing 34 total offspring), and the F2 High-Fat lineage through the paternal line [2HF(P), Chow females crossed with 1HF males, four litters producing 28 total offspring]. Three breeding combinations were used to produce F3: the F3 Chow lineage (Chow females crossed with Chow males, seven litters producing 54 total offspring), the F3 High-Fat lineage through the maternal line [3HF(M), 2HF(P) females crossed with Chow males, four litters producing 27 total offspring], and the F3 High-Fat lineage through the paternal line [3HF(P), Chow females crossed with 2HF(P) males, seven litters producing 56 total offspring (Fig. 1)]. F3 animals through the 1HF maternal line were not included in this study. We set litter size thresholds between five and nine pups to prevent the possibility of gestational or lactational under- or overnutrition. Two F1 High Fat and three F2 Chow litters fell outside of the range and were not used for experiments or breeding. Pups were weaned from their mothers at 4 wk of age onto a chow diet, and all animals subsequent to this point were maintained on chow. The room was kept at a temperature of 22 C and relative humidity of 42%, and food and water was provided throughout the study ad libitum. All studies were done according to experimental protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Fig. 1.
Breeding scheme. F1 and F2 of both the Chow and High Fat (HF) lineages were bred as previously described (28). For the current experiment, we focused our study on the transgenerational transmission through the F2 paternal line [2HF(P), bred from 1HF males]. Both male and female 2HF(P) offspring were bred to produce the F3, producing litters from the F3 paternal [3HF(P)) and F3 maternal (3HF(M)] lines, respectively. The Chow lineage was bred for each successive generation in parallel with the HF lineage (data not shown).
Diet
The 4.73 kcal/g high-fat diet (HF) used in these studies was obtained from Research Diets, Inc. (New Brunswick, NJ) and consisted of (by kcal): 20% protein (casein + l-cystine), 35% carbohydrate (corn starch, maltodextrin, sucrose), 45% fat (lard, soybean oil), and essential vitamins and minerals. The 4.0 kcal/g breeder chow diet (Autoclaveable Rodent Diet 5010) was obtained from Purina Lab Diets (St. Louis, MO) and consisted of (by kilocalories) 27.5% protein (soybean meal, fish meal), 13.5% fat (lard, soybean oil), and 59% carbohydrate (corn starch, wheat).
Body weight and length
Body weights and lengths were obtained between 1000 h and 1300 h. Length measurements were performed ventrally from the tip of the nose to the anus while mice were under anesthesia. Measurements for same-sex littermates were averaged and counted as a single data point. Litter size was four to seven for F3 (averaged from 13–31 total offspring).
Glucose tolerance test (GTT)
Mice were fasted overnight. A solution of sterile 0.3 g/ml glucose was freshly prepared in 0.9% sterile saline. Beginning at 0900 h, baseline glucose levels from whole blood removed from tail snips were analyzed using the OneTouch Ultra (Johnson & Johnson, new Brunswick, NJ) system. Animals were then given an ip glucose injection of 2 mg/g body weight. Blood glucose measurements were taken from tail blood at 0, 15, 30, 60, and 120 min after injection. GTT were performed on F3 offspring between 10 and 11 wk of age. Measurements for same-sex littermates were averaged and counted as a single data point. Litter size was three to six (averaged from six to 11 total offspring per group).
Insulin tolerance
Mice were fasted for 4 h before the experiment. Baseline glucose measurements were performed from tail blood before ip injection of insulin (Humulin R; Eli Lilly & Co., Indianapolis, IN) diluted to 0.08 mU/μl in sterile saline for a final delivery of 0.8 mU/g body weight. Blood glucose measurements were performed at 0, 15, 30, 60, and 120 min after injection, and readings at all time points were normalized to the 0-min baseline. If blood glucose were to fall below 30 mg/dl, animals were rescued with ip injected 4.5 mg/g glucose in 0.9% sterile saline and were removed from the experiment. Insulin tolerance tests (ITT) were performed on F3 offspring between 12 and 13 wk of age. Measurements for same-sex littermates were averaged and counted as a single data point. Litter size was three to six (averaged from six to 11 total offspring per group).
Quantitative RT-PCR-based TaqMan array
Livers were dissected at 20 wk of age and frozen on dry ice. Frozen tissue (100 mg) was put into 1 ml Trizol (Invitrogen, Carlsbad, CA) and sonicated at medium-low power for 10 sec. Samples were allowed to incubate at room temperature for 5 min, after which 0.2 ml chloroform was added and shaken vigorously. After 5 min at room temperature, samples were spun at 12,000 rpm for 10 min at 4 C. The aqueous phase was mixed with 0.5 ml isopropanol, and then centrifuged for 12,000 rpm for 10 min at 4 C. Pellets were washed in 1 ml 75% ethanol made with diethylpyrocarbonate, allowed to dry, and then resuspended in 100 μl diethylpyrocarbonate. RNA (2 μg) was converted to cDNA using a high-capacity reverse transcriptase synthesis kit (Applied Biosystems, Carlsbad, CA). Quantitative real-time PCR was performed using TaqMan probes arranged onto a customized 96-well array. Two samples were run per plate, and all genes were normalized to glyceradehyde-3-phosphate dehydrogenase. Genes are listed in Table 1(litter, n = 4–7).
Table 1.
Expression levels and assay identifications for imprinted gene array
| Plate no. | Expressing parent | Gene | Assay identification (ID) | 3HF(M) Expression | 3HF(P) Expression |
|---|---|---|---|---|---|
| A1 | Paternal | Nnat | Nnat-Mm00731416_s1 | −0.213 | 0.037 |
| B1 | Paternal | Igf2 | Igf2-Mm00439564_m1 | 0.131 | 0.207 |
| C1 | Paternal | Gnas | Gnas-Mm00530548_m1 | −0.090 | −0.226 |
| D1 | Paternal | Dio3 | Dio3-Mm00548953_s1 | −0.060 | 0.184 |
| A2 | Paternal | Ndn | Ndn-Mm02524479_s1 | 0.048 | −0.085 |
| B2 | Paternal | Nap1l5 | Nap1l5-Mm02526917_s1 | −0.121 | −0.143 |
| C2 | Paternal | Mest | Mest-Mm00485003_m1 | 0.080 | −0.154 |
| D2 | Paternal | Magel2 | Magel2-Mm00844026_s1 | −0.538 | −0.603 |
| A3 | Paternal | Peg3 | Peg3-Mm00493299_s1 | 0.110 | −0.123 |
| B3 | Paternal | Peg12 | Peg12-Mm00844053_s1 | −0.340 | −0.854 |
| C3 | Paternal | Peg10 | Peg10-Mm01167724_m1 | 0.207 | 0.726 |
| D3 | Paternal | Ins1 | Ins1-Mm01259683_g1 | −0.305 | −0.698 |
| A4 | Paternal | Sgce | Sgce-Mm00448714_m1 | 0.187 | 0.161 |
| B4 | Paternal | Rtl1 | Rtl1-Mm02392620_s1 | −0.151 | −0.319 |
| C4 | Paternal | Rasgrf1 | Rasgrf1-Mm00441097_m1 | 0.053 | 1.399 |
| D4 | Paternal | Plagl1 | Plagl1-Mm00494250_m1 | −0.142 | 0.015 |
| B5 | Paternal | Xist | Xist-Mm01232884_m1 | −0.050 | −0.228 |
| C5 | Paternal | Snrpn; Snurf | Snrpn;Snurf-Mm01310473_g1 | −0.209 | −0.167 |
| D5 | Paternal | Slc38a4 | Slc38a4-Mm00459056_m1 | −0.123 | −0.164 |
| A5 | Maternal | Asb4 | Asb4-Mm00480830_m1 | −0.193 | 0.177 |
| A6 | Maternal | Ascl2 | Ascl2-Mm01268891_g1 | 0.895 | 0.671 |
| B6 | Maternal | Cdkn1c | Cdkn1c-Mm01272135_g1 | −0.168 | −0.130 |
| C6 | Maternal | Atp10a | Atp10a-Mm00437724_m1 | −0.094 | 0.061 |
| D6 | Maternal | Copg2 | Copg2-Mm00444398_m1 | −0.168 | −0.185 |
| A7 | Maternal | Igf2r | Igf2r-Mm00439576_m1 | 0.024 | 0.075 |
| B7 | Maternal | H19 | H19-Mm00469706_g1 | 0.361 | 0.056 |
| C7 | Maternal | Gatm | Gatm-Mm00491879_m1 | 0.018 | −0.268 |
| D7 | Maternal | Dcn | Dcn-Mm03003496_s1 | 0.154 | 0.121 |
| A8 | Maternal | Ube3a | Ube3a-Mm00839910_m1 | −0.138 | −0.136 |
| B8 | Maternal | Tnfrsf23 | Tnfrsf23-Mm00656375_m1 | 0.010 | −0.162 |
| C8 | Maternal | Phlda2 | Phlda2-Mm00493899_g1 | 0.100 | 0.488 |
| D8 | Maternal | Kcnq1 | Kcnq1-Mm00434640_m1 | −0.002 | 0.114 |
Gene expression values are normalized to Chow control (3Ch) expression.
Statistical measures
Results were calculated by multifactor ANOVA using maternal diet as a factor and separating by sex. Individual group statistics were then calculated upon statistical significance of the ANOVA using post hoc Student's two-tailed t tests. Measures for longitudinal weights and GTT and ITT were analyzed by repeated-measures ANOVA over time followed by post hoc t tests. Quantitative real-time PCR array data were assessed by logistic regression in which any gene that exceeded either 50% up or down-regulation from baseline were coded as nominal variables, and effects of genotype were analyzed by a χ2 test. All analyses were carried out using JMP statistical software (SAS, Cary, NC).
Results
F3 female offspring from the paternal lineage have an increased body size
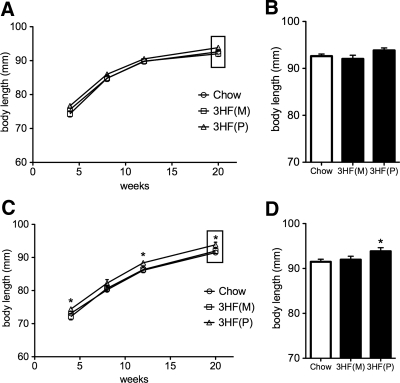
We previously reported that mHFD results in increased body length across two generations of offspring through both maternal and paternal lineages (28). To determine whether this effect was due in part to a germline-based contribution, we examined the effect of mHFD in F3 offspring. Male offspring did not exhibit increased body length at any time point measured from either maternal or paternal lineages (Fig. 2, A and B; F (3, 19) = 2.35, P = 0.12), although female offspring from the paternal lineage displayed an increased body length phenotype at 4, 12, and 20 wk of age [Fig. 2, C and D; main effect of group F (3, 19) = 3.31, P = 0.041; 3HF(P) different from Chow at 4 (P = 0.037), 12 (P = 0.032), and 20 wk (P = 0.027) time points]. The transgenerational effect of increased body length did not pass through the maternal lineage to either male or female offspring (P values between 0.27 and 0.65 for all time points).
Fig. 2.
Exposure to maternal high fat diet results in increased body length in F3 female, but not male, offspring only through the paternal lineage. A and C, Longitudinal body length for F3 (A) male and (C) female offspring. F3 male offspring of dams exposed to high fat diet [3HF(M), offspring produced from maternal transmission; 3HF(P), offspring produced from paternal transmission] do not exhibit an increased body length phenotype relative to control offspring (Chow, 12% fat, white bars, all points, P > 0.05). 3HF(P) female offspring exhibit increased body length at 4 (P = 0.037), 12 (P = 0.032), and 20 wk (P = 0.027) time points when compared with Chow. B and D, Time point (20 wk) for F3 (B) male and (D) female offspring. Body length differences in 3HF(P) females relative to Chow at 20 wk. (*, P < 0.05). Data are mean ± sem.
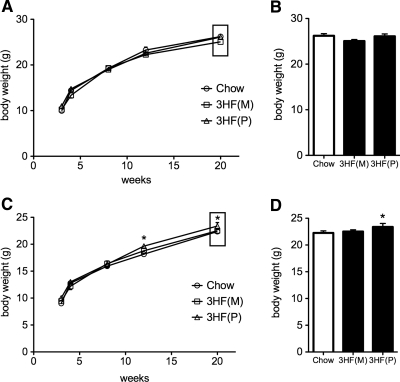
Similar to body length, increased body weight did not transmit to male offspring through either lineage (Fig. 3, A and B; F (3, 19) = 1.09, P = 0.355), although it did transmit to F3 female offspring through the paternal lineage [Fig. 3, C and D; main effect of group F (3, 19) = 3.12, P = 0.039, 3HF(P) different from Chow at 12 (P = 0.047) and 20 wk (P = 0.021) time points]. An increased body weight phenotype was not detected in either sex of F3 offspring through the maternal lineage (P values between 0.25 and 0.31 for all time points).
Fig. 3.
Exposure to a maternal high fat diet results in increased body weight in F3 female, but not male, offspring through the paternal lineage. A and C, Longitudinal body weight for F3 (A) male and (C) female offspring. F3 male offspring of dams exposed to high fat diet [3HF(M), offspring from a maternal transmission; 3HF(P), offspring from a paternal transmission] do not exhibit an increased body weight phenotype relative to control offspring (Chow, 12% fat; white bars, P > 0.05 for all time points). 3HF(P) female offspring exhibit increased body weight at 12 (P = 0.047) and 20 wk (P = 0.021) time points compared with Chow controls. B and D, Time point (20 wk) for F3 (B) male and (D) female offspring. Body weight differences at 20 wk in 3HF(P) females relative to 3HF(M) and Chow groups (*, P < 0.05). Data are mean ± sem.
F3 male offspring have improved glucose tolerance
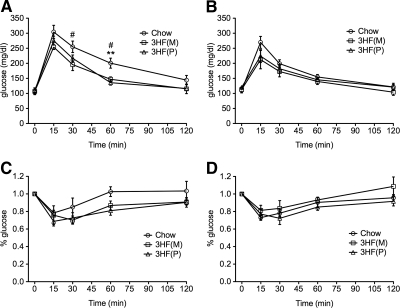
Because we previously demonstrated that mHFD results in normal glucose but altered insulin sensitivity across two generations (28), we examined responses in GTT and ITT in F3 adult offspring. Surprisingly, male offspring from maternal and paternal lineages exhibited improved glucose clearance during the GTT as analyzed by area under the curve analysis, characterized by a reduced rise in plasma glucose and more rapid return to baseline relative to controls [Fig 4A; main effect of group, F (3, 13) = 4.053, P = 0.046, 3HF(M) different from Chow at 30 (P = 0.05) and 60 min (P = 0.024); 3HF(P) different from Chow at 60 min, P = 0.0052]. Although there was a trend suggesting reduced maximal rise in plasma glucose, we did not detect a significant difference by area under the curve analysis for female offspring from either maternal or paternal lineages (Fig 4B; F (3, 16) = 1.33, P = 0.279).
Fig. 4.
3HF(P) and 3HF(M) male offspring exhibit enhanced glucose tolerance and normal insulin sensitvity. GTT (A and B) and ITT (C and D) for male (A and C) and female (B and D) F3 offspring. Area under the curve analysis reveals a main effect of group in male (A, P = 0.046) but not female (B, P = 0.279) offspring of the maternal [3HF(M)] and paternal [3HF(P)] lineages relative to controls (Chow) in glucose clearance in the GTT. Male offspring from maternal [3HF(M)] and paternal [3HF(P)] lineages have significantly improved plasma glucose clearance at 30 and 60 min after injection [#, 3HF(M) different from Chow controls; *, 3HF(P) different from Chow controls; **, P = 0.005; #, P < 0.05]. Neither male (C) nor female (D) offspring from either lineage exhibit significantly impaired glucose uptake after insulin bolus relative to Chow controls as measured by area under the curve analysis in the ITT (males, P = 0.16; females, P = 0.29). ITT points are normalized to baseline glucose levels at 0 min. Data are mean ± sem.
F3 offspring have normal insulin sensitivity
F3 male offspring from both maternal and paternal lineages demonstrated a trend toward improved insulin sensitivity in the ITT, although this effect was not statistically significant by area under the curve analysis (Fig 4C; F (3, 11) = 2.12, P = 0.16). No differences were detected in female offspring from maternal and paternal lineages in insulin sensitivity (Fig 4D; F (3, 13) = 1.35, P = 0.29).
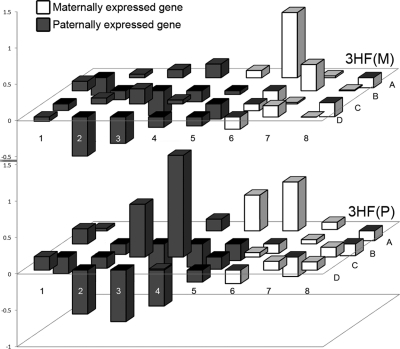
Paternally expressed genes are dynamically regulated in F3 female offspring from a paternal transmission
Imprinted loci are regulated and expressed in a sex-specific manner (30). Because we observed parent-of-origin specificity in the ability to transmit the body size phenotype in the F3, we analyzed the expression of imprinted genes involved in growth as a potential mechanism for the paternal-specific transmission of the body length and weight phenotypes. Paternally expressed genes from a paternal transmission exhibited a trend of greater volatility of expression than those of a maternal transmission as measured by logistic regression analysis (Fig. 5A and B; χ2 = 2.61, P = 0.106; list of genes in Table 1). Volatility was defined as genes displaying greater than a 50% deviation from baseline in either direction. This effect was not observed for maternally expressed genes from either parental transmission (χ2 = 0, P = 1.0).
Fig. 5.
Paternally expressed genes exhibit a more dynamic regulation in a paternal transmission. Parent-specific gene expression in adult F3 female offspring of both maternal [top panel, 3HF(M)] and paternal [bottom panel, 3HF(P)] lineages. Analysis by logistic regression of imprinted genes revealed a trend (P = 0.106) that paternally expressed genes (black bars) exhibited more dynamic regulation in 3HF(P) relative to 3HF(M) when compared with maternally expressed genes (white bars). Gene identifications are presented in Table 1.
Discussion
Epigenetic responses to a changing environment can program genes, resulting in a more rapid escalation in phenotypes predisposing toward diseases such as obesity or diabetes in future generations. Because transgenerational epigenetics play a role in population-wide responses to environmental challenges, an analysis of the conditions necessary for a trait to pass from one generation to the next will aid in our understanding of epidemiology dynamics. We have previously reported that mHFD consumption during pregnancy and lactation results in increased body length and impaired insulin sensitivity in F1 and F2 male and female offspring through both maternal and paternal lineages (28). However, as direct mHFD programming of F1 somatic tissues and F2 primordial germ cells can occur during this in utero exposure, it is necessary to examine transmission to F3 to determine whether the phenotype is carried by a stable germline-based epigenetic mark. Therefore, we have examined phenotypes in male and female F3 offspring from the F1 paternal lineage through the F2 maternal and paternal lineages.
Our results demonstrate specificity in modes of phenotype transmission as well as of inheritance. In our examination of F3 HFD offspring, only females showed a persistence of the F2 phenotype of increased body length. Interestingly, the F2 paternal, but not maternal, lineage passed on the increased body length and body weight phenotype to F3 females, an effect not detected in F3 male offspring. However, it should be noted that reduced statistical power stemming from a smaller experimental N in the 3HF(M) line (n = 4 litters, 27 total offspring) in our study may be masking a transgenerational phenotype persisting through the maternal lineage as well. Regardless, these results are intriguing because they confirm that mHFD is sufficient to stably affect specific traits through the paternal germline. This paternal to female offspring inheritance pattern was also recently described in a case of β-cell dysfunction as a result of preadolescent to adult paternal HFD exposure in rats (31). Paternal specific transmission patterns have also been found in humans, as males exposed to increased food availability in the prepubescent period pass on an increased risk for diabetes-related mortality to their grand-offspring (1, 2). What remains unclear in these studies is whether programming occurs as a direct result of increased dietary fat, a reduction in protein and/or carbohydrate that are a consequence of a diet proportionally high in fat, or through differences in micronutrient composition.
Maternal obesity and HFD exposure have been linked to increased rates of offspring diabetes in humans and reduced insulin sensitivity in an array of animal and comparative models (5–7, 9, 15, 18, 20–23, 32–37). We previously reported reduced insulin sensitivity in F1 and F2 HFD offspring (28). In F3 offspring, we analyzed the dynamics of glucose responses by glucose and insulin tolerance tests. Surprisingly, not only did the F2 phenotype of reduced insulin sensitivity not transmit to the F3 offspring, but F3 males actually showed an improved capacity to clear glucose relative to controls. The mechanism for such a response is not currently known; however it is possible that although the epigenetic marks responsible for body size appear to be stable into F3, epigenetic regulation of genes involved in glucose homeostasis may be more plastic, responding to the present environment in the F1 and F2. The organism may transgenerationally compensate for an altered glucose metabolism in earlier generations to preserve homeostatic conditions, resulting in phenotypes whereby an insulin-insensitive animal transmits an insulin-sensitive phenotype. Similarly counterintuitive fluctuations in phenotypes across generations have been reported previously (41). These results speak to the complexities of transgenerational transmission, and additional experiments are required to determine the cause of variances in phenotype across generations.
Because our studies detected an F2 paternal transmission of body size only to F3 females, we hypothesized that mHFD exposure had produced a stable epigenetic mark that would likely involve imprinted genes (38, 39). Effects on imprinted genes across multiple generations have been described after exposure to the endocrine disruptor vinclozolin (40). We analyzed the expression pattern of paternally and maternally imprinted genes in F3 female adult livers, comparing paternal and maternal transmission. In these data, we detected a nonstatistically significant trend toward a greater dynamic change in expression of paternally expressed genes when transmitted to female offspring only through the F2 paternal but not maternal lineage, fitting with our phenotypic endpoint. These data may support a novel involvement of imprinted genes in the sex-dependent transgenerational control of growth, although further study is clearly required to determine their ultimate role in the process, the specific tissues involved, and the developmental time window in which changes may occur.
It is becoming clear that epigenetic transmission may encompass multiple routes, rendering transgenerational inheritance patterns less straightforward than a collection of traits passing verbatim from one generation to the next (41–43). Traits can pass down a lineage through a combination of behavioral reinforcement (e.g. maternal behavior affecting offspring), direct somatic programming (e.g. in utero exposure to maternal environment), and through both transient and stable programming of the germline (29, 44). Some traits remain confined to a single generation, because our F1 offspring exhibited an increased adiposity phenotype that was not detected in F2 offspring (28). Traits can also skip generations, because F1 female offspring were not insulin insensitive but passed this trait onto their male and female F2 offspring (28, 41). Finally, phenotypes are transmitted or inherited in a sex-specific manner, as evidenced by previous studies in addition to our current report of an F2 paternal to F3 female transmission (8, 9, 27, 41, 45–51). This complexity may be related to the changing programming environment affecting each generation. Each founder dam represents an important experimental unit for a transgenerational epigenetic study because all future generations are influenced by this initial exposure. In addition to this component, each generation encounters secondary programming influences such as the presence of somatic phenotypes and epigenetic reprogramming between generations, contributing a unique component to each generation of animals.
Here, we provide evidence that mHFD programs a true germline-based transgenerational phenotype in the male gametes. This phenotype does not require further reinforcement through repeated experience to persist (29). Confirmation of a germline-based mechanism requires both the analysis of F3 to rule out any direct effects of mHFD programming and the transmission through the paternal lineage to avoid the confounding contributions of maternal factors such as altered in utero environment or behavior (29, 44). In our breeding, males do not have a direct physiological or behavioral interaction with their offspring, contributing only sperm to the next generation. Therefore, a phenotype passing from F2 to F3 through the paternal lineage provides compelling evidence that the mode of the transmission is through a stable mark in the germline. The increased body size phenotype that persists into F3 female offspring through the paternal lineage is statistically significant, yet small in magnitude similar to the F2 phenotype. Studies examining transgenerational effects necessitate the analysis of an entire litter as an N of 1; thus we averaged male or female offspring within a litter to constitute each N. This strategy reduces variability stemming from the mixed genetic background, although the overall statistical power of the experiment is reduced. Despite these considerations, we have detected small body length increases seen in F2 in multiple separate experiments, suggesting that our findings are due to a transgenerational effect and not unintentional selection bias.
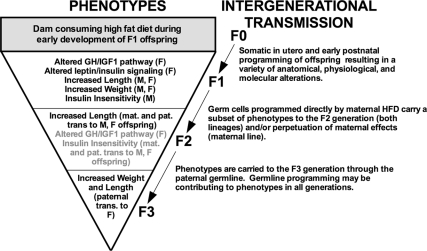
In our model, phenotypes that began in the F1 as a combination of increased adiposity, enhanced growth, insulin insensitivity, and GH/IGF-1 axis programming narrowed through the generations to the very specific phenotype of increased body length and weight in F3 females as transmitted by the F2 paternal lineage (Fig. 6). That some traits extinguish whereas others persist clearly suggests that divergent mechanisms of transmission are involved and that those traits that do persist are capable of being carried by the male germline. Males may consolidate and transmit their epigenetic influence through stable programming of their germ cells to compensate for the lack of direct interaction that females have during offspring development, ensuring that offspring receive important adaptive information.
Fig. 6.
Summary figure illustrating broad programming after mHFD exposure that narrows over generations, resulting in a specific epigenetic transmission to F3 female offspring. F1 offspring undergo extensive somatic programming as a result of mHFD exposure, resulting in traits such as increased adiposity that do not transmit to subsequent generations (28). A subset of the F1 phenotype is passed to F2 through both maternal and paternal lines through either perpetuating maternal effects (maternal line) or transient programming of primordial germ cells by mHFD (maternal and paternal lines). The gametic marks responsible for these transmissions may be transient, in which case they terminate in F2. Increased body length and weight, as transmitted through the paternal lineage to F3 female offspring, are stably programmed in the germline. (M, male offspring; F, female offspring; mat., maternal lineage; pat., paternal lineage).
Acknowledgments
We thank Kaitlyn Hellner-Burris and Adrienne Adler (both University of Pennsylvania) for technical assistance.
This work was supported by PubMed Central-National Institutes of Health Grant DK019525.
Disclosure Summary: The authors declare no conflict of interest.
Footnotes
- F1
- First generation
- F2
- second generation
- F3
- third generation
- GTT
- glucose tolerance test
- HFD
- high-fat diet
- ITT
- insulin tolerance test
- mHFD
- maternal high-fat diet.
References
- 1. Kaati G, Bygren LO, Edvinsson S. 2002. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. Eur J Hum Genet 10:682–688 [DOI] [PubMed] [Google Scholar]
- 2. Bygren LO, Kaati G, Edvinsson S. 2001. Longevity determined by paternal ancestors' nutrition during their slow growth period. Acta Biotheor 49:53–59 [DOI] [PubMed] [Google Scholar]
- 3. Roseboom T, de Rooij S, Painter R. 2006. The Dutch famine and its long-term consequences for adult health. Early Hum Dev 82:485–491 [DOI] [PubMed] [Google Scholar]
- 4. Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic Transgenerational Actions of Endocrine Disruptors and Male Fertility. Science 308:1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, Grove KL. 2009. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest 119:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pentinat T, Ramon-Krauel M, Cebria J, Diaz R, Jimenez-Chillaron JC. 2010. Transgenerational inheritance of glucose intolerance in a mouse model of neonatal overnutrition. Endocrinology 151:5617–5623 [DOI] [PubMed] [Google Scholar]
- 7. Guo F, Jen KL. 1995. High-fat feeding during pregnancy and lactation affects offspring metabolism in rats. Physiol Behav 57:681–686 [DOI] [PubMed] [Google Scholar]
- 8. Pinheiro AR, Salvucci ID, Aguila MB, Mandarim-de-Lacerda CA. 2008. Protein restriction during gestation and/or lactation causes adverse transgenerational effects on biometry and glucose metabolism in F1 and F2 progenies of rats. Clin Sci 114:381–392 [DOI] [PubMed] [Google Scholar]
- 9. Jimenez-Chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, Patti ME. 2009. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes 58:460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. James WPT. 2008. The epidemiology of obesity: the size of the problem. J Intern Med 263:336–352 [DOI] [PubMed] [Google Scholar]
- 11. Bruce KD, Hanson MA. 2010. The developmental origins, mechanisms, and implications of metabolic syndrome. J Nutr 140:648–652 [DOI] [PubMed] [Google Scholar]
- 12. de Beer H. 2004. Observations on the history of Dutch physical stature from the late-Middle Ages to the present. Econ Hum Biol 2:45–55 [DOI] [PubMed] [Google Scholar]
- 13. Gustafsson A, Werdelin L, Tullberg BS, Lindenfors P. 2007. Stature and sexual stature dimorphism in Sweden, from the 10th to the end of the 20th century. Am J Hum Biol 19:861–870 [DOI] [PubMed] [Google Scholar]
- 14. Wu Q, Suzuki M. 2006. Parental obesity and overweight affect the body-fat accumulation in the offspring: the possible effect of a high-fat diet through epigenetic inheritance. Obes Rev 7:201–208 [DOI] [PubMed] [Google Scholar]
- 15. Khan IY, Dekou V, Douglas G, Jensen R, Hanson MA, Poston L, Taylor PD. 2005. A high-fat diet during rat pregnancy or suckling induces cardiovascular dysfunction in adult offspring. Am J Physiol Regul Integr Comp Physiol 288:R127–R133 [DOI] [PubMed] [Google Scholar]
- 16. Khan IY, Taylor PD, Dekou V, Seed PT, Lakasing L, Graham D, Dominiczak AF, Hanson MA, Poston L. 2003. Gender-linked hypertension in offspring of lard-fed pregnant rats. Hypertension 41:168–175 [DOI] [PubMed] [Google Scholar]
- 17. Mitra A, Alvers KM, Crump EM, Rowland NE. 2009. Effect of high-fat diet during gestation, lactation, or postweaning on physiological and behavioral indexes in borderline hypertensive rats. Am J Physiol Regul Integr Comp Physiol 296:R20–R28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parente LB, Aguila MB, Mandarim-de-Lacerda CA. 2008. Deleterious effects of high-fat diet on perinatal and postweaning periods in adult rat offspring. Clin Nutr 27:623–634 [DOI] [PubMed] [Google Scholar]
- 19. Wu Q, Mizushima Y, Komiya M, Matsuo T, Suzuki M. 1998. Body fat accumulation in the male offspring of rats fed high-fat diet. J Clin Biochem Nutr 25:71–79 [Google Scholar]
- 20. Elahi MM, Cagampang FR, Mukhtar D, Anthony FW, Ohri SK, Hanson MA. 2009. Long-term maternal high-fat feeding from weaning through pregnancy and lactation predisposes offspring to hypertension, raised plasma lipids and fatty liver in mice. Br J Nutr 102:514–519 [DOI] [PubMed] [Google Scholar]
- 21. White CL, Purpera MN, Morrison CD. 2009. Maternal obesity is necessary for the programming effect of a high-fat diet on offspring. Am J Physiol Regul Integr Comp Physiol 296:R1464–R1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shankar K, Harrell A, Liu X, Gilchrist JM, Ronis MJ, Badger TM. 2008. Maternal obesity at conception programs obesity in the offspring. Am J Physiol Regul Integr Comp Physiol 294:R528–R538 [DOI] [PubMed] [Google Scholar]
- 23. Gniuli D, Calcagno A, Caristo ME, Mancuso A, Macchi V, Mingrone G, Vettor R. 2008. Effects of high-fat diet exposure during fetal life on type 2 diabetes development in the progeny. J Lipid Res 49:1936–1945 [DOI] [PubMed] [Google Scholar]
- 24. Zambrano E, Bautista CJ, Deás M, Martínez-Samayoa PM, González-Zamorano M, Ledesma H, Morales J, Larrea F, Nathanielsz PW. 2006. A low maternal protein diet during pregnancy and lactation has sex- and window of exposure-specific effects on offspring growth and food intake, glucose metabolism and serum leptin in the rat. J Physiol 571:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thamotharan M, Garg M, Oak S, Rogers LM, Pan G, Sangiorgi F, Lee PW, Devaskar SU. 2007. Transgenerational inheritance of the insulin-resistant phenotype in embryo-transferred intrauterine growth-restricted adult female rat offspring. Am J Physiol Endocrinol Metab 292:E1270–E1279 [DOI] [PubMed] [Google Scholar]
- 26. Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. 2007. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 97:435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benyshek DC, Johnston CS, Martin JF. 2006. Glucose metabolism is altered in the adequately-nourished grand-offspring (F3 generation) of rats malnourished during gestation and perinatal life. Diabetologia 49:1117–1119 [DOI] [PubMed] [Google Scholar]
- 28. Dunn GA, Bale TL. 2009. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology 150:4999–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Youngson NA, Whitelaw E. 2008. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet 9:233–257 [DOI] [PubMed] [Google Scholar]
- 30. Trasler JM. 2006. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod Fertil Dev 18:63–69 [DOI] [PubMed] [Google Scholar]
- 31. Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. 2010. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467:963–966 [DOI] [PubMed] [Google Scholar]
- 32. Armitage JA, Khan IY, Taylor PD, Nathanielsz PW, Poston L. 2004. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J Physiol 561:355–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. 2009. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 18:4046–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. 2008. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 115:1243–1249 [DOI] [PubMed] [Google Scholar]
- 35. Gorski JN, Dunn-Meynell AA, Hartman TG, Levin BE. 2006. Postnatal environment overrides genetic and prenatal factors influencing offspring obesity and insulin resistance. Am J Physiol Regul Integr Comp Physiol 291:R768–R778 [DOI] [PubMed] [Google Scholar]
- 36. Simeoni U, Barker DJ. 2009. Offspring of diabetic pregnancy: long-term outcomes. Semin Fetal Neonatal Med 14:119–124 [DOI] [PubMed] [Google Scholar]
- 37. Nathanielsz PW, Poston L, Taylor PD. 2007. In utero exposure to maternal obesity and diabetes: animal models that identify and characterize implications for future health. Clin Perinatol 34:515–526 [DOI] [PubMed] [Google Scholar]
- 38. Daxinger L, Whitelaw E. 2010. Transgenerational epigenetic inheritance: More questions than answers. Genome Res 20:1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gluckman PD, Hanson MA, Beedle AS. 2007. Non-genomic transgenerational inheritance of disease risk. Bioessays 29:145–154 [DOI] [PubMed] [Google Scholar]
- 40. Stouder C, Paoloni-Giacobino A. 2010. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction 139:373–379 [DOI] [PubMed] [Google Scholar]
- 41. Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, Vizi S, Mansuy IM. 2010. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry 68:408–415 [DOI] [PubMed] [Google Scholar]
- 42. Kim HK, Capaldi DM, Pears KC, Kerr DC, Owen LD. 2009. Intergenerational transmission of internalising and externalising behaviours across three generations: gender-specific pathways. Crim Behav Mental Health 19:125–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vigé A, Gallou-Kabani C, Junien C. 2008. Sexual dimorphism in non-Mendelian inheritance. Pediatr Res 63:340–347 [DOI] [PubMed] [Google Scholar]
- 44. Jirtle RL, Skinner MK. 2007. Environmental epigenomics and disease susceptibility. Nat Rev Genet 8:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wolff GL, Kodell RL, Moore SR, Cooney CA. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 12:949–957 [PubMed] [Google Scholar]
- 46. Cooney CA, Dave AA, Wolff GL. 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 132:2393S–2400S [DOI] [PubMed] [Google Scholar]
- 47. Waterland RA, Jirtle RL. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Waterland RA, Travisano M, Tahiliani KG. 2007. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J 21:3380–3385 [DOI] [PubMed] [Google Scholar]
- 49. Cropley JE, Suter CM, Beckman KB, Martin DIK. 2006. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc Natl Acad Sci USA 103:17308–17312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zambrano E, Martínez-Samayoa PM, Bautista CJ, Deás M, Guillén L, Rodríguez-González GL, Guzmán C, Larrea F, Nathanielsz PW. 2005. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J Physiol 566:225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, Golding J. ALSPAC Study Team 2006. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 14:159–166 [DOI] [PubMed] [Google Scholar]